Submitted:
30 May 2024
Posted:
31 May 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Materials and Methods
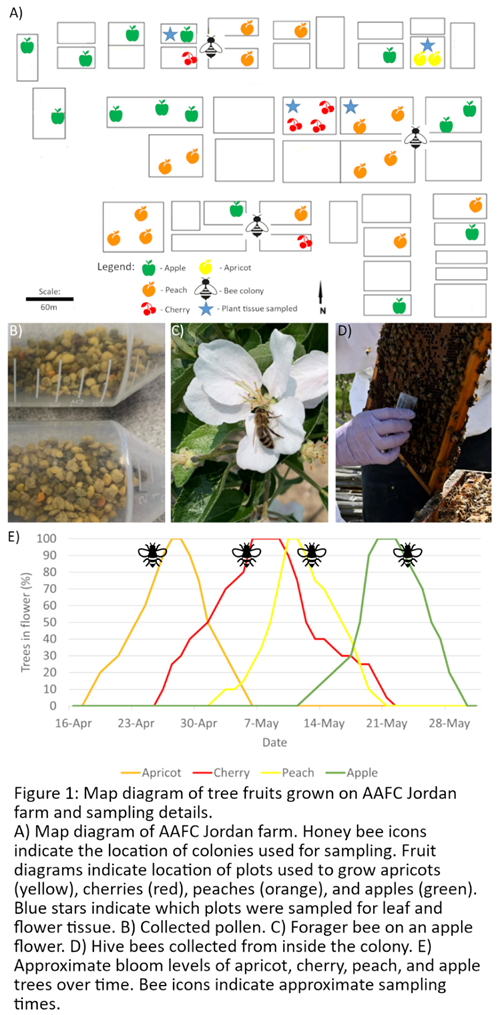
Farm Site and Sampling Details
RNA Extraction and Sequencing
Bioinformatics and Phylogenetics
Results
Virus Detection during Tree Fruit Bloom

Virus Species Diversity in Different Sample Types and Sampling Times
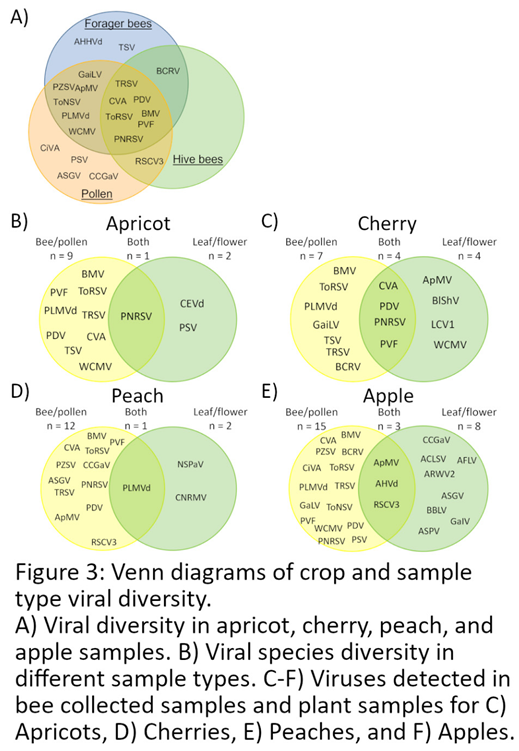
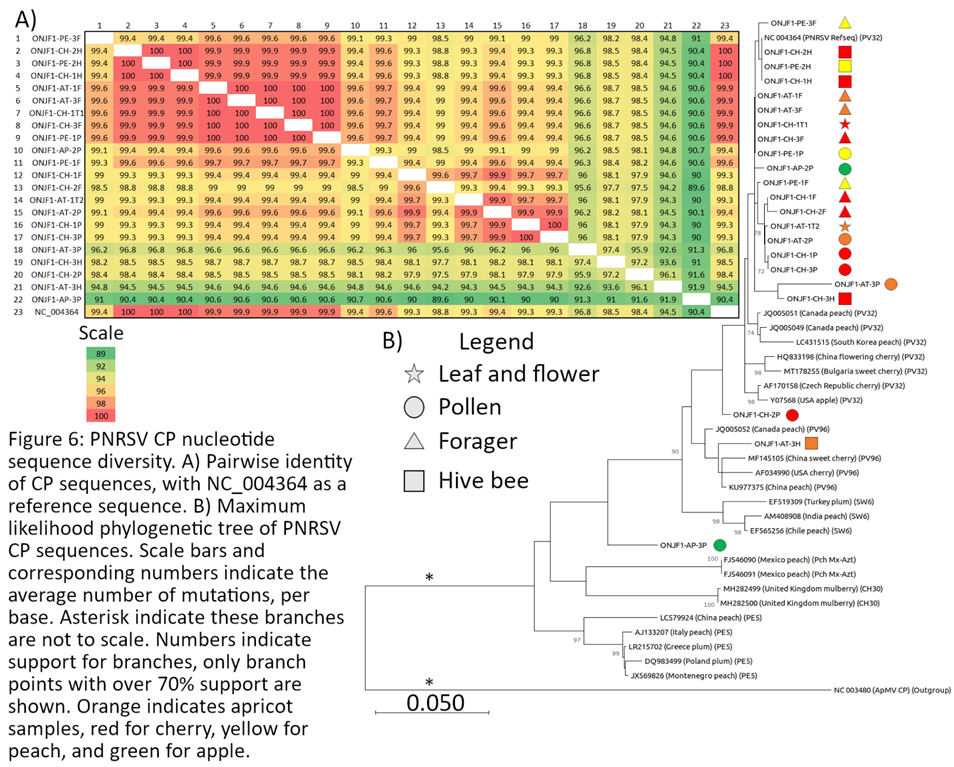
Coat Protein Sequence Diversity of CVA, PDV, and PNRSV


| Virus species | Genus or family | Frequency (%) | Total Detections(n) | Average frequency of detection (%) | Average Genome Coverage (%) | Average VRPM | |||
| Apricot | Cherry | Peach | Apple | ||||||
| Plant tissue | |||||||||
| n=2 | n=2 | n=2 | n=2 | ||||||
| Prunus necrotic ringspot virus | Ilarvirus | 100 | 50 | 3 | 38 | 70.8 | 2041 | ||
| Apple mosaic virus | Ilarvirus | 50 | 50 | 2 | 25 | 20.6 | 0 | ||
| Cherry virus A | Capillovirus | 100 | 2 | 25 | 99.6 | 1181 | |||
| Prune dwarf virus | Ilarvirus | 100 | 2 | 25 | 15.7 | 1 | |||
| Apple chlorotic leaf spot virus | Trichovirus | 100 | 2 | 25 | 78.7 | 500 | |||
| Apple flat limb virus | Rubodvirus | 100 | 2 | 25 | 39.65 | 4 | |||
| Apple rubbery wood virus 2 | Rubodvirus | 100 | 2 | 25 | 91.5 | 107 | |||
| Apple stem pitting virus | Foveavirus | 100 | 2 | 25 | 67.6 | 225 | |||
| Prunus virus F | Fabavirus | 100 | 2 | 25 | 97.1 | 67 | |||
| Little cherry virus 1 | Velarivirus | 100 | 2 | 25 | 99.95 | 1314 | |||
| Nectarine stem pitting-associated virus | Luteovirus | 100 | 2 | 25 | 82.2 | 223 | |||
| Cherry necrotic rusty mottle virus | Robigovirus | 100 | 2 | 25 | 95.05 | 58 | |||
| Raphanus sativus cryptic virus 3 | Unclassified Partitiviridae | 50 | 1 | 13 | 18.9 | 0 | |||
| Apple hammerhead viroid | Pelamoviroid | 50 | 1 | 13 | 100 | 2410 | |||
| Apple stem grooving virus | Capillovirus | 50 | 1 | 13 | 99.6 | 7528 | |||
| Citrus concave gum associated virus | Coguvirus | 50 | 1 | 13 | 99.2 | 417 | |||
| Peach latent mosaic viroid | Pelmaviroid | 50 | 1 | 13 | 99.6 | 27 | |||
| Blueberry latent virus | Amalgavirus | 50 | 1 | 13 | 13.2 | 4 | |||
| Blueberry shock virus | Ilarvirus | 50 | 1 | 13 | 11.1 | 7 | |||
| Citrus excordis viroid | Pospiviroidae | 50 | 1 | 13 | 96.4 | 169 | |||
| Grapevine associated ilarvirus | Ilarvirus | 50 | 1 | 13 | 24.6 | 1 | |||
| Peanut stunt virus | Cucumovirus | 50 | 1 | 13 | 16.9 | 0 | |||
| White clover mosaic virus | Potexvirus | 50 | 1 | 13 | 26.7 | 0 | |||
Discussion
Metagenomics Based Detection of Plant Viruses through Pollen
Viral Diversity in Bee-Collected Pollen Samples
Sequence Diversity of Identified Viruses
Conclusion
Competing Interests
Author Contributions
Funding
Acknowledgements
References
- Soundararajan, P.; Won, S.Y.; Kim, J.S. Insight on Rosaceae Family with Genome Sequencing and Functional Genomics Perspective. BioMed Res. Int. 2019, 2019, 1–12, . [CrossRef]
- AAFC - Agriculture and Agri-Food Canada. Statistical overview of the Canadian Fruit Industry 2021. Horticulture Section, Crops and Horticulture Division (2022). https://agriculture.canada.ca/sites/default/files/documents/2022-12/Fruit%20Report_2021_ENG.pdf.
- Umer, M.; Liu, J.; You, H.; Xu, C.; Dong, K.; Luo, N.; Kong, L.; Li, X.; Hong, N.; Wang, G.; et al. Genomic, Morphological and Biological Traits of the Viruses Infecting Major Fruit Trees. Viruses 2019, 11, 515, . [CrossRef]
- Fetters, A.M.; Cantalupo, P.G.; Wei, N.; Robles, M.T.S.; Stanley, A.; Stephens, J.D.; Pipas, J.M.; Ashman, T.-L. The pollen virome of wild plants and its association with variation in floral traits and land use. Nat. Commun. 2022, 13, 1–11, . [CrossRef]
- Smadi, M.; Lee, E.; Phelan, J.; Wang, A.; Bilodeau, G.J.; Pernal, S.F.; Guarna, M.M.; Rott, M.; Griffiths, J.S. Plant virus diversity in bee and pollen samples from apple (Malus domestica) and sweet cherry (Prunus avium) agroecosystems. Front. Plant Sci. 2024, 15, 1335281, . [CrossRef]
- Chandel V, Rana T, Handa A, Thakur PD, Hallan V, Zaidi AA. Incidence of Prunus necrotic ringspot virus on Malus domestica in India. J. Phytopath., 2008, 156, 382-384.
- Hu, G.J.; Dong, Y.F.; Zhang, Z.P.; Fan, X.D.; Ren, F.; Li, Z.N.; Zhou, J. First Report of Prunus necrotic ringspot virus Infection of Apple in China. Plant Dis. 2016, 100, 1955, . [CrossRef]
- Kinoti, W.M.; Constable, F.E.; Nancarrow, N.; Plummer, K.M.; Rodoni, B. Generic Amplicon Deep Sequencing to Determine Ilarvirus Species Diversity in Australian Prunus. Front. Microbiol. 2017, 8, 1219, . [CrossRef]
- Çelik, A.; Ertunç, F. First report of prunus necrotic ringspot virus infecting apple in Turkey. J. Plant Pathol. 2019, 101, 1227–1227, . [CrossRef]
- Xiao, H.; Hao, W.; Storoschuk, G.; MacDonald, J.L.; Sanfaçon, H. Characterizing the Virome of Apple Orchards Affected by Rapid Decline in the Okanagan and Similkameen Valleys of British Columbia (Canada). Pathogens 2022, 11, 1231, . [CrossRef]
- Kozieł, E.; Bujarski, J.J.; Otulak, K. Molecular Biology of Prune Dwarf Virus—A Lesser Known Member of the Bromoviridae but a Vital Component in the Dynamic Virus–Host Cell Interaction Network. Int. J. Mol. Sci. 2017, 18, 2733, . [CrossRef]
- Kesanakurti, P.; Belton, M.; Saeed, H.; Rast, H.; Boyes, I.; Rott, M. Screening for plant viruses by next generation sequencing using a modified double strand RNA extraction protocol with an internal amplification control. J. Virol. Methods 2016, 236, 35–40, . [CrossRef]
- Bag, S.; Al Rwahnih, M.; Li, A.; Gonzalez, A.; Rowhani, A.; Uyemoto, J.K.; Sudarshana, M.R. Detection of a New Luteovirus in Imported Nectarine Trees: A Case Study to Propose Adoption of Metagenomics in Post-Entry Quarantine. Phytopathology® 2015, 105, 840–846, . [CrossRef]
- Villamor, D.E.V.; Mekuria, T.A.; Pillai, S.S.; Eastwell, K.C.; Ho, T.; Al Rwahnih, M.; Martin, R.R.; Tzanetakis, I.E.; Green, K.J.; Mollov, D.; et al. High-Throughput Sequencing Identifies Novel Viruses in Nectarine: Insights to the Etiology of Stem-Pitting Disease. Phytopathology® 2016, 106, 519–527, . [CrossRef]
- Wright, A.A.; Cross, A.R.; Harper, S.J. A bushel of viruses: Identification of seventeen novel putative viruses by RNA-seq in six apple trees. PLOS ONE 2020, 15, e0227669, . [CrossRef]
- McLeish MJ, Fraile A, Farcia-Arsenal F. Ecological complexity in plant virus host range evolution. Adv. Virus Res., 2018, 101: 293-339.
- McLeish, M.J.; Fraile, A.; García-Arenal, F. Evolution of plant–virus interactions: host range and virus emergence. Curr. Opin. Virol. 2019, 34, 50–55, . [CrossRef]
- Hamelin, F.M.; Allen, L.J.; Prendeville, H.R.; Hajimorad, M.R.; Jeger, M.J. The evolution of plant virus transmission pathways. J. Theor. Biol. 2016, 396, 75–89, . [CrossRef]
- Gallet, R.; Michalakis, Y.; Blanc, S. Vector-transmission of plant viruses and constraints imposed by virus–vector interactions. Curr. Opin. Virol. 2018, 33, 144–150, . [CrossRef]
- McLaughlin, A.A.; Hanley-Bowdoin, L.; Kennedy, G.G.; Jacobson, A.L. Vector acquisition and co-inoculation of two plant viruses influences transmission, infection, and replication in new hosts. Sci. Rep. 2022, 12, 1–13, . [CrossRef]
- Card, S.D.; Pearson, M.N.; Clover, G.R.G. Plant pathogens transmitted by pollen. Australas. Plant Pathol. 2007, 36, 455–461, . [CrossRef]
- Jones RAC. Plant and insect viruses in managed and natural environments: novel and neglected transmission pathways. Adv. Virus Res., 2018, 101: 149-187.
- Amari, K.; Burgos, L.; Pallas, V.; Sanchez-Pina, M.A. Prunus necrotic ringspot virusEarly Invasion and Its Effects on Apricot Pollen Grain Performance. Phytopathology® 2007, 97, 892–899, . [CrossRef]
- Amari, K.; Burgos, L.; Pallás, V.; Sánchez-Pina, M.A. Vertical transmission of Prunus necrotic ringspot virus: hitch-hiking from gametes to seedling. J. Gen. Virol. 2009, 90, 1767–1774, . [CrossRef]
- Isogai, M.; Yoshida, T.; Nakanowatari, C.; Yoshikawa, N. Penetration of pollen tubes with accumulated Raspberry bushy dwarf virus into stigmas is involved in initial infection of maternal tissue and horizontal transmission. Virology 2014, 452-453, 247–253, . [CrossRef]
- Shipp, J.; Buitenhuis, R.; Stobbs, L.; Wang, K.; Kim, W.; Ferguson, G. Vectoring of Pepino mosaic virus by bumble-bees in tomato greenhouses. Ann. Appl. Biol. 2008, 153, 149–155, . [CrossRef]
- Levitzky, N.; Smith, E.; Lachman, O.; Luria, N.; Mizrahi, Y.; Bakelman, H.; Sela, N.; Laskar, O.; Milrot, E.; Dombrovsky, A. The bumblebee Bombus terrestris carries a primary inoculum of Tomato brown rugose fruit virus contributing to disease spread in tomatoes. PLOS ONE 2019, 14, e0210871, . [CrossRef]
- Tayal, M.; Wilson, C.; Cieniewicz, E. Bees and thrips carry virus-positive pollen in peach orchards in South Carolina, United States. J. Econ. Èntomol. 2023, 116, 1091–1101, . [CrossRef]
- Oliver, J.E.; Freer, J.; Andersen, R.L.; Cox, K.D.; Robinson, T.L.; Fuchs, M. Genetic Diversity of Prunus necrotic ringspot virus Isolates Within a Cherry Orchard in New York. Plant Dis. 2009, 93, 599–606, . [CrossRef]
- Abdallah, D.; Baraket, G.; Perez, V.; Hannachi, A.S.; Hormaza, J.I. Self-compatibility in peach [Prunus persica (L.) Batsch]: patterns of diversity surrounding the S-locus and analysis of SFB alleles. Hortic. Res. 2020, 7, 170, . [CrossRef]
- Piri S, Kiani E, Sedaghathoor S. Study on fruitset and pollen-compatibility status in sweet cherry (Prunus avium L.) cultivars. Erwebs-Obstbau, 2022, 64: 165-170.
- Delaplane K S, Mayer D F. Crop Pollination by Bees. CABI Publishing: New York. 2000.
- Beekman, M.; Ratnieks, F.L.W. Long-range foraging by the honey-bee, Apis mellifera L.. Funct. Ecol. 2000, 14, 490–496, . [CrossRef]
- Steffan-Dewenter, I.; Kuhn, A. Honeybee foraging in differentially structured landscapes. Proc. R. Soc. B: Biol. Sci. 2003, 270, 569–575, . [CrossRef]
- Roberts, J.M.K.; Ireland, K.B.; Tay, W.T.; Paini, D. Honey bee-assisted surveillance for early plant virus detection. Ann. Appl. Biol. 2018, 173, 285–293, . [CrossRef]
- Lee, E.; Vansia, R.; Phelan, J.; Lofano, A.; Smith, A.; Wang, A.; Bilodeau, G.J.; Pernal, S.F.; Guarna, M.M.; Rott, M.; et al. Area Wide Monitoring of Plant and Honey Bee (Apis mellifera) Viruses in Blueberry (Vaccinium corymbosum) Agroecosystems Facilitated by Honey Bee Pollination. Viruses 2023, 15, 1209, . [CrossRef]
- Cunningham, M.M.; Tran, L.; McKee, C.G.; Polo, R.O.; Newman, T.; Lansing, L.; Griffiths, J.S.; Bilodeau, G.J.; Rott, M.; Guarna, M.M. Honey bees as biomonitors of environmental contaminants, pathogens, and climate change. Ecol. Indic. 2021, 134, 108457, . [CrossRef]
- Hong, C.; Manimaran, S.; Shen, Y.; Perez-Rogers, J.F.; Byrd, A.L.; Castro-Nallar, E.; A Crandall, K.; Johnson, W.E. PathoScope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome 2014, 2, 33–33, . [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359, . [CrossRef]
- Xiang, Y.; Belton, M.; Saeed, H.; Hayes, S.; Lawrence, T.; Birch, C.; Bhagwat, B.; et al. Application of Next Generation Sequencing for Diagnostic Testing of Tree Fruit Viruses and Viroids. Plant Dis. 2017, 101, 1489–1499, . [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027, . [CrossRef]
- Gao, R.; Xu, Y.; Candresse, T.; He, Z.; Li, S.; Ma, Y.; Lu, M. Further insight into genetic variation and haplotype diversity of Cherry virus A from China. PLOS ONE 2017, 12, e0186273, . [CrossRef]
- Simkovich, A.J.; Li, Y.; Kohalmi, S.E.; Griffiths, J.S.; Wang, A. Molecular Identification of Prune Dwarf Virus (PDV) Infecting Sweet Cherry in Canada and Development of a PDV Full-Length Infectious cDNA Clone. Viruses 2021, 13, 2025, . [CrossRef]
- Kinoti, W.M.; Constable, F.E.; Nancarrow, N.; Plummer, K.M.; Rodoni, B. The Incidence and Genetic Diversity of Apple Mosaic Virus (ApMV) and Prune Dwarf Virus (PDV) in Prunus Species in Australia. Viruses 2018, 10, 136, . [CrossRef]
- Kamenova, I.; Borisova, A.; Popov, A. Incidence and genetic diversity of Prune dwarf virus in sweet and sour cherry in Bulgaria. Biotechnol. Biotechnol. Equip. 2019, 33, 980–987, . [CrossRef]
- Glassa M, Betinova E, Kudela O, Subr Z. Biological and molecular characterization of Prunus necrotic ringspot virus isolates and possible approaches to their phylogenetic typing. Ann. App. Biol., 2002, 140, 215-329.
- Free, J.B. The Flower Constancy of Honeybees. J. Anim. Ecol. 1963, 32, 119–131, . [CrossRef]
- Danner, N.; Molitor, A.M.; Schiele, S.; Härtel, S.; Steffan-Dewenter, I. Season and landscape composition affect pollen foraging distances and habitat use of honey bees. Ecol. Appl. 2016, 26, 1920–1929, . [CrossRef]
- Jones, L.; Lowe, A.; Ford, C.R.; Christie, L.; Creer, S.; de Vere, N. Temporal Patterns of Honeybee Foraging in a Diverse Floral Landscape Revealed Using Pollen DNA Metabarcoding of Honey. Integr. Comp. Biol. 2022, 62, 199–210, . [CrossRef]
- Lowe, A.; Jones, L.; Brennan, G.; Creer, S.; Christie, L.; de Vere, N. Temporal change in floral availability leads to periods of resource limitation and affects diet specificity in a generalist pollinator. Mol. Ecol. 2022, 32, 6363–6376, . [CrossRef]
- Tremblay, .D.; Duceppe, M.; Thurston, G.B.; Gagnon, M.; Côté, M.; Bilodeau, G.J. High-resolution biomonitoring of plant pathogens and plant species using metabarcoding of pollen pellet contents collected from a honey bee hive. Environ. DNA 2019, 1, 155–175, . [CrossRef]
- McKinnon, A.C.; Collins, L.; Wood, J.L.; Murphy, N.; Franks, A.E.; Steinbauer, M.J. Precision Monitoring of Honey Bee (Hymenoptera: Apidae) Activity and Pollen Diversity during Pollination to Evaluate Colony Health. Insects 2023, 14, 95, . [CrossRef]
- Leponiemi, M.; Freitak, D.; Moreno-Torres, M.; Pferschy-Wenzig, E.-M.; Becker-Scarpitta, A.; Tiusanen, M.; Vesterinen, E.J.; Wirta, H. Honeybees’ foraging choices for nectar and pollen revealed by DNA metabarcoding. Sci. Rep. 2023, 13, 1–15, . [CrossRef]
- Leontidou, K.; Vokou, D.; Sandionigi, A.; Bruno, A.; Lazarina, M.; De Groeve, J.; Li, M.; Varotto, C.; Girardi, M.; Casiraghi, M.; et al. Plant biodiversity assessment through pollen DNA metabarcoding in Natura 2000 habitats (Italian Alps). Sci. Rep. 2021, 11, 1–12, . [CrossRef]
- Hasiow-Jaroszewska B, Boezen D, Zwart MP. Metagenomic studies of viruses in weeds and wild plants: a powerful approach to characterize variable virus communities. Viruses, 2021, 13: 1939.
- Bell, K.L.; de Vere, N.; Keller, A.; Richardson, R.T.; Gous, A.; Burgess, K.S.; Brosi, B.J.; Van der Bank, M.; Adamowicz, S.J.; Chain, F.J.; et al. Pollen DNA barcoding: current applications and future prospects. Genome 2016, 59, 629–640, . [CrossRef]
- Milla, L.; Schmidt-Lebuhn, A.; Bovill, J.; Encinas-Viso, F. Monitoring of honey bee floral resources with pollen DNA metabarcoding as a complementary tool to vegetation surveys. Ecol. Solutions Évid. 2022, 3, e12120, . [CrossRef]
- Hoffmann, V.; Paul, B.; Falade, T.; Moodley, A.; Ramankutty, N.; Olawoye, J.; Djouaka, R.; Lekei, E.; de Haan, N.; Ballantyne, P.; et al. A one health approach to plant health. CABI Agric. Biosci. 2022, 3, 1–7, . [CrossRef]
- Fetters, A.M.; Ashman, T. The pollen virome: A review of pollen-associated viruses and consequences for plants and their interactions with pollinators. Am. J. Bot. 2023, 110, e16144, . [CrossRef]
- Pallas V, Aparicio F, Herranz MC, Sanches-Navarro JA, Scott SW. The molecular biology of ilarviruses. Adv. Virus Res., 2013, 87: 139-81.
- Pallas, V.; Aparicio, F.; Herranz, M.C.; Amari, K.; Sanchez-Pina, M.A.; Myrta, A.; Sanchez-Navarro, J.A. Ilarviruses of Prunus spp.: A Continued Concern for Fruit Trees. Phytopathology® 2012, 102, 1108–1120, . [CrossRef]
- Tzanetakis IE, Gergerich RC, Martin RR. A new Ilarvirus found in rose. Plant Pathol., 2006, 55(4): 568–568.
- Tzanetakis IE, Postman JD,, Martin RR. First report of blackberry chlorotic ringspot virus in Rubus sp. in the United States. Plant dis., 2007, 91: 463.
- Poudel, B.; Ho, T.; Laney, A.; Khadgi, A.; Tzanetakis, I.E. Epidemiology of Blackberry chlorotic ringspot virus. Plant Dis. 2014, 98, 547–550, . [CrossRef]
- Bratsch, S.A.; Grinstead, S.; Creswell, T.C.; Ruhl, G.E.; Mollov, D. Characterization of Tomato Necrotic Spot Virus, a Subgroup 1 Ilarvirus Causing Necrotic Foliar, Stem, and Fruit Symptoms in Tomatoes in the United States. Plant Dis. 2019, 103, 1391–1396, . [CrossRef]
- Bonilla, F.O.R.; Cieniewicz, E. Distribution and Diversity of Prunus Necrotic Ringspot Virus, Prune Dwarf Virus, and Peach Latent Mosaic Viroid in Wild Prunus spp. in South Carolina and Georgia. Phytofrontiers™ 2022, 2, 363–370, . [CrossRef]
- Fuchs, M.; Hily, J.-M.; Petrzik, K.; Sanfaçon, H.; Thompson, J.R.; van der Vlugt, R.; Wetzel, T.; ICTV Report Consortium ICTV Virus Taxonomy Profile: Secoviridae 2022. J. Gen. Virol. 2022, 103, 001807, . [CrossRef]
- Thompson JR, Dasgupta I, Fuchs M, Iwanami T, Karasev AV, Petrzik K, Sanfacon H, Tzanetakis I, van der Vlugt R, Wetzel T, Yoshikana N, and ICTV Report Consortium. ICTV virus taxonomy profile: Secoviridae. J. Gen. Virol., 2017, 98: 529-531.
- Koloniuk, I.; Sarkisova, T.; Petrzik, K.; Lenz, O.; Přibylová, J.; Fránová, J.; Špak, J.; Lotos, L.; Beta, C.; Katsiani, A.; et al. Variability Studies of Two Prunus-Infecting Fabaviruses with the Aid of High-Throughput Sequencing. Viruses 2018, 10, 204, . [CrossRef]
- Sasaya, T.; Palacios, G.; Briese, T.; Di Serio, F.; Groschup, M.H.; Neriya, Y.; Song, J.-W.; Tomitaka, Y. ICTV Virus Taxonomy Profile: Phenuiviridae 2023. J. Gen. Virol. 2023, 104, 001893, . [CrossRef]
- Wunsch, A.; Hoff, B.; Sazo, M.M.; van Zoeren, J.; Lamour, K.H.; Hurtado-Gonzales, O.P.; Fuchs, M. Viruses of Apple Are Seedborne but Likely Not Vertically Transmitted. Viruses 2024, 16, 95, . [CrossRef]
- Simkovich, A.; Kohalmi, S.E.; Wang, A. First Report of Little Cherry Virus 1 Infecting Sweet Cherry in Ontario, Canada. Plant Dis. 2021, 105, 4173–4173, . [CrossRef]
- Kesanakurti, P.; Belton, M.; Saeed, H.; Rast, H.; Boyes, I.; Rott, M. Comparative analysis of cherry virus A genome sequences assembled from deep sequencing data. Arch. Virol. 2017, 162, 2821–2828, . [CrossRef]
- Beaver-Kanuya, E.; Harper, S. Detection and quantification of four viruses in Prunus pollen: Implications for biosecurity. J. Virol. Methods 2019, 271, 113673, . [CrossRef]
- Cui H, Hong N, Wang G, Wang A. Detection and genetic diversity of Prunus necrotic ringspot virus in the Niagara fruit belt, Canada. Can. J. plant pathol., 2012, 34, 104-113.
- Kamenova, I.; Borisova, A. Biological and molecular characterization of Prunus necrotic ringspot virus isolates from sweet and sour cherry. Biotechnol. Biotechnol. Equip. 2021, 35, 567–575, . [CrossRef]
| Crop | Number of Plots | Total Area (ha) | Variety | Rootstock | Total number of trees | Age of trees (years) |
|---|---|---|---|---|---|---|
| Apricot | 1 | 0.57 | Haroblush, Harlayne | Krymsk 1, Krymsk 86, Krymsk 99 | 208 | 2 |
| Cherry | 3 | 0.96 | Vista, Vogue, Hedelfingen, Tehranivee, Stella, Vandalay, Montmorency | Mazzard, Mahaleb, Gisela 5 | 317 | 14-50 |
| Peach | 10 | 3.849 | Redhaven, Vivid, Nectarine, Harrow Diamond, Cresthaven | Halford, Bailey, TLC, Bailey Field | 2832 | 2-17 |
| Apple | 11 | 3.83 | Brookfield Gala, Empire, Red Fuji, Ambrosia, Honey Crisp, Silken, Morspur McIntosh, Golden Delicious, Royal Gala, Courtland, Delicious Red Chief, McIntosh Marshall, Jonagold, McIntosh | G41, M26, BUD 9, M9, Mark | 3123 | 3-26 |
| Virus | Genus | Frequency of detection (%) | Total detections(n) | Average frequency of detection (%) | Average Genome Coverage (%) | Average VRPM | |||||||||||
| Apricot | Cherry | Peach | Apple | ||||||||||||||
| Forager | Hive | Pollen | Forager | Hive | Pollen | Forager | Hive | Pollen | Forager | Hive | Pollen | ||||||
| n=3 | n=3 | n=3 | n=3 | n=3 | n=3 | n=3 | n=3 | n=3 | n=3 | n=3 | n=3 | ||||||
| Prune dwarf virus | Ilarvirus | 33 | 33 | 100 | 67 | 100 | 100 | 33 | 100 | 67 | 33 | 100 | 23 | 64 | 62 | 32548 | |
| Cherry virus A | Capillovirus | 67 | 67 | 100 | 67 | 100 | 100 | 33 | 33 | 33 | 100 | 21 | 58 | 70 | 5042 | ||
| Prunus necrotic ringspot virus | Ilarvirus | 67 | 33 | 67 | 100 | 100 | 100 | 67 | 33 | 33 | 100 | 21 | 58 | 54 | 26401 | ||
| Prunus virus F | Fabavirus | 33 | 33 | 67 | 67 | 100 | 100 | 33 | 100 | 16 | 44 | 63 | 2920 | ||||
| Tobacco ringspot virus | Nepovirus | 33 | 33 | 67 | 67 | 67 | 100 | 100 | 67 | 16 | 44 | 47 | 11657 | ||||
| Tomato ringspot virus | Nepovirus | 67 | 67 | 100 | 33 | 33 | 33 | 100 | 67 | 33 | 16 | 44 | 36 | 4000 | |||
| Brome mosaic virus | Bromovirus | 67 | 67 | 100 | 33 | 33 | 67 | 33 | 67 | 33 | 15 | 42 | 29 | 1743 | |||
| Peach latent mosaic viroid | Pelamoviroid | 67 | 67 | 67 | 100 | 67 | 11 | 31 | 37 | 0 | |||||||
| Apple mosaic virus | Ilarvirus | 67 | 67 | 4 | 11 | 33 | 135 | ||||||||||
| White clover mosaic virus | Potexvirus | 33 | 33 | 67 | 4 | 11 | 13 | 343 | |||||||||
| Blackberry chlorotic ringspot virus | Ilarvirus | 33 | 33 | 33 | 3 | 8 | 34 | 5762 | |||||||||
| Pelargonium zonate spot virus | Anulavirus | 33 | 67 | 3 | 8 | 23 | 1449 | ||||||||||
| Tobacco streak virus | Ilarvirus | 67 | 33 | 3 | 8 | 16 | 117 | ||||||||||
| Gaillardia latent virus | Carlavirus | 33 | 33 | 2 | 6 | 18 | 4 | ||||||||||
| Raphanus sativus cryptic virus 3 | Unclassified Partitiviridae | 33 | 33 | 2 | 6 | 47 | 53 | ||||||||||
| Tomato necrotic spot virus | Ilarvirus | 33 | 33 | 2 | 6 | 40 | 296 | ||||||||||
| Apple hammerhead viroid | Pelamoviroid | 33 | 1 | 3 | 20 | 0 | |||||||||||
| Apple stem grooving virus | Capillovirus | 33 | 1 | 3 | 66 | 279 | |||||||||||
| Citrus concave gum associated virus | Coguvirus | 33 | 1 | 3 | 67 | 1408 | |||||||||||
| Citrus virus A | Coguvirus | 33 | 1 | 3 | 28 | 39 | |||||||||||
| Peanut stunt virus | Cucumovirus | 33 | 1 | 3 | 10 | 0 | |||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





