Introduction of Breast Cancer
Breast cancer is still a complicated and common health issue that affects millions of people globally. The most common cancer in women globally is breast cancer, which is treatable in about 70–80% of cases when detected early and without metastases. With the existing therapeutic options, advanced breast cancer with distant organ metastases that is distribution of cancer cells from one place to another place with regarded as incurable [
1]. The prevalence of breast cancer has been steadily rising in both industrialized and developing nations over the past few decades, with a notable tendency toward even younger ages. Currently, the most common treatments for breast cancer are surgery, radiation therapy, chemotherapy, hormone therapy, and targeted therapy. Chemotherapy is one of them that is frequently used to control the population of tumor cells following surgery or in cases when surgery is not possible. Chemotherapy, however, can frequently result in a variety of negative side effects, including alopecia, nausea, exhaustion, early ovarian failure, weight gain, and heart dysfunction, among others, which lowers patients' quality of life (QOL). Drug resistance is typically brought on by prolonged exposure to chemotherapy drugs, which can both lower the therapeutic benefit and cause patient death. To improve the clinical treatment of breast cancer, it is therefore imperative to create new anti-breast cancer medications [
2]. The cost of healthcare is heavily impacted by breast cancer. 35–50% of the entire expense of treating breast cancer is devoted to surgery, with the majority of that amount coming from the hospital stay. Hospital stays have dropped from 10–14 days to 5–7 days during the 1990s, and day care admissions have also declined [
3]. Worldwide, breast cancer is the most common cause of cancer-related deaths among women. Globally, rates vary by around five times, yet in areas where the disease was not previously prevalent, rates are rising. Estrogens are associated with numerous identified risk factors. In postmenopausal women, risk factors include obesity, late menopause, and early menarche. Prospective studies have also demonstrated a correlation between increased risk and high levels of endogenous oestradiol. Having children lowers risk; more babies and an early first birth provide better protection; nursing also likely offers some protection. The little increase in breast cancer risk associated with oral contraceptives and hormone therapy for menopause tends to decrease with cessation of usage. While physical activity is probably beneficial, alcohol increases risk. Although they only account for a small percentage of cases, certain gene mutations significantly raise the risk of breast cancer [
4]. Gene function is abnormally caused by genetic and epigenetic changes that contribute to breast cancer. The development of novel approaches for breast cancer diagnosis and therapy has been greatly influenced by the recent advances in the field of epigenomics, which have also had a significant impact on our understanding of the mechanisms behind breast cancer. Three mutually interacting processes have been identified as being involved in epigenetic regulation: nucleosomal remodeling, histone changes, and DNA methylation. These mechanisms alter the structure of chromatin to produce heterochromatin or euchromatin, which either stimulates or inhibits the expression of a gene. Changes in the expression of important genes due to abnormal epigenetic control in breast cells have the potential to cause, promote, and sustain carcinogenesis. They may also contribute to the development of treatment resistance. We now go over the roles that the epigenetic machinery is known to play in the onset and recurrence of breast cancer. We also emphasize the importance of epigenetic modifications as novel targets for anticancer therapy and as prognostic biomarkers [
5].
Types and Management of Breast Cancer
As most cancers are called after the portion of the body in which they first appeared, breast cancer describes the irregular growth and division of cells that start in the breast tissue. The glandular tissues and the stromal (supporting) tissues are the two primary tissue types that make up the breast. The lobules, or milk-producing glands, and the ducts, or milk tubes, are housed in the glandular tissues, whereas the stromal tissues are the fatty and fibrous connective tissues of the breast. Additionally, lymphatic and immune system tissue that eliminates waste products and cellular fluids makes up the breast. There are various tumor kinds that can appear in the breast in different places. The majority of tumors are caused by benign, or non-cancerous, alterations in the breast. For instance, women with fibrocystic change, a non-cancerous disorder, may experience lumpiness, fibrosis (a formation of connective tissue that resembles scars), cysts (accumulated packets of fluid), and areas of thickening, tenderness, or breast pain. The cells lining the ducts are where most breast malignancies start (ductal tumors). A tiny percentage originate in other tissues, but some (lobular cancers) start in the cells lining the lobules [
6].
Types of Cancer According to Site
1)Non-Invasive Breast Cancer: Breast cancer cells that stay within the ducts and don't spread to the surrounding fatty and connective tissues. Ninety percent of non-invasive breast cancer cases are ductal carcinoma in situ (DCIS). Less frequently occurring lobular carcinoma in situ (LCIS) is thought to be a sign of an elevated risk of breast cancer.
2)Invasive Breast Cancer: Breast cancer cells that spread to the surrounding fatty and connective tissues of the breast after breaching the duct and lobular wall. It is possible for cancer to be invasive without also spreading to other organs or lymph nodes.
Breast cancer that occurs frequently:
1)Lobular neoplasia, also known as lobular carcinoma in situ (LCIS): Cancer that is "in situ" means that it hasn't progressed outside of the original site of development. A substantial rise in the number of cells within the breast's milk glands, or lobules, is known as LCIS.
2)In situ ductal carcinoma (DCIS): The most prevalent kind of non-invasive breast cancer, known as DCIS, is limited to the breast ducts. For example ductal comedocarcinoma.
3)Invasive lobular carcinoma (ILC): Another name for ILC is infiltrating lobular carcinoma. ILC starts in the breast's milk glands, or lobules, but it frequently spreads (metastasized) to other parts of the body. 10% to 15% of cases of breast cancer are caused by ILC.
4)Invasive ductal carcinoma (IDC):Another name for infiltrating ductal carcinoma (IDC). IDC starts in the breast's milk ducts, enters the duct, and spreads to the fatty tissue there as well as potentially to other parts of the body. Eighty percent of cases of breast cancer are diagnosed with IDC, the most frequent kind of the disease.
Breast Cancer Treatment:
Breast cancer risk can be determined using risk assessment methods, and individuals who are at a higher risk may be eligible for medication that lowers their risk. The menopausal state affects the pharmaceutical selection. Treatment for breast cancer varies according to stage. Ductal carcinoma in situ, or stage 0, is a benign condition that, in up to 40% of cases, develops into invasive malignancy. Mastectomy or radiation therapy along with a lumpectomy is the treatment for in situ ductal cancer. Patients who have estrogen receptor-positive ductal carcinoma in situ may also benefit from endocrine therapy. There are three therapy phases for nonmetastatic early invasive stages (I, IIa, IIb) and locally progressed stages (IIIa, IIIb, IIIc). When cancers express ERBB2 (estrogen), progesterone, or estrogen receptors, systemic endocrine or immunotherapies are used during the preoperative phase. The only treatment available when tumors lack any of the three receptors is preoperative chemotherapy. In terms of surgical procedures, there are two that have comparable survival rates: a mastectomy or a lumpectomy with radiotherapy if the tumor can be fully removed with acceptable cosmetic outcomes. In cases when nodal illness is suspected, a sentinel lymph node biopsy is also carried out. Chemotherapy, immunotherapy, endocrine therapy, and radiation are all part of the postoperative phase. Postoperative bisphosphonates ought to be made available to postmenopausal women as well. Breast cancer in stage IV (metastatic) can be managed but not cured. Increasing lifespan and quality of life are among the objectives of treatment [
7].
Drug Discovery-Target Identification
The discovery of a biological target (such as a receptor, enzyme, protein, gene, etc.) that is engaged in a biological process assumed to be malfunctioning in disease-bearing people is typically the first step toward the development of a new medication. Studies conducted in every therapeutic field show that it takes at least 12 years, and frequently considerably longer, to develop a new medication from target identification to marketing approval [
8]. Before a new medication can be found on the pharmacy shelf, it will often take ten to fifteen years and more than US
$2 billion. Natural products have historically been the primary source of novel drug entities for drug discovery; however, high-throughput synthesis and combinatorial chemistry-based development have since taken precedence [
9]. The first important steps in the drug discovery process are target identification and validation. Thus, this first step in the drug development process is inevitably of interest to researchers [
10]. In biomedical research, target identification is essential because it creates the framework for the creation of novel treatments and medications. For the purposes of drug development, precision medicine, reducing medication attrition, raising therapeutic efficacy, and ultimately revolutionizing patient care, target identification is crucial [
11].
Network Pharmacology
Network Pharmacology used to identify disease targets."Network pharmacology" was initially introduced by Hopkins et al. in 2007. Using system biology, this approach examines how medications can intervene and identify possible disease targets for treatment [
12]. Network pharmacology (NP) is an emerging discipline useful in drug discovery, which combines genomic technologies and system biology through computational biological tools. Network pharmacology is an approach capable of describing complex relationships among biological systems, drugs and diseases. It also clarifies the possible mechanisms of complex bio-actives through large data set analysis and determines the synergistic effects in cancer treatment [
13]. As the information age progresses, different network technologies are being developed on a constant basis. Network pharmacology is gaining momentum as an integration of pharmacology and information network, based on system biology, bioinformatics, and high-throughput histology. The approach integrates network biology and polypharmacology, drawing from the limited effectiveness of highly selective single-target medications. We can quickly find medications and disease targets from a vast quantity of data by using network pharmacology, and we can also comprehend the mechanisms and connections that exist between them. It's a useful approach [
14]. The "one gene, one drug, one disease" paradigm has characterized drug discovery for decades, with a primary focus on creating ligands that are highly selective in order to avoid undesirable side effects [
15]. The biomolecular network's "network target" refers to the important molecules, pathways, or modules that play a role in mechanism-based relationships between medications and illnesses. These associations quantitatively show the drugs' overall regulatory processes [
16]. The development of network pharmacology has created new opportunities for comprehending the complex bioactive elements present in a wide range of therapeutic plants. Network pharmacology presents a range of active substances, associated methods, resources, and databases, as well as applications for drug discovery and development [
17]. It is well-known that, in order to deal with issues like the ineffectiveness and developing resistance of single-targeted molecules, drug discovery frequently necessitates a systems-level polypharmacology method. Approaches to network pharmacology are being developed and used more often to discover novel therapeutic prospects and to repurpose current medications. The field of natural products did not benefit from these recent advancements very quickly. Here, we make the case that a network pharmacology approach would make it possible to map the target space of natural products effectively, which would offer a methodical way to increase the druggable space of proteins linked to a variety of complex disorders [
18]. Network pharmacology has become a viable strategy to expedite drug discovery and clarify the modes of action of various target constituents. The basic idea of network pharmacology is ideally suited for the analysis of drugs with multiple targets [
19]. In order to identify the active chemical, a new science called network pharmacology combines network analysis, multiple drug target prediction, and oral bioavailability prediction [
20].
Material and Method
1)Prediction of Target Genes Associated with Breast Cancer:
Gene Related to Breast cancer were obtained from the UniprotKB database by text mining (
https://www.uniprot.org/) through searching “Breast Cancer”. After filtering with the term “Homo Sapiens” 2658 genes associated with breast cancer were identified. Amongst that 165 gene were selected after
2)GeneCodis and Pathway Analysis:
We used GeneCodis (
https://genecodis.genyo.es/) database for GO enrichment analysis by KEGG. That provided a systematic and comprehensive annotation information on biological functions, molecular functions,Biological components. The list of 165 gene ewre set to geneCodis and the species was limited to “Homo Sapiens”. GO and KEGG pathway analysis were performed.
3)Protein Protein Interaction and Visualization Data:
STRING database (
https://string-db.org/) contain known and predicted Protein protein interaction. The 10 pathway were selected in with GO and KEGG that paste in string with species was defined as Homo Sapiens. The PPI data was obtained. The interaction network was drawn and the network was analyzed by cytosacpe. In that the degree and betweenness centrality was measured.
4)Molecular Docking:
The computer-generated three-dimensional structure of tiny ligands is positioned into a receptor structure using a process called molecular docking in a range of orientations, conformations, and locations. Medicinal chemistry and drug discovery can benefit from this approach, which offers insights on molecular recognition. Computer-Aided Drug Design and Discovery now includes docking as a crucial component (CADDD) [
28]. Molecular docking is commonly used in modern drug design for understanding drug-receptor interaction [
29]. In present study we used tulsi extract(Ocimum Sanctum) and they have different active constituents like eugenol, camphor, etc).
They act as ligand and bind with macromolecules(protein, gene, enzymes) the structure of gene taken from protein database (
https://www.rcsb.org/) and the molecular interaction shown by Pyrex software and the binding affinity energy kcal/mol between protein and ligand determined.
Results:
1)Prediction of Target Associated with Breast cancer:
UniprotKB is a unique database that is used to select targets associated with gene in that total 2,658 were selected after filtering and deleting duplicates. In final total we selected 165 genes (In
Table 1).
2)Gene Ontology KEGG Analysis:
By using UniprotKB 5658 gene identified breast cancer about that we selected 165 and by GeneCodis we got pathways and that we selected 9 pathways related to breast cancer (
Table 2).
Figure A.
KEGG Pathway with potential pathway targets.
Figure A.
KEGG Pathway with potential pathway targets.
3)Breast Cancer PPI Network
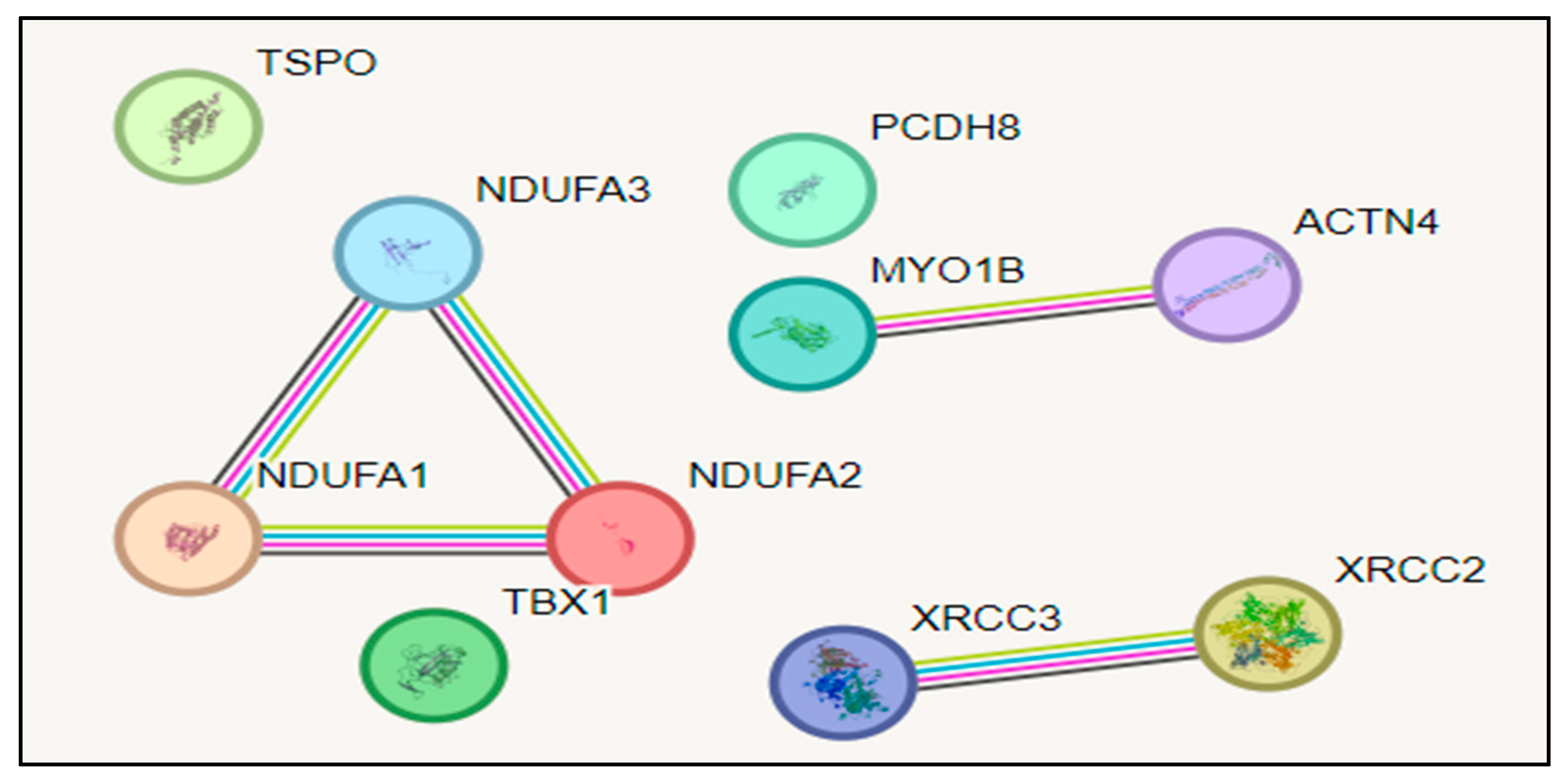
By using STRING we got a PPI network in that there is interaction between NDUFA1,NDUFA2 and NDUFA3 one network then interaction between MYO1B second network and ACTN4 then XRCC3 and XRCC2 is third network (Figure B). In that Green lines: Indicate interactions detected by gene neighborhood, meaning the genes encoding the proteins are located near each other in the genome. Red lines: Represent interactions identified through gene fusions, indicating the proteins are part of a fusion protein in another species. Blue lines: Signify interactions from gene co-occurrence, suggesting the proteins are frequently observed together across different species. Black lines: Denote interactions detected through co-expression, meaning the proteins are often produced at the same time in the same tissues or conditions. Purple lines: Represent interactions identified through experimental data. Light blue lines: Indicate interactions derived from curated databases.
Figure B.
Protein-Protein Interaction Network.
Figure B.
Protein-Protein Interaction Network.
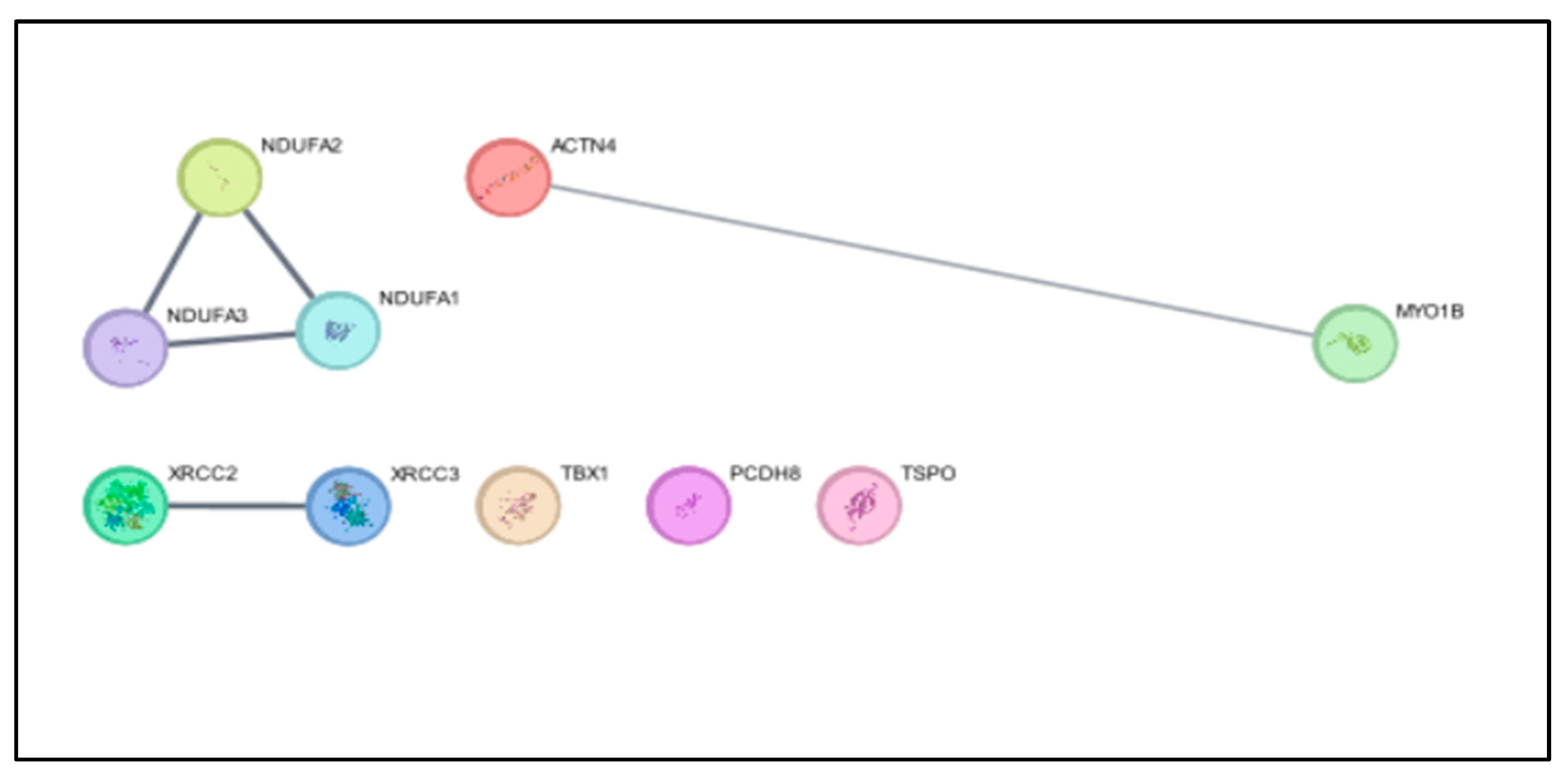
PPI Network Analyzed by CytoScape:
In CytoScape we did the analysis of PPI and detected their Degree and Betweenness by using the CentiScape app (In Figure C).
Figure C.
PPI in CytoScape Analysis.
Figure C.
PPI in CytoScape Analysis.
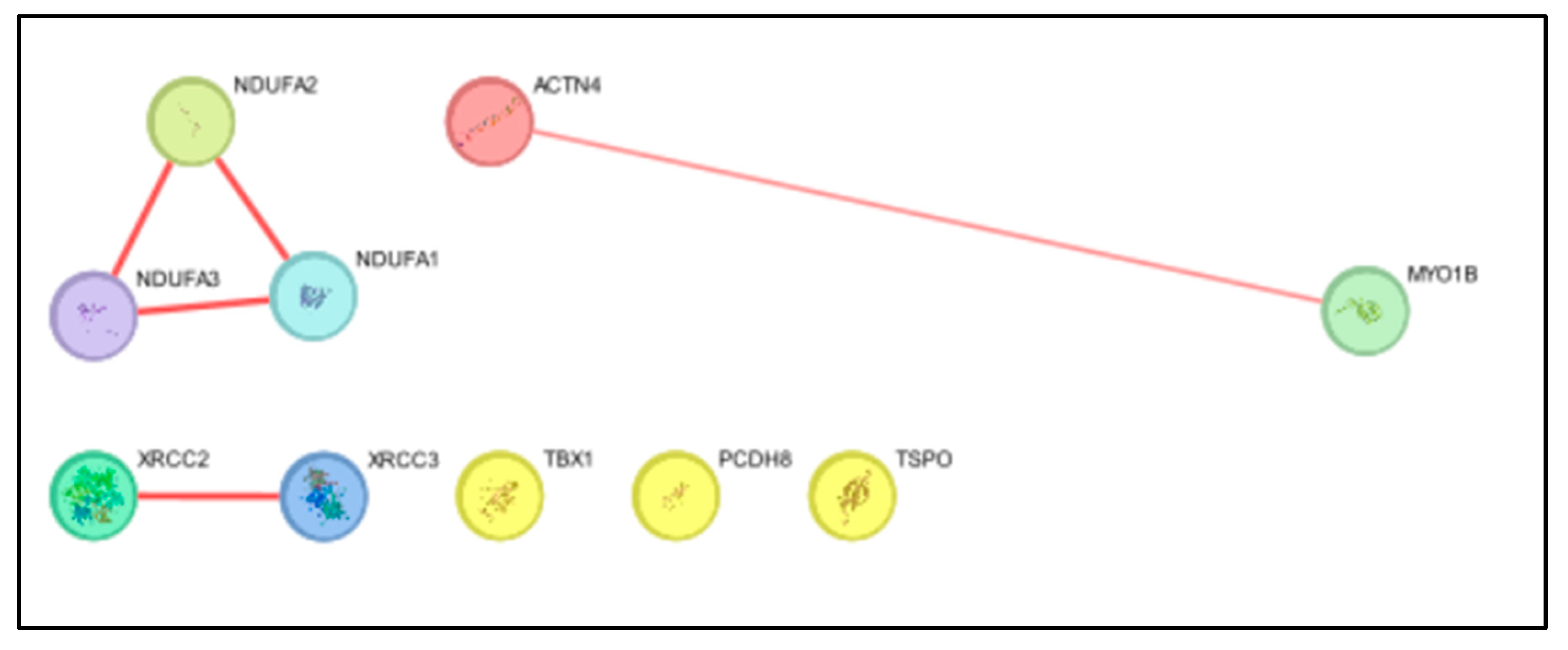
Degree and Betweenness Analysis:
A Cytoscape application called CentiScape is intended exclusively for network centrality metric analysis. For the purpose of computing and visualizing different centrality metrics in biological networks, including protein-protein interaction network."Degree" and "betweenness centrality" are two metrics in Cytoscape that are used to examine node attributes in a network (such a network of protein-protein interactions) that commonly represent genes or proteins. The term "degree" describes how many edges, or connections, a node in a network has. A protein node's degree in a network of protein-protein interactions indicates how many other proteins it directly interacts with. Higher degree proteins are thought to be more linked within the network. The term Betweenness Centrality is The frequency with which a node in a network is on the shortest path between other nodes is measured by betweenness centrality. Within a network of protein-protein interactions, a protein node possessing a high betweenness centrality serves as an intermediary between distinct segments of the network. It suggests that the protein is important for bridging various modules or pathways within the network (In Figure D).
Figure D.
Degree and Betweenness centrality in CytoScape.
Figure D.
Degree and Betweenness centrality in CytoScape.
4)Molecular Docking:
In that we determine the binding affinity of ligand and protein and the criteria of binding affinity generally the the more negative the binding energy has the stronger the binding affinity. The targeted proteins are MYO1B,TBX1,XRCC3 bind with different active constituents. In that DPHP shows the maximum binding affinity with different targets. MYO1 with DPHP is -9.2 kcal/mol,TBX1 with DPHP is -9.2kcal/mol,XRCC2 with DPHP is -10.1.
Table 3.
List of Chemical Constituents (Ligand) of Ocimum Sanctum.
Table 3.
List of Chemical Constituents (Ligand) of Ocimum Sanctum.
| BORNEOL |
EUGENOL |
| BOSABOLANE |
FARNESENE |
| CAMPHENE |
SALINENE |
| COPAENE |
TERPINEN-4-OL |
| CARVACROL |
URSOLIC ACID |
| DPHP |
|
| ESTRAGOLE |
|
Table 3.
Binding affinity (kcal/mol) Breast Cancer target protein and Ocimum Sanctum chemical constituents.
Table 3.
Binding affinity (kcal/mol) Breast Cancer target protein and Ocimum Sanctum chemical constituents.
| Ligand with Protein |
BindingAffinity
(kcal/mol) |
Ligand with Protein |
Binding Affinity
(kcal/mol) |
| MYO1_BORNEOL |
-5 |
TBX1_BORNEOL |
-5.5 |
| MYO1_BOSABOLANE |
-5.3 |
TBX1_BOSABOLANE |
-5.5 |
| MYO1B_CAMPHENE |
-5 |
TBX1_CAMPHENE |
-4.9 |
| MYO1B_COPAENE |
-6.3 |
TBX1_COPAENE |
-6.2 |
| MYO1B_CORVACROL |
-5.6 |
TBX1_CORVACROL |
-5.4 |
| MYO1B_DPHP |
-9.2 |
TBX1_DPHP |
-9.2 |
| MYO1B_ESTRAGOL |
-5.6 |
TBX1_ESTRAGOL |
-4.9 |
| MYO1B_EUGENOL |
-5.2 |
TBX1_EUGENOL |
-5.3 |
| MYO1B_FERNESENE |
-5.5 |
TBX1_FERNESENE |
-5.6 |
| MYO1B_SALINENE |
-6.3 |
TBX1_SALINENE |
-6.4 |
| MYO1B_TERPINE4OL |
-5.2 |
TBX1_TERPINE4OL |
-5.7 |
| MYO1B_UROSOLIC ACID |
-7.5 |
TBX1_UROSOLIC ACID |
-8.2 |
| XRCC3_BORNEOL |
-5.5 |
XRCC2_DPHP |
-10.1 |
| XRCC3_BOSABOLANE |
-6.5 |
XRCC3_ESTRAGOL |
-5.5 |
| XRCC3_CAMPHENE |
-5.3 |
XRCC3_EUGENOL |
-5.5 |
| XRCC3_COPAENE |
-6.4 |
XRCC3_FERNESENE |
-6 |
| XRCC3_CORVACROL |
-5.9 |
XRCC3_SALINENE |
-6.7 |
| XRCC3_TERPINE4OL |
-5.9 |
XRCC3_UROSOLIC ACID |
-8.7 |
Conclusion
In this paper we identified breast cancer target and determine protein protein interaction and their affinities with different chemical compounds of ocimum sanctum. The protein protein interaction of different protein from 165 we are selected but not repeated genes we take. About 10 proteins we taken about that (NDUFA1,NDUFA2,NDUFA3,ACTN4,MYO1B,XRCC2,XRCC3) 7 protein shows PPI. At last we check binding affinity with chemical compound with different gene and mostly three gene bind with different compounds. The compound DPHP shows the maximum binding affinity -9.2kcal/mol with MYOB1,-9.2kcal/mol with TBX1 and -10.1kcal/mol with XRCC3.
Reference
- Obeagu EI, Obeagu GU. Breast cancer: A review of risk factors and diagnosis. Medicine. 2024 Jan 19;103(3):e36905. [CrossRef]
- Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J, Cardoso F. Breast cancer. Nature reviews Disease primers 5: 66.
- de Kok M, Frotscher CN, van der Weijden T, Kessels AG, Dirksen CD, van de Velde CJ, Roukema JA, Bell AV, van der Ent FW, von Meyenfeldt MF. Introduction of a breast cancer care programme including ultra short hospital stay in 4 early adopter centers: framework for an implementation study. BMC cancer. 2007 Dec;7:1-9. [CrossRef]
- Key TJ, Verkasalo PK, Banks E. Epidemiology of breast cancer. The Lancet oncology. 2001 Mar 1;2(3):133-40.
- Lo PK, Sukumar S. Epigenomics and breast cancer. [CrossRef]
- Sharma GN, Dave R, Sanadya J, Sharma P, Sharma K. Various types and management of breast cancer: an overview. Journal of advanced pharmaceutical technology & research. 2010 Apr 1;1(2):109-26.
- Trayes KP, Cokenakes SE. Breast cancer treatment. American family physician. 2021 Aug;104(2):171-8.
- Mohs RC, Greig NH. Drug discovery and development: Role of basic biological research. Alzheimer's & Dementia: Translational Research & Clinical Interventions. 2017 Nov 1;3(4):651-7.. [CrossRef]
- Berdigaliyev N, Aljofan M. An overview of drug discovery and development. Future medicinal chemistry. 2020 Feb;12(10):939-47. [CrossRef]
- Wang S, Sim TB, Kim YS, Chang YT. Tools for target identification and validation. Current opinion in chemical biology. 2004 Aug 1;8(4):371-7. [CrossRef]
- Keerthana N, Koteeswaran K. Target identification and validation in research. World Journal of Biology Pharmacy and Health Sciences. 2024;17(3):107-17.
- Dong Y, Tao B, Xue X, Feng C, Ren Y, Ma H, Zhang J, Si Y, Zhang S, Liu S, Li H. Molecular mechanism of Epicedium treatment for depression based on network pharmacology and molecular docking technology. BMC complementary medicine and therapies. 2021 Dec;21:1-3.. [CrossRef]
- Sakle NS, More SA, Mokale SN. A network pharmacology-based approach to explore potential targets of Caesalpinia pulcherima: An updated prototype in drug discovery. Scientific reports. 2020 Oct 14;10(1):17217. [CrossRef]
- Zhou Z, Chen B, Chen S, Lin M, Chen Y, Jin S, Chen W, Zhang Y. Applications of network pharmacology in traditional Chinese medicine research. Evidence-Based Complementary and Alternative Medicine. 2020 Oct;2020. [CrossRef]
- Zhang GB, Li QY, Chen QL, Su SB. Network pharmacology: a new approach for Chinese herbal medicine research. Evidence-based complementary and alternative medicine. 2013 Oct;2013. [CrossRef]
- Li S. Network pharmacology evaluation method guidance-draft. World Journal of Traditional Chinese Medicine. 2021 Jan 1;7(1):146-54. [CrossRef]
- Noor F, Tahir ul Qamar M, Ashfaq UA, Albutti A, Alwashmi AS, Aljasir MA. Network pharmacology approach for medicinal plants: review and assessment. Pharmaceuticals. 2022 May 4;15(5):572. [CrossRef]
- Kibble M, Saarinen N, Tang J, Wennerberg K, Mäkelä S, Aittokallio T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Natural product reports. 2015;32(8):1249-66. [CrossRef]
- Lee WY, Lee CY, Kim YS, Kim CE. The methodological trends of traditional herbal medicine employing network pharmacology. Biomolecules. 2019 Aug 13;9(8):362. [CrossRef]
- Zhang W, Chen Y, Jiang H, Yang J, Wang Q, Du Y, Xu H. Integrated strategy for accurately screening biomarkers based on metabolomics coupled with network pharmacology. Talanta. 2020 May 1;211:120710. [CrossRef]
- Yang Y, Adelstein SJ, Kassis AI. Target discovery from data mining approaches. Drug discovery today. 2012 Feb 1;17:S16-23.
- Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome biology. 2007 Jan;8:1-8. [CrossRef]
- Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic acids research. 2009 Jul 1;37(suppl_2):W317-22. [CrossRef]
- Mering CV, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic acids research. 2003 Jan 1;31(1):258-61. [CrossRef]
- Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT, Pyysalo S, Bork P. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic acids research. 2023 Jan 6;51(D1):D638-46. [CrossRef]
- Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: network analysis and visualization of proteomics data. Journal of proteome research. 2018 Nov 19;18(2):623-32. [CrossRef]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K. Integration of biological networks and gene expression data using Cytoscape. Nature protocols. 2007 Oct;2(10):2366-82. [CrossRef]
- Jakhar R, Dangi M, Khichi A, Chhillar AK. Relevance of molecular docking studies in drug designing. Current Bioinformatics. 2020 May 1;15(4):270-8. [CrossRef]
- Vijesh AM, Isloor AM, Telkar S, Arulmoli T, Fun HK. Molecular docking studies of some new imidazole derivatives for antimicrobial properties. Arabian Journal of Chemistry. 2013 Apr 1;6(2):197-204. [CrossRef]
Table 1.
The 165 breast cancer gene from UniprotKB were identified.
Table 1.
The 165 breast cancer gene from UniprotKB were identified.
| APOL1 APOL |
UNC13B UNC13 |
TSPAN4 NAG2 TM4SF7 |
| RASGRF2 GRF2 |
AIM2 |
IKBKB IKKB |
| HAT1 KAT1 |
PLD2 |
EREG |
| TFEC BHLHE34 TCFEC TFECL |
AURKA AIK AIRK1 ARK1 AURA AYK1 BTAK IAK1 STK15 STK6 |
ZNF646 KIAA0296 |
| SPTBN2 KIAA0302 SCA5 |
PER2 KIAA0347 |
KPNA5 |
| MDM4 MDMX |
TRIM24 RNF82 TIF1 TIF1A |
EPHB6 |
| CASC3 MLN51 |
NDUFA1 |
SPTLC1 LCB1 |
| FANCG XRCC9 |
PPM1D WIP1 |
GRM6 GPRC1F MGLUR6 |
| SIVA1 SIVA |
EIF3D EIF3S7 |
YY2 ZNF631 |
| BIRC5 API4 IAP4 |
GRIN2D GluN2D NMDAR2D |
GYG2 |
| CLOCK BHLHE8 KIAA0334 |
PDPK1 PDK1 |
PER1 KIAA0482 PER RIGUI |
| KDM6A UTX |
ERVK-18 |
DHX15 DBP1 DDX15 |
| PLXNB1 KIAA0407 PLXN5 SEP |
SIPA1L1 E6TP1 KIAA0440 |
PHGDH PGDH3 |
| ADAM12 MLTN UNQ346/PRO545 |
CRX CORD2 |
KLK10 NES1 PRSSL1 |
| ZNHIT1 CGBP1 ZNFN4A1 |
PRODH PIG6 POX2 PRODH2 |
EFS CASS3 |
| SRGAP3 ARHGAP14 KIAA0411 KIAA1156 MEGAP SRGAP2 |
CTIF KIAA0427 |
MSI1 |
| HOXA3 HOX1E |
GRID2 GLURD2 |
TBX1 |
| SUV39H1 KMT1A SUV39H |
RAD51C RAD51L2 |
ATP8B1 ATPIC FIC1 PFIC |
| XRCC3 |
XRCC2 |
DENR DRP1 H14 |
| HR |
DNPH1 C6orf108 RCL |
SNAI2 SLUG SLUGH |
| CHMP2A BC2 CHMP2 |
NDUFA2 |
ACTN4 |
| SPOP |
MYO1B |
SSNA1 NA14 |
| RRP9 RNU3IP2 U355K |
AKAP8 AKAP95 |
TLL1 TLL |
| PRICKLE3 LMO6 |
EXTL3 EXTL1L EXTR1 KIAA0519 |
KALRN DUET DUO HAPIP TRAD |
| MED14 ARC150 CRSP2 CXorf4 DRIP150 EXLM1 RGR1 TRAP170 |
KLK8 NRPN PRSS19 TADG14 UNQ283/PRO322 |
ST18 KIAA0535 ZNF387 |
| ZEB2 KIAA0569 SIP1 ZFHX1B ZFX1B HRIHFB2411 |
MCM3AP GANP KIAA0572 MAP80 |
NUPR1 COM1 |
| GSDME DFNA5 ICERE1 |
BRINP1 DBC1 DBCCR1 FAM5A IB3089A |
ACSL4 ACS4 FACL4 LACS4 |
| CUBN IFCR |
UGT1A9 GNT1 UGT1 |
PQBP1 NPW38 JM26 |
| PRAF2 JM4 |
CTSV CATL2 CTSL2 CTSU UNQ268/PRO305 |
NBN NBS NBS1 P95 |
| ERVK-19 |
FKTN FCMD |
CBFA2T3 MTG16 MTGR2 ZMYND4 |
| FZD7 |
ASTN2 KIAA0634 |
CPNE3 CPN3 KIAA0636 |
| RAB11FIP3 ARFO1 KIAA0665 |
ANKRD17 GTAR KIAA0697 |
HMMR IHABP RHAMM |
| PDCD6 ALG2 |
ZNF217 ZABC1 |
BCAS1 AIBC1 NABC1 |
| SH3BGRL |
FLNB FLN1L FLN3 TABP TAP |
TACC1 KIAA1103 |
| POLQ POLH |
TECTA |
EED |
| WBP4 FBP21 FNBP21 |
SCGB2A1 LIPHC MGB2 UGB3 |
RPS6KA5 MSK1 |
| SPAG6 PF16 |
SNRNP200 ASCC3L1 BRR2 HELIC2 KIAA0788 |
RPS6KA4 MSK2 |
| UTP20 DRIM |
RAD51D RAD51L3 |
BCAR3 NSP2 SH2D3B UNQ271/PRO308 |
| SERPINI2 MEPI PI14 |
IDH1 PICD |
ALDH1L1 FTHFD |
| DYSF FER1L1 |
BCAS2 DAM1 |
AKAP3 AKAP110 SOB1 |
| NCR1 LY94 |
CDKL5 STK9 |
PBR |
| SNCG BCSG1 PERSYN PRSN |
CCN5 CT58 CTGFL WISP2 UNQ228/PRO261 |
ND2 MTND2 |
| AQP8 |
BAIAP3 KIAA0734 |
UFL1 KIAA0776 MAXER NLBP RCAD |
| SASH1 KIAA0790 PEPE1 |
ZNF432 KIAA0798 |
SLCO2B1 KIAA0880 OATP2B1 OATPB SLC21A9 |
| TRIM37 KIAA0898 MUL POB1 |
HEXIM1 CLP1 EDG1 HIS1 MAQ1 |
ZNF202 ZKSCAN10 |
| LAGE-2 |
NDUFA3 |
GAS8-AS1 C16orf3 |
| UNC5C UNC5H3 |
PCDH8 |
CCNDBP1 DIP1 GCIP HHM |
| LYPD3 C4.4A UNQ491/PRO1007 |
TACC2 |
ZBTB7A FBI1 LRF ZBTB7 ZNF857A |
| LYPLA2 APT2 |
MAP3K6 ASK2 MAPKKK6 MEKK6 |
CCN4 WISP1 |
| BMP10 |
PSMG1 C21LRP DSCR2 PAC1 |
CLDN7 CEPTRL2 CPETRL2 |
| ABCA1 ABC1 CERP |
ADGRL2 KIAA0786 LEC1 LPHH1 LPHN2 |
YEATS4 GAS41 |
| NFATC1 NFAT2 NFATC |
EYA4 |
LDOC1 BCUR1 |
| AP2A1 ADTAA CLAPA1 |
TTC4 My044 |
NDST3 HSST3 UNQ2544/PRO4998 |
| CLDN1 CLD1 SEMP1 UNQ481/PRO944 |
EML2 EMAP2 EMAPL2 |
LATS1 WARTS |
| SNAI1 SNAH |
FADS2 |
OLFM2 NOE2 |
Table 2.
Pathways associated with breast cancer with including protein.
Table 2.
Pathways associated with breast cancer with including protein.
| Description |
Genes Count |
Pval adj |
Relative enrichment |
Genes |
Mitochondrial electron transport, NADH to ubiquinone |
Mar-38
|
7.62E-03
|
52.64 |
NDUFA1,
NDUFA2,
NDUFA3 |
Vesicle transport along actin filament |
Feb-19
|
2.83E-02 |
70.19
|
MYO1B,ACTN4 |
Retinoic acid receptor signaling pathway |
Feb-18 |
2.83E-02 |
74.09
|
TBX1
|
| Somitogenesis |
Feb-48 |
3.73E-02 |
27.78 |
PCDH8,XRCC2 |
Regulation of cardiac muscle hypertrophy in response to stress |
01-Jan |
3.73E-02
|
666.78
|
BMP10
|
| Negative regulation of autophagy of mitochondria |
01-Feb
|
3.73E-02 |
333.39 |
PBR
|
| Telomere maintenance via telomere trimming |
01-Feb |
3.73E-02 |
333.39
|
XRCC3 |
| Type IV hypersensitivity |
01-Feb
|
3.73E-02
|
333.39
|
EPHB6
|
| Positive regulation of phosphorylation |
Feb-35 |
3.73E-02
|
38.1 |
FZD7,EREG
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).