Submitted:
29 May 2024
Posted:
30 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
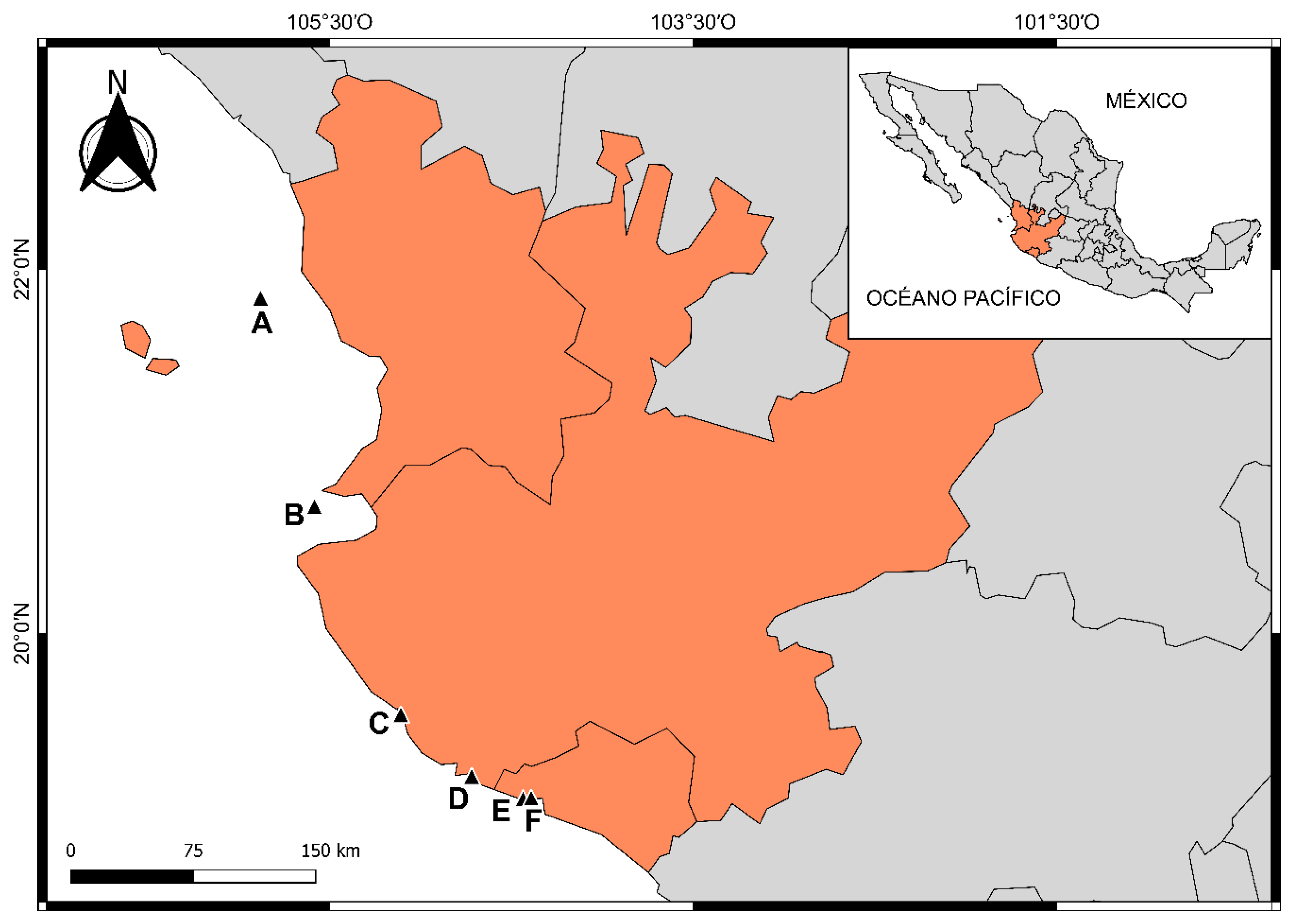
2.1. Study Area
2.2. Sampling Collection
2.3. Sequence Extraction and Analysis
2.4. Statistical Analysis
3. Results
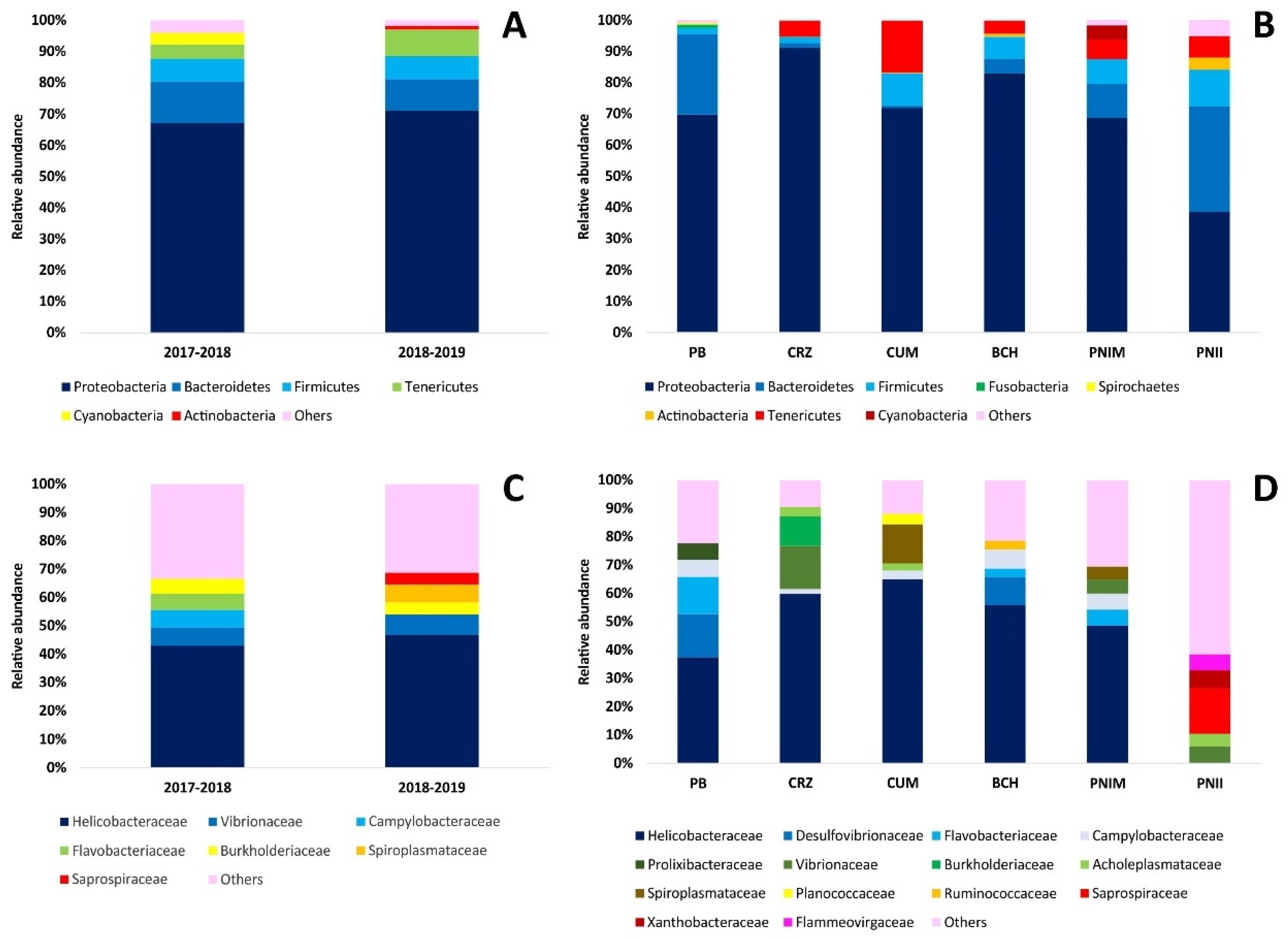
3.1. Characterization of the Bacterial Assembly of T. roseus
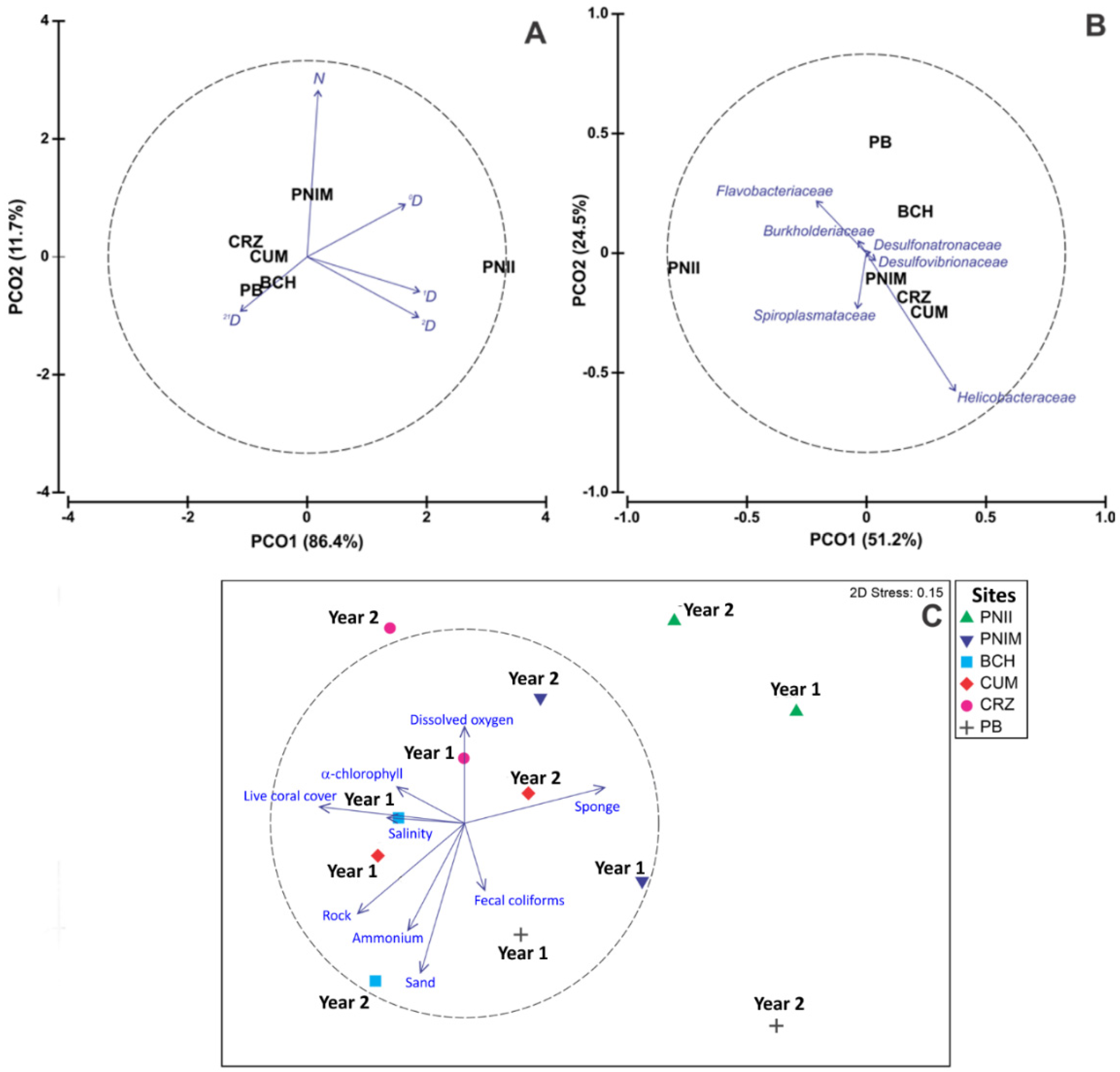
3.2. Alpha and Beta Diversity
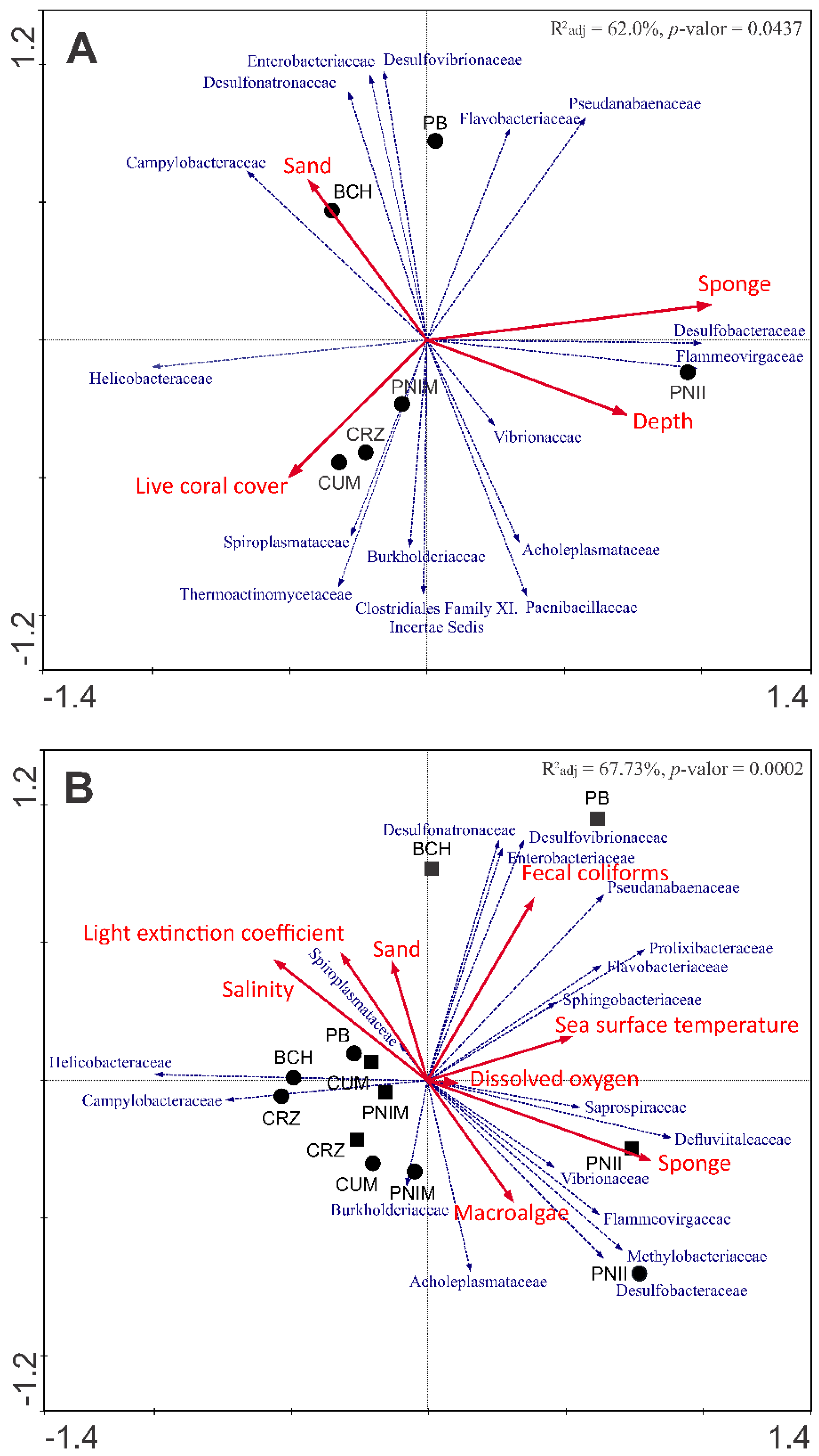
3.3. Relation of Bacterial Assemblage and Environmental Variables
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Steele, J. A., P. D. Countway, L. Xia, P. D. Vigil, J. M. Beman, D. Y. Kim & J. M Rose, 2011. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. The ISME Journal 5(9):1414. [CrossRef]
- Buttigieg, P. L., E. Fadeev, C. Bienhold, L. Hehemann, P. Offre & A. Boetius, 2018. Marine microbes in 4D—using time series observation to assess the dynamics of the ocean microbiome and its links to ocean health. Current Opinion in Microbiology 43: 169–185. [CrossRef]
- Devine, S. P., K. N. Pelletreau & M. E. Rumpho, 2012. 16S rDNA-based metagenomic analysis of bacterial diversity associated with two populations of the kleptoplastic sea slug Elysia chlorotica and its algal prey Vaucheria litorea. The Biological Bulletin 223(1): 138–154.
- Gordon, J., N. Knowlton, D. A. Relman, F. Rohwer & M. Youle, 2013. Superorganisms and holobionts. Microbe 8(4): 152–153. [CrossRef]
- Bosch, T. C. & D. J. Miller, 2016. The holobiont imperative. Vienna: Springer 10:978–3.
- Cleary, D. F., A. R. M. Polónia, Y. M. Huang & T. Swierts, 2020. Compositional variation between high and low prokaryotic diversity coral reef biotopes translates to different predicted metagenomic gene content. Antonie van Leeuwenhoek 113(4): 563–587. [CrossRef]
- Ostria-Hernández, M.L., J. Hernández-Zulueta, O. Vargas-Ponce, L. Díaz-Pérez, R. Araya, A.P. Rodríguez-Troncoso, E. Ríos-Jara & F.A. Rodríguez-Zaragoza, 2022. Core microbiome of corals Pocillopora damicornis and Pocillopora verrucosa in the north-eastern tropical Pacific. Marine Ecology 43: 6 e12729. [CrossRef]
- Amelia, T. S. M., N. S. Lau, A. A. A. Amirul & Bhubalan, K, 2020. Metagenomic data on bacterial diversity profiling of high-microbial-abundance tropical marine sponges Aaptos aaptos and Xestospongia muta from waters off terengganu, South China Sea. Data in brief 31. [CrossRef]
- Hernández-Zulueta, J., L. Díaz-Pérez, J.Q. García-Maldonado, G.G. Nava-Martínez, M.A. García- Salgado & F.A. Rodríguez-Zaragoza, 2022. Bacterial assemblages associated with Acropora palmata affected by white band disease in the Mexican region of the Caribbean and Gulf of Mexico. Journal of Sea Research 185: 102230. [CrossRef]
- Hermosillo-Nuñez, B., F. Rodríguez-Zaragoza, M. Ortiz, C. Galván-Villa, A. Cupul-Magaña & E. Ríos-Jara, 2015. Effect of habitat structure on the most frequent echinoderm species inhabiting coral reef communities at Isla Isabel National Park (Mexico). Community ecology 16(1): 125–134. [CrossRef]
- Hakim, J. A., H. Koo, R. Kumar, E. J. Lefkowitz, C. D. Morrow, M. L. Powell, S.A. Watts & A. K. Bej, 2016. The gut microbiome of the sea urchin, Lytechinus variegatus, from its natural habitat demonstrates selective attributes of microbial taxa and predictive metabolic profiles. FEMS Microbiology Ecology 92(9): fiw146. [CrossRef]
- Brothers, C. J., W. J. Van Der Pol, C. D. Morrow, J. A. Hakim, H. Koo & J. B. McClintock, 2018. Ocean warming alters predicted microbiome functionality in a common sea urchin. Proceedings of the Royal Society B: Biological Sciences 285(1881): 20180340. [CrossRef]
- Brink, M., C. Rhode, B. M. Macey, K. W. Christison & R. Roodt-Wilding, 2019. Metagenomic assessment of body surface bacterial communities of the sea urchin, Tripneustes gratilla. Marine Genomics 47: 1–11. [CrossRef]
- Hakim, J. A., J. B. Schram, A. W. Galloway, C. D. Morrow, M. R. Crowley, S. A. Watts & A. K. Bej, 2019. The Purple Sea Urchin Strongylocentrotus purpuratus demonstrates a compartmentalization of gut bacterial microbiota, predictive functional attributes, and taxonomic co-occurrence. Microorganisms 7(2):35.
- Becker, P. T., S. Samadi, M. Zbinden, C. Hoyoux, P. Compère & C. De Ridder, 2009. First insights into the gut microflora associated with an echinoid from wood falls environments. Cahiers de Biologie Marine 50(4): 343.
- Zhang, F., Y. Tian, F. Gao, S. Chen, D. Li & Y. Zhou, 2014. Bacterial community in the intestine of the sea urchin Strongylocentrotus intermedius during digestion of Macrocystis pyrifera. Marine and Freshwater Behaviour and Physiology 47(2): 117–127. [CrossRef]
- Thorsen, M. S., A., Wieland, H., Ploug, C., Kragelund, & P. H. Nielsen, 2003. Distribution, identity and activity of symbiotic bacteria in anoxic aggregates from the hindgut of the sea urchin Echinocardium cordatum. Ophelia 57(1): 1–12.
- Granja-Fernández, R., B. Maya-Alvarado, A.L. Cupul-Magaña, A.P. Rodríguez- Troncoso, F.A. Solís-Marín & R.C. Sotelo-Casas, 2021. Echinoderms (Echinodermata) from the Central Mexican Pacific. Revista de Biología Tropical 69(S1):219–253. [CrossRef]
- Hermosillo-Núñez, B. B., F. A. Rodríguez-Zaragoza, M. Ortiz, L. E. Calderon-Aguilera & A. L. Cupul-Magaña, 2016. Influence of the coral reef assemblages on the spatial distribution of echinoderms in a gradient of human impacts along the tropical Mexican Pacific. Biodiversity and Conservation 25(11): 2137–2152. [CrossRef]
- Sotelo-Casas, R. C., A. L. Cupul-Magaña, F. A. Rodríguez-Zaragoza, F. A. Solís-Marín & A. P. Rodríguez-Troncoso, 2018. Structural and environmental effects on an assemblage of echinoderms associated with a coral community. Marine Biodiversity 48(3):1401–1411. [CrossRef]
- Glynn, P. W., 1988. El Niño warming, coral mortality y reef framework destruction by echinoid bioerosion in the eastern Pacific. Galaxea 7(2): 129–160.
- James, D. W., 2000. Diet, movement, and covering behavior of the sea urchin Toxopneustes roseus in rhodolith beds in the Gulf of California, México. Marine Biology 137(5-6):913–923. [CrossRef]
- Nakagawa, H., H. Hashimoto, H. Hayashi, M. Shinohara, K. Ohura, E. Tachikawa & T. Kashimoto, 1996. Isolation of a novel lectin from the globiferous pedicellariae of the sea urchin Toxopneustes pileolus. Natural Toxins 391(2): 213–223.
- Satoh, F., H. Nakagawa, H. Yamada, K. Nagasaka, T. Nagasaka, Y. Araki, Y. Tomihara, M. Nozaki, H. Sakuraba, T. Ohshima, T. Hatakeyama & H. Aoyagi, 2002. Fishing for bioactive substances from scorpionfish and some sea urchins. Journal of Natural Toxins 11(4): 297–304.
- Edo, K., H. Sakai, H. Nakagawa, T. Hashimoto, M. Shinohara, & K. Ohura, 2012. Immunomodulatory activity of a pedicellarial venom lectin from the toxopneustid sea urchin Toxopneustes pileolus. Toxin Reviews 31(3-4): 54–60. [CrossRef]
- Spalding, M. D., H. E. Fox, G. R. Allen, N. Davidson, Z. A. Ferdaña, M. Finlayson, B.S. Halpern, M.A. Jorge, A. Lombana, S. Lourie, K.D. Martin, E. McManus, J. Molnar, C.A. Recchia & J. Robertson, 2007. Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. BioScience 57(7): 573–583. [CrossRef]
- Lavín, M.F., R. Castro, E. Beier, V.M. Godínez, A. Amador & P. Guest, 2009. SST, thermohaline structure, and circulation in the southern Gulf of California in June 2004 during the North American Monsoon Experiment. Journal of Geophysical Research: Oceans 114 (C2): C02025. [CrossRef]
- Pantoja, D. A., S. G. Marinone, A. Parés-Sierra & F. Gómez-Valdivia, 2012. Modelación numérica de la hidrografía y circulación estacional y de mesoescala en el Pacífico Central Mexicano. Ciencias marinas 38(2): 363–379.
- Pérez-de Silva, C. V., A. L. Cupul-Magaña, F. A. Rodríguez-Zaragoza & A. P. Rodríguez-Troncoso, 2023. Temporal oceanographic variation using satellite imagery data in the central Mexican Pacific convergence zone. Ciencias Marinas 49. [CrossRef]
- Hernández-Zulueta, J., F.A. Rodriguez-Zaragoza, R. Araya, O. Vargas, A.M. Rodríguez-Troncoso,A. L. Cupul-Magaña, L. Diaz-Perez, E. Rios-Jara & M. Ortiz, 2017. Multi-scale analysis of hermatypic coral assemblages at Mexican Central Pacific. Scientia Marina 81(1): 91–102. [CrossRef]
- Hernández-Zulueta, J., Díaz-Pérez, L., Galván-Villa, C. M., Ayón-Parente, M., Gómez-Petersen, P., Godínez-Domínguez, E. & F. A. Rodríguez-Zaragoza, 2021. Structure and spatial variation of the hermatypic coral assemblages of the southern coast of Jalisco, in the Mexican Central Pacific. Journal of Sea Research 170: 102010. [CrossRef]
- Strickland, J.D.H. & T.R. Parsons, 1968. Determination of reactive nitrate. A practical handbook of seawater analysis. Bulletin of the Fisheries Research Board of Canada167:71–75.
- Poole, H.H, & W.R.G. Atkins, 1929. Photo-electric measurement of submarine illumination troughout the year. Journal of the Marine Biological Association of the United Kingdom 16: 297–324.
- Rodríguez-Zaragoza, F. A., A. L. Cupul-Magaña, C. M. Galván-Villa, E. Ríos-Jara, M. Ortiz, E. G. Robles-Jarero & J. E. Arias-González, 2011. Additive partitioning of reef fish diversity variation: a promising marine biodiversity management tool. Biodiversity and Conservation 20(8):1655–1675. [CrossRef]
- Aroson, R.B., P.J. Edmunds, W.F. Precht, D.W. Swanson & D.R. Levitan, 1994. Large scale, long-term monitoring of Caribbean coral reefs: simple, quick, inexpensive techniques. Atoll research bulletin 421:1–19. [CrossRef]
- DeSantis, T.Z., P. Hugenholtz, N. Larsen, M. Rojas, E.L. Brodie, K. Keller, T. Huber, D. Dalevi, G. Hu & L. Andersen, 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology 72: 5069–5072. [CrossRef]
- Gotelli, N. J. & R. K. Colwell, 2001. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecology letters 4(4): 379–391. [CrossRef]
- Chao, A., C. H. Chiu & L. Jost, 2014. Unifying species diversity, phylogenetic diversity, functional diversity, and related similarity and differentiation measures through Hill numbers. Annual review of ecology, evolution, and systematics 45: 297–324. [CrossRef]
- Anderson, M.J., R.N. Gorely & K.R. Clarke, 2008. PERMANOVA+Primer: Guide to Software and Statistical Methods. PRIMER-E Ltd., Plymouth, 214.
- Legendre, P. & E. D. Gallagher, 2001. Ecologically meaningful transformations for ordination of species data. Oecologia 129: 271–280. [CrossRef]
- Clarke, K. R. & R.N. Gorley, 2006. Primer v6: user manual/tutorial. Primer-E Ltd., Plymouth, UK.
- ter Braak, C.J.F. & P. Šmilauer, 2012. CANOCO referent manual and CanoDraw for Windows user’s guide: software for Canonical Community Ordination (version 5.0). Microcomputer Power. Ithaca, NY.
- Hernández-Zulueta, J., R. Araya, O. Vargas-Ponce, L. Díaz-Pérez, A. P. Rodríguez-Troncoso, J. Ceh, E. Ríos-Jara & F. A. Rodríguez-Zaragoza, 2016. First deep screening of bacterial assemblages associated with corals of the Tropical Eastern Pacific. FEMS Microbiology Ecology 92(12): 1–12. [CrossRef]
- Wyrtki, K., 1966. Oceanography of the eastern equatorial Pacific Ocean. Oceanography and Marine Biology: An Annual Review 4: 33–68.
- Filonov, A., C. Monzon & I. Tereshschenko, 1996. On the conditions of internal tide wave generation along the west coast of Mexico. Ciencias Marinas 22: 255–272. [CrossRef]
- Littman, R. A., B. L. Willis, C. Pfeffer & D. G. Bourne, 2009. Diversities of coral associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbiology Ecology 68: 152–163.
- Carlos, C., T. T. Torres & L. Ottoboni, 2013. Bacterial communities and species-specific associations with the mucus of Brazilian coral species. Scientific Reports 3:1624. [CrossRef]
- Baldassarre, L., H. Ying, A.M. Reitzel, S. Franzenburg & S. Fraune, 2022. Microbiota mediated plasticity promotes thermal adaptation in the sea anemone Nematostella vectensis. Nature Communications 13: 3804. [CrossRef]
- Maire, J. & M.J.H. van Oppen, 2022. A role for bacterial experimental evolution in coral bleaching mitigation? Trends in Microbiology 30: 217–228.10.1016/j.tim.2021.07.006. [CrossRef]
- Ríos-Jara, E., C. M. Galván-Villa & F. A. Solís-Marín, 2008. Equinodermos del Parque Nacional Isla Isabel, Nayarit, México. Revista mexicana de biodiversidad 79(1): 131–141.
- Ahmed, H. I., Herrera, M., Liew, Y. J., & Aranda, M. (2019). Long-term temperature stress in the coral model Aiptasia supports the “Anna Karenina principle” for bacterial microbiomes. Frontiers in microbiology, 10, 975. [CrossRef]
- Lloyd, M. M. & M. H. Pespeni, 2018. Microbiome shifts with onset and progression of Sea Star Wasting Disease revealed through time course sampling. Scientific reports 8(1): 1–12. [CrossRef]
- Yang, F., Z. Xiao, Z. Wei & Long, L, 2021. Bacterial communities associated with healthy and bleached crustose coralline alga Porolithon onkodes. Frontiers in Microbiology12: 1365. [CrossRef]
- Xie, R. & X. Fang, 2020. The unusual 2014–2016 El Niño events: Dynamics, prediction and enlightenments. Science China Earth Sciences 63: 626–633. [CrossRef]
- Schwob, G., L. Cabrol, E. Poulin & J. Orlando, 2020. Characterization of the gut microbiota of the Antarctic heart urchin (Spatangoida) Abatus agassizii. Frontiers in microbiology 11: 308. [CrossRef]
- Miller, P. M., T. Lamy, H. M. Page & R. J. Miller, 2021. Sea urchin microbiomes vary with habitat and resource availability. Limnology and Oceanography Letters 6(3): 119–126. [CrossRef]
- Kroeker, K. J., L. E. Bell, E. M. Donham, U. Hoshijima, S. Lummis, J. A. Toy & E. Willis-Norton, 2020. Ecological change in dynamic environments: Accounting for temporal environmental variability in studies of ocean change biology. Global change biology 26(1): 54–67. [CrossRef]
- Pantos, O., P. Bongaerts, P. G. Dennis, G. W. Tyson & O. Hoegh-Guldberg, 2015. Habitat-specific environmental conditions primarily control the microbiomes of the coral Seriatopora hystrix. The ISME journal 9(9): 1916–1927. [CrossRef]
- Tanrattanapitak, N. & S. Pairohakul, 2018. Bacterial Community in Gut Contents of the Sea Urchin Diadema setosum (Leske, 1778) and the Ambient Sediments from Sichang Island using Metagenomics Approaches. NU. International Journal of Science 15(1): 117–125.
- Carrier, T. J., S. Dupont & A. M. Reitzel, 2019. Geography, not food availability, reflects compositional differences in the bacterial communities associated with larval sea urchins. bioRxiv 394486.
- Yao, Q., K. Yu, J. Liang, Y. Wang, B. Hu, X. Huang, B. Chen & Z. Qin, 2019. The composition, diversity and predictive metabolic profiles of bacteria associated with the gut digesta of five sea urchins in Luhuitou fringing reef (northern South China Sea). Frontiers in microbiology 10: 1168. [CrossRef]
- Faddetta, T., F. Ardizzone, F. Faillaci, C. Reina, E. Palazzotto, F. Strati, C. De Filippo, G. Spinelli, A. M. Puglia, G. Gallo & V. Cavalieri, 2020. Composition and geographic variation of the bacterial microbiota associated with the coelomic fluid of the sea urchin Paracentrotus lividus. Scientific reports 10(1): 1–12. [CrossRef]
- Sotelo-Casas, R. C., A. P. Rodríguez-Troncoso, F. A. Rodríguez-Zaragoza, F. A. Solís-Marín, E. Godínez-Domínguez & A. L. Cupul-Magaña, 2019. Spatial-temporal variations in echinoderm diversity within coral communities in a transitional region of the northeast of the eastern pacific. Estuarine, Coastal and Shelf Science 227: 106346. [CrossRef]
- Hernández-Zulueta, J., L. Díaz-Pérez, R. Araya, O. Vargas-Ponce, A. P. Rodríguez-Troncoso, E. Ríos-Jara, M. Ortiz & F. A. Rodríguez-Zaragoza, 2017. Bacterial assemblages associated with coral species of the Mexican Central Pacific. Revista de Biología Marina y Oceanografía 52: 201–218. [CrossRef]
- Saad, S. M., Hassnien, F. S., Abdel-Aal, M. M., Zakar, A. H., & Elshfey, S. A. (2018). Enterobacteriaceae in some fresh and marine fish. Benha Veterinary Medical Journal, 34(1), 261-268. [CrossRef]
- Shore, A., Sims, J. A., Grimes, M., Howe-Kerr, L. I., Grupstra, C.G.B., Doyle, S.M., Stadler, L., Sylvan, J.B., Shamberger, K.E.F., Davies, S.W., Santiago-Vázquez, L.Z., & Correa A.M.S (2020). On a reef far, far away: Offshore transport of floodwaters following extreme storms impacts sponge health and associated microbial communities. Front. Mar. Sci. 8:608036. [CrossRef]
- Navarrete-Euan, H., Rodríguez-Escamilla, Z., Pérez-Rueda, E., Escalante-Herrera, K., & Martínez-Núñez, M. A. (2021). Comparing Sediment Microbiomes in Contaminated and Pristine Wetlands along the Coast of Yucatan. Microorganisms, 9(4), 877. [CrossRef]
- Kim, M., Heo, E., Kang, H., & Adams, J. (2013). Changes in soil bacterial community structure with increasing disturbance frequency. Microbial ecology, 66(1), 171-181. [CrossRef]
- Lee, J., Jung, Y. J., Lee, H. K., Hong, S. G., & Kim, O. S. (2016). Complete genome sequence of Pedobacter cryoconitis PAMC 27485, a CRISPR-Cas system-containing psychrophile isolated from Antarctica. Journal of biotechnology, 226, 74-75. [CrossRef]
- Wu, S., Li, Y., Wang, P., Zhong, L., Qiu, L., & Chen, J. (2016). Shifts of microbial community structure in soils of a photovoltaic plant observed using tag-encoded pyrosequencing of 16S rRNA. Applied microbiology and biotechnology, 100(8), 3735-3745. [CrossRef]
- Barneah, O., Ben-Dov, E., Kramarsky-Winter, E., & Kushmaro, A. (2007). Characterization of black band disease in Red Sea stony corals. Environmental microbiology, 9(8), 1995-2006. [CrossRef]
- McDevitt-Irwin, J. M., Baum, J. K., Garren, M., & Vega Thurber, R. L. (2017). Responses of coral-associated bacterial communities to local and global stressors. Frontiers in Marine Science, 4, 262. [CrossRef]
| Factors | Pseudo-F | P-value | C.V.% |
| Alpha diversity | |||
| Year | 2.0658 | 0.1336 | 14.4 |
| Site | 3.3813 | 0.0127 | 37.3 |
| Year*Site | 0.34989 | 0.9788 | 0.0 |
| Residuals | 48.3 | ||
| Beta diversity | |||
| Year | 0.89 | 0.0283 | 16.1 |
| Site | 0.62 | 0.0277 | 19.5 |
| Year*Site | 0.47 | 0.1463 | 17.9 |
| Residuals | 0.36 | 46.5 | |
| Environmental Variables | |||
| Year | 1.8421 | 0.1162 | 17.9 |
| Site | 2.0416 | 0.0032 | 34.4 |
| Residuals | 47.7 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





