Submitted:
31 May 2024
Posted:
03 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Hydrolysate Generation from Irish and Norwegian Mesopelagic Trawls and Proximate Compositional Analysis of Resulting Hydrolysates
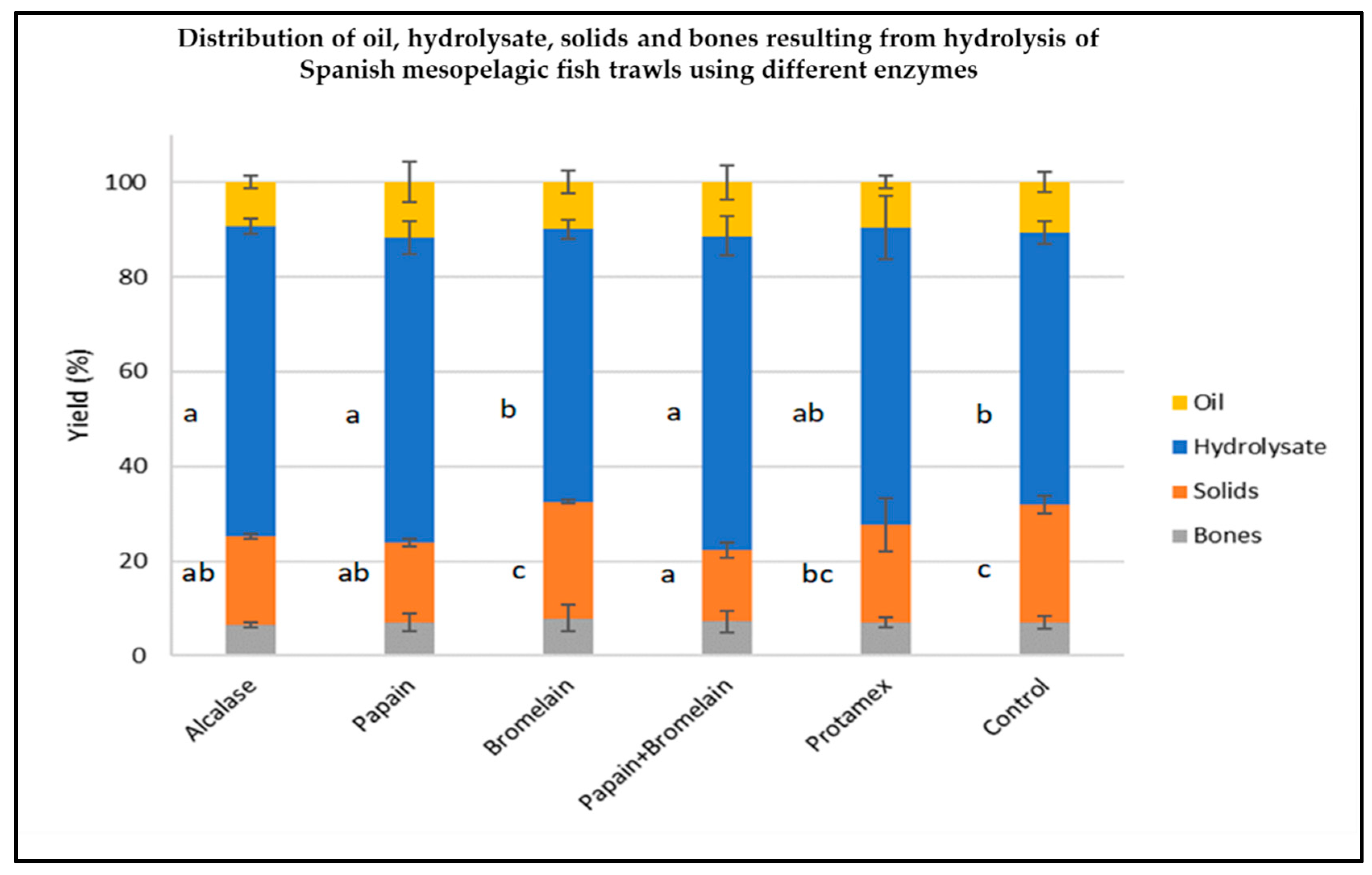
2.2. Hydrolysate Generation from Spanish Mesopelagic Fish Trawl
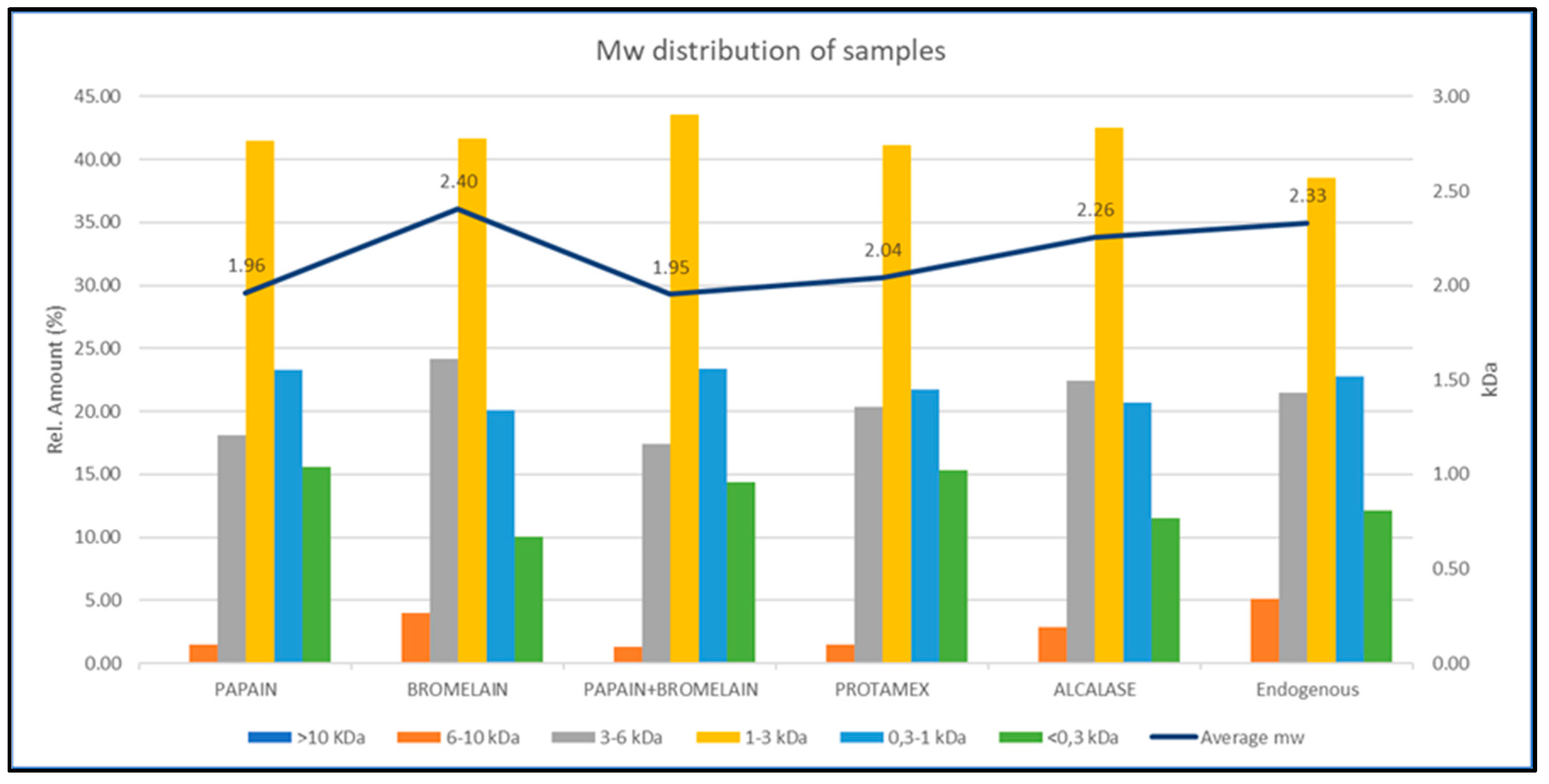
2.3. Molecular Weight Distribution and the Degree of Hydrolysates Generated from M. muelleri Captured on Spanish Trawls
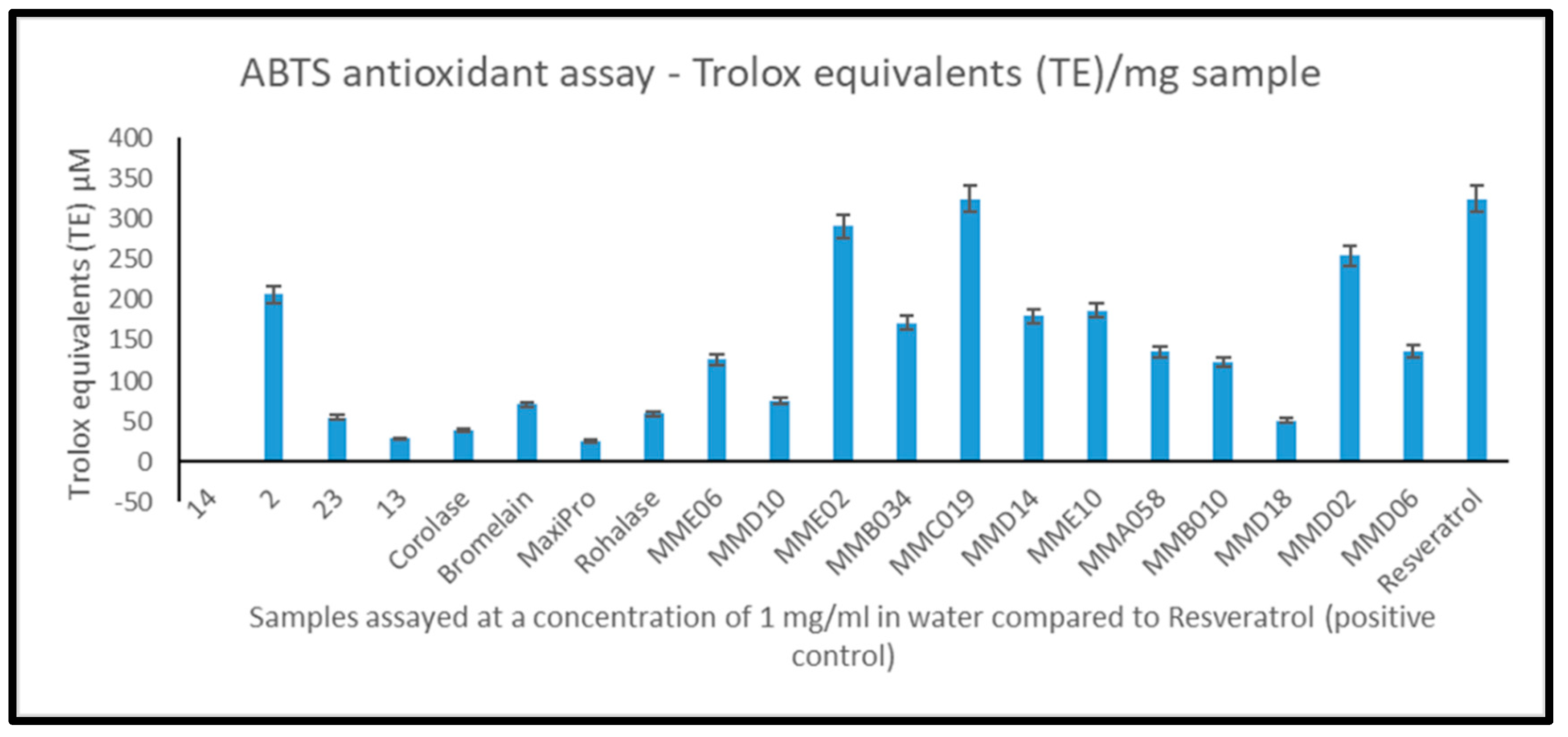
2.4. In Vitro Bioactivity Screening of Generated Mesopelagic Fish Hydrolysates
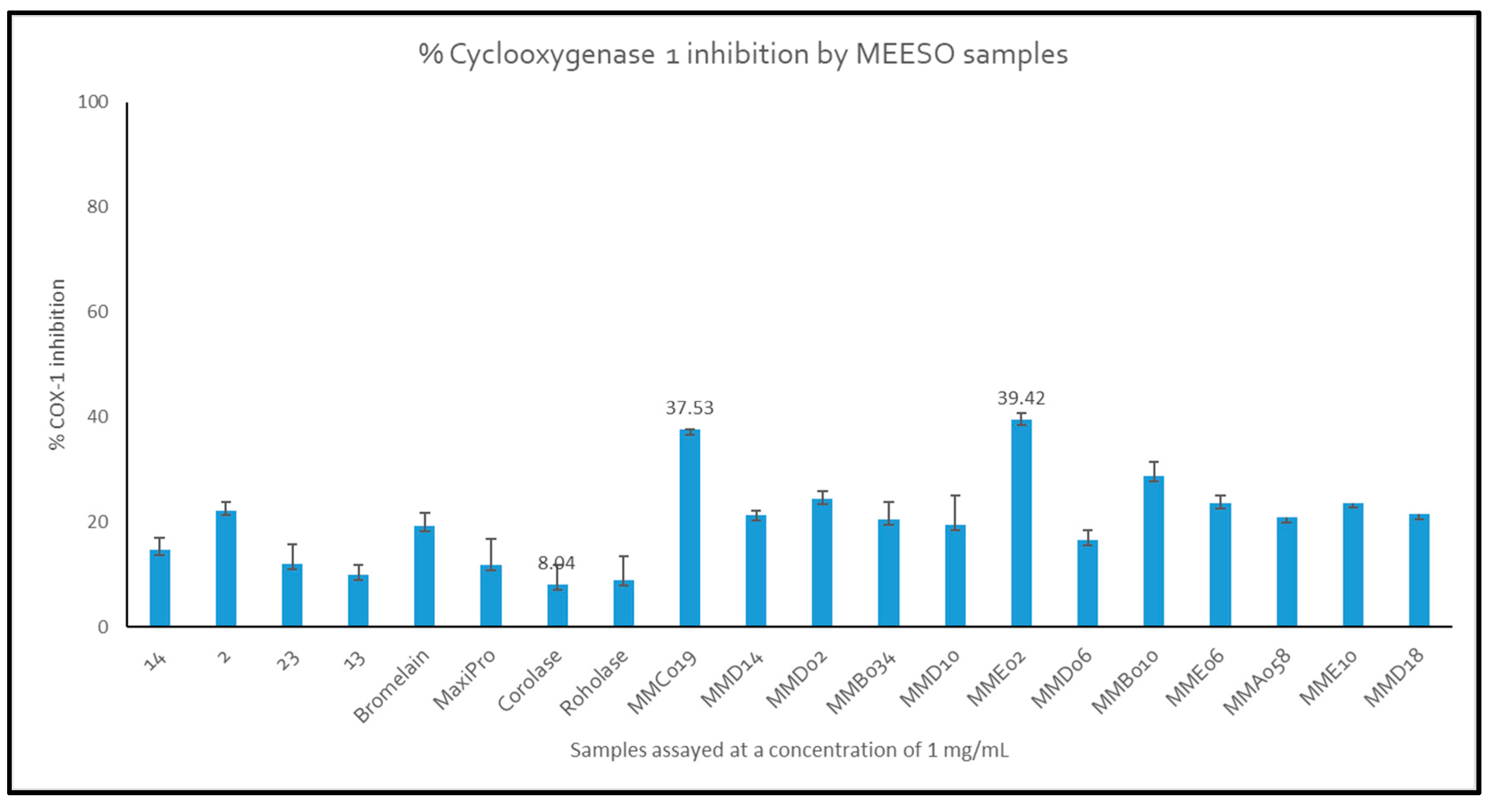
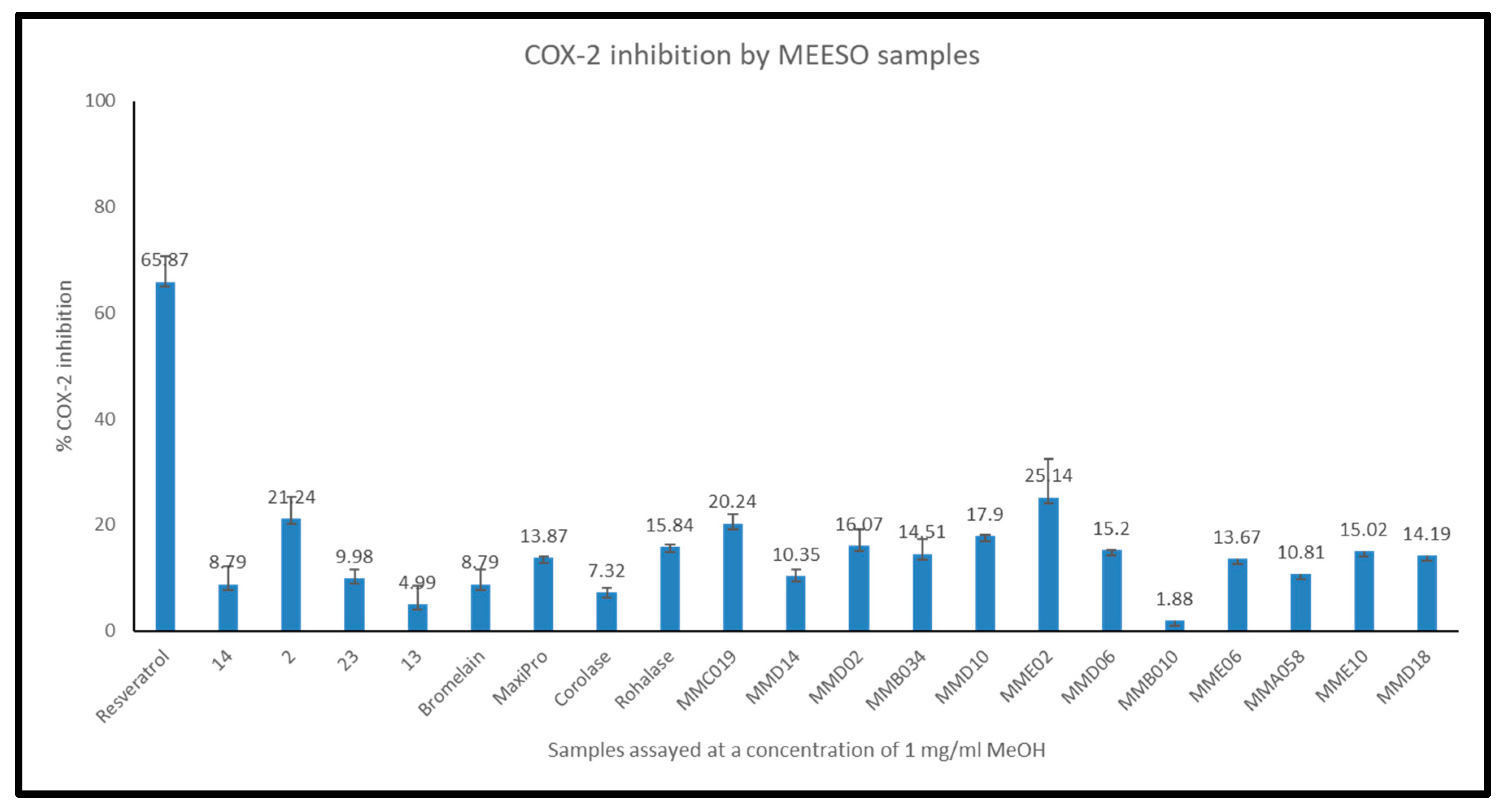
2.4.1. Cyclooxygenase (COX) Enzyme Inhibition by Generated Hydrolysates
2.4.2. Monoacylglycerol Lipase (MAGL) Enzyme Inhibition by Generated Hydrolysates
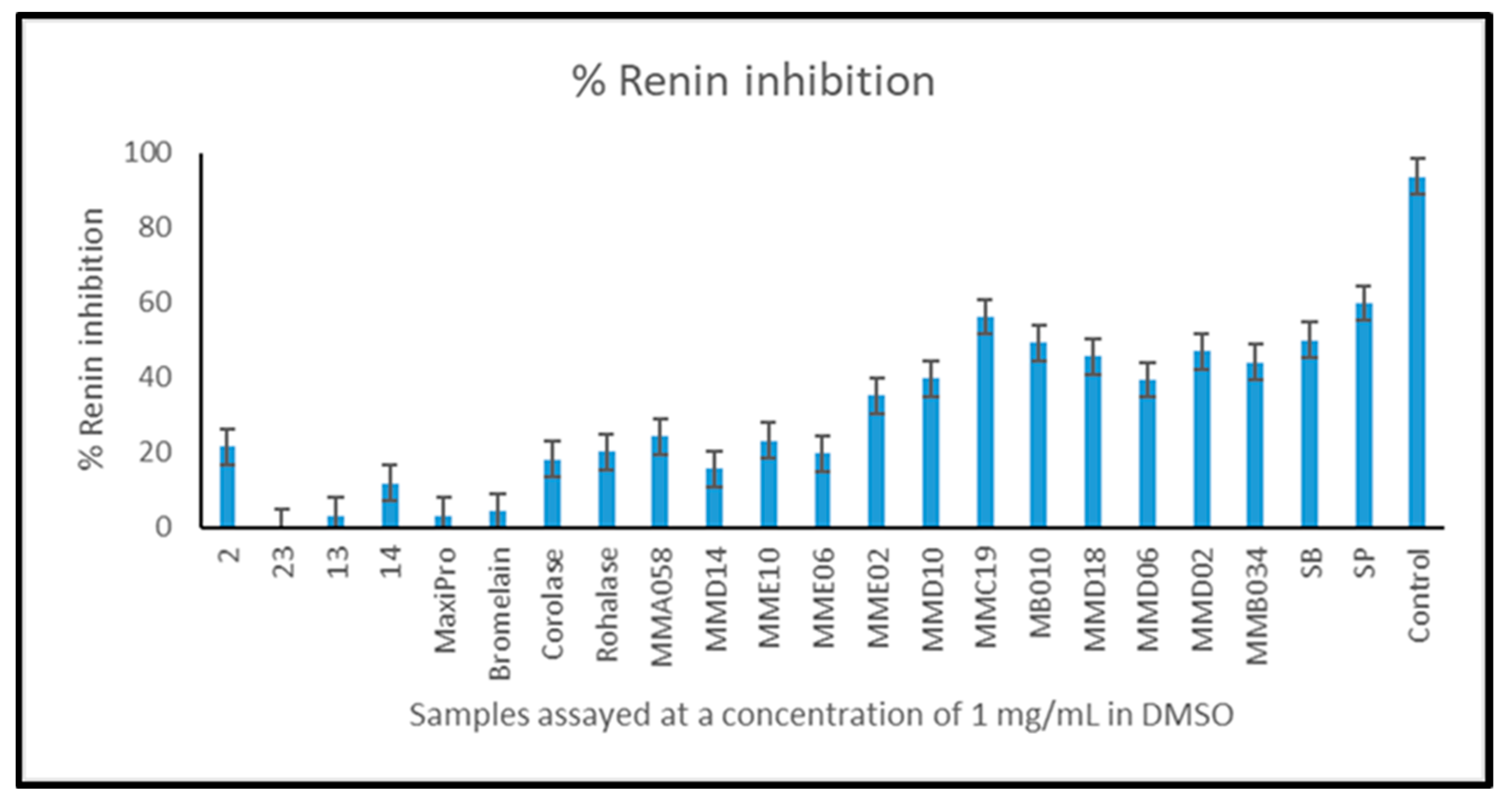
2.4.4. Angiotensin-1-Converting Enzyme (ACE-1) and Renin Inhibition by Mesopelagic Hydrolysates
2.4.5. Dipeptidyl Peptidase IV Inhibition by Mesopelagic Hydrolysates
2.5. Identification of Bioactive Peptides Using Mass Spectrometry and In Silico Analysis
2.6. Chemical Synthesis and Confirmation of Anti-Inflammatory Activity of Peptides Using In Vitro COX and MAGL Inhibition Bioassays
3. Discussion
4. Materials and Methods
4.1. Supply and Processing of Raw Materials
4.1.1. Irish and Norwegian Samples

Enzymatic Hydrolysis of Irish and Norwegian Samples
Hydrolysate Enrichment Using Molecular Weight Cut-Off (MWCO) Filtration
Proximate Compositional Analysis of Mesopelagic Species
4.1.2. Spanish Samples
Enzymatic Hydrolysis of Spanish Samples
Analysis of Spanish Samples
4.2. Bioactivity Screening of Hydrolysates and Permeates
4.2.1. Cyclooxygenase (COX; EC E.C. 1.14. 99.1) Inhibition COX-1 and COX-2
4.2.2. Monoacylglycerol Lipase (MAGL; EC 3.1. 1. 23) Inhibition
4.2.3. ACE-I Inhibition Assay
4.2.4. DPP-IV Inhibition Assay
4.2.5. Antioxidant Capacity: ABTS Assay
4.2.6. Renin Inhibition Activity
4.3. Peptides Identified Using Mass Spectrometry
4.4. Assessment of Bioactive Potential of Identified Peptides
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Staby, A. , Røstad, A., Kaartvedt, S., Long-term acoustical observations of the mesopelagic fish Maurolicus muelleri reveal novel and varied vertical migration patterns. Marine Ecology Progress Series 2011, 441, 241–255. [Google Scholar]
- Naik, A.S., Whitaker, R.D., Albrektsen, S., Solstad, R.G., Thoresen, L., Hayes, M. Mesopelagic fish protein hydrolysates and extracts: A source of novel anti-hypertensive and anti-diabetic peptides. Front. Mar. Sci., 2021, 8, 1-9. [CrossRef]
- Abdulbasit, A. , Biochemical composition and bioactive properties of mesopelagic fish-derived protein hydrolysate from Northern Krill (Meganyctiphanes norvegica) and Muller’s Pearlside (Maurolicus muelleri) for possible utilization of novel marine resources, 2022, Master’s thesis in Biotechnology, Norwegian University of Science and Technology, Faculty of Natural Sciences, 1-70.
- Madina, M.A.; Grimaldo, E.; Grimsmo, L.; Toldnes, B.; Slizyte, R.; Carvajal, A.K.; Schei, M.; Selnes, M.; Falch, E. Exploring the Potential of Atlantic Mesopelagic Species Processed on Board Commercial Fishing Vessels as a Source of Dietary Lipids. Foods 2024, 13, 1094. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.H. , Kriest, I. , Getzlaff, J., Diving deeper: Mesopelagic fish biomass estimates comparison using two different models. Front. Mar. Sci. 2023, 10. [Google Scholar] [CrossRef]
- Calcinai L, Bonomini MG, Leni G, Faccini A, Puxeddu I, Giannini D, Petrelli F, Prandi B, Sforza S, Tedeschi, T. Effectiveness of enzymatic hydrolysis for reducing the allergenic potential of legume by-products. Sci Rep. 2022 Oct 7;12(1):16902. [CrossRef] [PubMed] [PubMed Central]
- Jiang, X.; Rao, Q. Effect of Processing on Fish Protein Antigenicity and Allergenicity. Foods 2021, 10, 969. [Google Scholar] [CrossRef]
- Gianfranceschi, G.L. , Gianfranceschi, G., Quassinti, L., Bramucci, M., Biochemical requirements of bioactive peptides for nutraceutical efficacy. Journal of Functional Foods, 2018; 47, 252–263Get rights and content. [Google Scholar] [CrossRef]
- Minkiewicz P, Iwaniak A, Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides. Current Opportunities. Int J Mol Sci. 2019, 20. [CrossRef] [PubMed] [PubMed Central]
- Lund, A., Knop, F.K., Vilsbøll, T., Glucagon-like peptide-1 receptor agonist for the treatment of type 2 diabetes: Differences and similarities. European Journal of Internal Medicine, 2014, 25, 407-414. [CrossRef]
- Magon, N. Gonadotropin releasing hormone agonists: Expanding vistas. Indian J Endocrinol Metab. 2011, 15, 261–267. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rossino, G., Marchese, E., Galli, G., Verde, F., Finizio, M., Serra, M., Linciano, P., Collina, S. Peptides as Therapeutic Agents: Challenges and Opportunities in the Green Transition Era. Molecules. 2023, 28, 7165. [CrossRef] [PubMed]
- Chen, L., Deng, H., Cu,i H., Fang, J., Zuo, Z., Deng, J., Li, Y., Wang, X., Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget. 2017, 9, 7204-7218. [CrossRef] [PubMed]
- Placha, D., Jampilek, J. Chronic Inflammatory Diseases, Anti-Inflammatory Agents and Their Delivery Nanosystems. Pharmaceutics. 2021, 13, 1, 64. [CrossRef] [PubMed] [PubMed Central]
- Bu, J., Ding, R., Zhou, L., Chen, X., Shen, E. Epidemiology of Psoriasis and Comorbid Diseases: A Narrative Review. Front Immunol. 2022, 10;13:880201. [CrossRef] [PubMed] [PubMed Central]
- Zhou, W.B.S., Meng, J., Zang, J. Does low-grade systemic inflammation have a role in chronic pain? Front. Mol. Neurosci., 2021, 14. [CrossRef]
- Bindu, S., Mazumder, S., Bandyopadhyay, U. Non-steroidal anti-inflammatory drugs (NSAIDs) and organ damage: A current perspective. Biochem Pharmacol. 2020, 180, 114147. Epub 2020 Jul 10. [CrossRef] [PubMed]
- Smith, C.J., Zhang, Y., Koboldt, C.M., Muhammad, J., Zweifel, B.S., Shaffer, A., Talley, J.J., Masferrer, J.L., Seibert, K., Isakson, P.C. Pharmacological analysis of cyclooxygenase-1 in inflammation. Proc Natl Acad Sci U S A. 1998, 95, 13313-8. [CrossRef] [PubMed]
- Curtis, E., Fuggle, N., Shaw, S., Spooner, L., Ntani, G., Parsons, C., Corp, N., Honvo, G., Baird, J., Maggi, S., Dennison, E., Bruyère, O., Reginster, J.Y., Cooper, C. Safety of Cyclooxygenase-2 Inhibitors in Osteoarthritis: Outcomes of a Systematic Review and Meta-Analysis. Drugs Aging. 2019 36, 1, 25-44. [CrossRef] [PubMed]
- Seeram, N.P. , Momin, R.A., Nair, M.G., Bourquin, L.D. Cyclooxygenase inhibitory and antioxidant cyanidin glycosides in cherries and berries. Phytomedicine. 2001, 8, 362–9. [Google Scholar] [CrossRef] [PubMed]
- Allaire, M. , Al Sayegh, R., Mabire, M., Hammoutene, A., Siebert, M., Caër, C., Cadoux, M., Wan, J., Habib, A., Le Gall, M., de la Grange, P., Guillou, H., Postic, C., Paradis, V., Lotersztajn, S., Gilgenkrantz, H. Monoacylglycerol lipase reprograms hepatocytes and macrophages to promote liver regeneration. JHEP Rep. 2023, 8, 100794. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liang, Q. , Chalamaiah, M., Liao, W., Ren, X., Ma, H., Wu, J. Zein hydrolysate and its peptides exert anti-inflammatory activity on endothelial cells by preventing TNF-α-induced NF-ĸβ activation. Journal of Functional Foods, 2022; 64, 103598. [Google Scholar] [CrossRef]
- Dia, V.P., Wang, W., Oh, V.L., de Lumen, B.O., Gonzalez de Mejia, E. Isolation, purification and characterisation of lunasin from defatted soybean flour and in vitro evaluation of its anti-inflammatory activity. Food Chem. 2009, 114, 1, 108-115. [CrossRef]
- Ahn, C.B., Cho, Y.S., Je, J.,Y. Purification and anti-inflammatory action of tripeptide from salmon pectoral fin byproduct protein hydrolysate. Food Chem. 2015, 1, 168:151-6. Epub 2014 Jun 2. 1, 168; 1. [CrossRef] [PubMed]
- Jang, H.L. , Shin, S.R., Yoon, K.Y. Hydrolysis conditions for antioxidant peptides derived from enzymatic hydrolysates of sandfish (Arctoscopus japonicus). Food Sci Biotechnol. 2017, 26, 5, 1191-1197. [CrossRef] [PubMed]
- Kemp, D.C. , Kwon, J.Y. Fish and Shellfish-Derived Anti-Inflammatory Protein Products: Properties and Mechanisms. Molecules, 2021, 26, 3225. [CrossRef] [PubMed]
- Mooney, C. , Haslam, N.J., Pollastri, G., Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE, 2012, 7, e45012. [CrossRef]
- Khatun, M.S. , Hasan, M.M., Kurata, H. PreAIP: Computational Prediction of Anti-inflammatory Peptides by Integrating Multiple Complementary Features. Front Genet. 2019, 129. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Qi, L., Du, J., Sun, Y., Xiong, Y., Zhao, X., Dan, D., Zhi, Y., Dang, Y., Gao, X. Umami-MRNN: Deep learning based prediction of umami peptide using RNN and MLP. Food Chem., 2023, 405, A, 134935. [CrossRef]
- Association of Official Analytical Chemists. Official Methods of Analysis of AOAC International; AOAC International: Rockville, MD, USA, 1998. [Google Scholar]
- Deng, H., Li, W. Monoacylglycerol lipase inhibitors: modulators for lipid metabolism in cancer malignancy, neurological and metabolic disorders. Acta Pharm Sin B. 2020, 10, 4, 582-602. Epub 2019 Oct 18. [CrossRef] [PubMed]
- Hayes, M., Mora, L., Lucakova, S. Identification of Bioactive Peptides from Nannochloropsis oculata Using a Combination of Enzymatic Treatment, in Silico Analysis and Chemical Synthesis. Biomolecules. 2022, Dec 2;12(12):1806. [CrossRef] [PubMed]
- Lafarga, T. , Aluko, R.E., Rai, D.K., O’Connor, P., Hayes, M. Identification of bioactive peptides from a papain hydrolysate of bovine serum albumin and assessment of an antihypertensive effect in Spontaneously Hypertensive Rats. Food Research International 2016, 81, 91–99. [Google Scholar] [CrossRef]
- Chen, X., Huang, J., He, B. AntiDMPpred: a web service for identifying anti-diabetic peptides. PeerJ. 2022, 10:e13581. [CrossRef] [PubMed]
- Ju, Z., Li, M., Xu, J., Howell, D.C., Li, Z., Chen, F.E. Recent development on COX-2 inhibitors as promising anti-inflammatory agents: The past 10 years. Acta Pharm Sin B. 2022, 12, 2790-2807. Epub 2022 Jan 11. [CrossRef] [PubMed]
- Rivera-Jiménez, J. , Berraquero-García, C., Pérez-Gálve, R., García-Moreno, P.J., Espejo-Carpio, F.J., Guadix, A., and Guadix, E.M. Peptides and protein hydrolysates exhibiting anti-inflammatory activity: sources, structural features and modulation mechanisms. Food and Function, 2022, 13, 12510. [Google Scholar]
- Shin, T.-S. , Choi, K.S., Chun, J., Kho, K.H., Son, S.A., Shim, S-Y. Anti-inflammatory effect of Abalone (aliotis discus Hannai) Viscera via inhibition of ROS production in LPS stimulated RAW 267.4 cells. Microbiol. Biotechnol. Lett. 2022, 50, 1, 21–30. [Google Scholar] [CrossRef]
- Ghanbari, R. , Ebahimpour, M., Zarei, A., Ismail, A., Abdul-Hamid, A., and Saari, N., Purification and Characterisation of Nitric Oxide Inhibitory peptides from Actinopyga lecanora through enzymatic hydrolysis. Food Biotechnol., 2016, 30, 263–277. [Google Scholar]
- Grancieri, M. , Martino, H.S.D., Gonzalez de Mejia, E. Digested total protein and protein fractions from chia seed (Salvia hispanica L.) had high scavenging capacity and inhibited 5-LOX, COX-1-2, and iNOS enzymes. Food Chem., 2019, 289, 204–214. [Google Scholar] [PubMed]
- Cao, H. , Yu, R., Choi, Y., Ma, Z.Z., Zhang, H., Xiang, W., Lee, D.Y., Berman, B.M., Moudgil, K.D., Fong, H.H., van Breemen, R., B. Discovery of cyclooxygenase inhibitors from medicinal plants used to treat inflammation. Pharmacol Res. 2010, 61, 6, 519–24. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jha, V., Biagi, M., Spinelli, V., Di Stefano, M., Macchia, M., Minutolo, F., Granchi, C., Poli, G., Tuccinardi, T. Discovery of Monoacylglycerol Lipase (MAGL) Inhibitors Based on a Pharmacophore-Guided Virtual Screening Study. Molecules. 2020 26;26(1):78. [CrossRef] [PubMed]
- Zhang, Y. , Venkitasamy, C., Pan, Z., Liu, W., Zhao, L. Novel Umami Ingredients: Umami peptides and their taste. Journal of Food Science 2017, 82, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Mei, S., Yao, S., Mo, J., Wang, Y., Tang, J., Li, W., Wu, T. Integration of cloud-based molecular networking and docking for enhanced umami peptide screening from Pixian douban. Food Chem X. 2023, 27, 21:101098. [CrossRef] [PubMed]
- Belloir, C. , Savistchenko, J., Neiers, F., Taylor, A.J., McGrane, S., Briand, L. Biophysical and functional characterization of the N-terminal domain of the cat T1R1 umami taste receptor expressed in Escherichia coli. PLoS ONE, 2017, 12(10): e0187051. [CrossRef]
- Association of Official Analytical Chemists. Official Methods of Analysis of AOAC International; AOAC International: Rockville, MD, USA, 1998. [Google Scholar]
- Hoyle, N.T.; Merritt, J.H. Quality of fish protein hydrolysates from herring (Clupea harengus). J. Food Sci. 1994, 59, 76–79. [Google Scholar] [CrossRef]
- Nielsen, P.H. , Petersen, D., Dambmann, C. Food Science, 2001, Improved method for determining food protein degree of hydrolysis. 66, 5, 642-646.
- Hayes, M.; Aluko, R.E.; Aurino, E.; Mora, L. Generation of Bioactive Peptides from Porphyridium sp. and Assessment of Their Potential for Use in the Prevention of Hypertension, Inflammation and Pain. Mar. Drugs 2023, 21, 422. [Google Scholar] [CrossRef]
- Mooney, C. , Haslam, N., Pollastri, G., Shields, D. Towards the improved discovery and design of functional peptides: common features of diverse classes permit generalized prediction of bioactivity. PloS One 2012, 7(10). [Google Scholar]
- Khatun MS, Hasan MM, Kurata, H. PreAIP: Computational Prediction of Anti-inflammatory Peptides by Integrating Multiple Complementary Features. Front Genet. 2019 Mar 5;10:129. [CrossRef] [PubMed]
- Minkiewicz, P. , Iwaniak, A., Darewicz, M., BIOPEP-UWM database of bioactive peptides: Current opportunities. International Journal of Molecular Sciences, 2019, 20, 5978. [CrossRef]
- Qi, L. , Du, J., Sun, Y., Xiong, Y., Zhao, X., Pan, D., Zhi, Y., Dang, Y., Gao, X. 2023, Umami-MRNN: Deep learning-based prediction of umami peptide using RNN and MLP. Food Cheimstry, 405, A, 134935.
| Hydrolysate | Peptide Sequence | Peptide Ranker Value | PreAIP RF combined values | Anti-diabetic prediction (AntiDMPpred) | BIOPEP* | Umami |
|---|---|---|---|---|---|---|
| CE21009 Haul 23 Maurolicus muelleri (Code 23) Alcalase hydrolysate. Irish sample. Freeze-dried. High Confidence AIP (0.482). | KTLRKMGKWCCHCFPCCRGSGKSNVGAW | 0.999 | High confidence AIP (0.731) | Low probability | Novel | umami, predicted threshold: 25.470665mmol/L |
| DGINVLGLIVFCLVLGIVIGRKWEKGQIL | 0.996 | High confidence AIP (0.560) | Low probability | Novel | umami, predicted threshold: 12.649109mmol/L | |
| Origin Thin-lipped mullet | FDAFLPM | 0.955 | Medium confidence AIP (0.392) | Low probability | Novel | Non-umami |
| GLGGMLF | 0.939 | Low confidence AIP (0.370) | Low probability | Novel | umami, predicted threshold: 35.458916mmol/L | |
| Origin Salmo salar – Atlantic salmon | QCPLHRPWAL | 0.932 | High confidence AIP (0.499) | Low probability | Novel | Non-umami |
| LACNCNLHARRCRFNM | 0.908 | High confidence AIP (0.629) | Low probability | Novel | umami, predicted threshold: 37.657642 mmol/L | |
| TFSWGFDDFSCC | 0.889 | High confidence AIP (0.496) | Low probability | Novel | umami, predicted threshold: 14.819101 mmol/L | |
| GINVLGLIVFCLVLGI | 0.888 | High confidence AIP (0.620) | Low probability | Novel | umami, predicted threshold: 35.082146mmol/L | |
| LLSSELQSLLIATTCLRELISCC | 0.873 | High confidence (0.614) | Low probability | Novel | umami, predicted threshold: 13.255066 mmol/L | |
| Origin - Makaira nigricans - Atlantic Blue Marlin | NVGEVVCIFLTAALGLPEALI | 0.868 | High confidence AIP (0.612) | Likely to be anti-diabetic (probability of 0.8) | Novel | umami, predicted threshold: 19.895546 mmol/L |
| Maurolicus muelleri (MMC019) Endogenous enzyme autolysis (Spanish sample), spray dried. High confidence AIP (0.514) | SFVPNGASLEDCHCNLPCLA | 0.874 | High confidence AIP (0.506) | Low probability | Novel | umami, predicted threshold: 30.64746 mmol/L |
| GFSAVNMRKFG | 0.797 | High confidence AIP (0.527) | Low probability | Novel | umami, predicted threshold: 30.85351 mmol/L | |
| Origin - Cypinus carpio - Common carp | IAGFEIFDFNSLEQLC | 0.734 | High confidence AIP (0.540) | Low probability | Novel | umami, predicted threshold: 36.002125 mmol/L |
| NLFKDCNF | 0.693 | Medium confidence AIP (0.467) | Low probability | Novel | umami, predicted threshold: 18.799038 mmol/L | |
| PFGAADQDPF | 0.677 | Low confidence AIP (0.370) | Low probability | Novel | Non-umami | |
| NSGAGILPSPSTPRFP | 0.621 | Medium confidence AIP (0.453) | Low probability | Novel | umami, predicted threshold: 25.949366 mmol/L | |
| DVEFLPPQLPSDKFKDDPVG | 0.601 | Medium confidence AIP (0.433) | Low probability | Novel | umami, predicted threshold: 20.447205 mmol/L | |
| Origin - Takifuga rubipes - Japanese puffer fish | GFAGDDAPR | 0.598 | Negative AIP (0.284) | Low probability | Novel | umami, predicted threshold: 1.6841054 mmol/L |
| FSPFGAAD | 0.58 | Low confidence AIP (0.346) | Low probability | Novel | umami, predicted threshold: 17.39304 mmol/L | |
| PSRILYG | 0.574 | Medium confidence AIP (0.412) | Low probability | Novel | Non-umami | |
| Maurolicus muelleri (MME02 - Spanish haul) (Medium confidence AIP - 0.446) | VFIPFNPL | 0.871 | Low confidence AIP (0.382) | Low probability | Novel | Non-umami |
| NDLPWEF | 0.861 | Low confidence AIP (0.349) | Low probability | Novel | Non-umami | |
| VLLFFYAPWCGQ | 0.846 | High confidence AIP (0.524) | Low probability | Novel | Non-umami | |
| CGRASCPVLCSG | 0.845 | High confidence AIP (0.480) | Low probability | Novel | umami, predicted threshold: 23.602087mmol/L | |
| Origin - Makaira nigricans - Atlantic Blue Marlin | GFNPPDLDIM | 0.828 | Low confidence AIP (0.382) | Low probability | Novel | non-umami |
| Origin - Cypinus carpio - Common carp | SDNAYQFMLT | 0.72 | Medium confidence AIP (0.412) | Low probability | Novel | umami, predicted threshold: 34.065384mmol/L |
| CLGSPNPLDII | 0.687 | Medium confidence AIP (0.408) | Low probability | Novel | umami, predicted threshold: 36.979355mmol/L | |
| RCPEALF | 0.672 | High confidence AIP (0.556) | Low probability | Novel | non-umami | |
| ADDEDADGESSGEPPGAPKQEEAI | 0.667 | High confidence AIP (0.469) | Low probability | Novel | umami, predicted threshold: 8.319466mmol/L | |
| DSFGRLT | 0.662 | Low confidence AIP (0.387) | Low probability | Novel | umami, predicted threshold: 12.873926mmol/L |
| Haul code and species composition | Date | Latitude start | Longitude start | Bottom depth (m) | Target depth (m) |
|---|---|---|---|---|---|
| CE21004 Haul2 (Code 2) Notocopelus elongtus kroyeri |
23/03/2021 | 52°32.31 | 14°42.21 | 418 | 400 |
| CE21004 Haul4 (Code 14) Benthosema glaciale |
24/03/2021 | 53°29.98 | 14°18.52 | 860 | 500 |
| CE21004 Haul13 (Code 13) Maurolicus muelleri |
02/04/2021 | 59°49.64 | 13°18.39 | 1092 | 220-280 |
| CE21009 Haul23 Maurolicus muelleri (Code 23) |
24/06/2021 | 50°40.48 | 11°18.72 | 1029 | 175 |
| Haul code | Date | Latitude start | Longitude start | Bottom depth (m) | Target depth (m) |
|---|---|---|---|---|---|
| 9007 | 07/09/2019 | 43°51.24 | 5°58.23 | 400 | 200 |
| 9009 | 08/09/2019 | 43°51.50 | 5°26.42 | 320 | 300 |
| 9014 | 13/09/2019 | 43°47.81 | 6°28.82 | 400 | 206 |
| 9010 | 07/09/2020 | 43°54.09 | 5°47.62 | 500 | 192 |
| 9020 | 10/09/2020 | 43°39.98 | 4°45.74 | 189 | 138 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





