Submitted:
03 June 2024
Posted:
03 June 2024
You are already at the latest version
Abstract
Keywords:
Introduction
"The goose/Guangdong-lineage of H5N1 avian influenza viruses first emerged in 1996 and has been causing outbreaks in birds since then. Since 2020, a variant of these viruses has led to an unprecedented number of deaths in wild birds and poultry in many countries. First affecting Africa, Asia and Europe, in 2021, the virus spread to North America, and in 2022, to Central and South America. From 2021 to 2022, Europe and North America observed their largest and most extended epidemic of avian influenza with unusual persistence of the virus in wild bird populations. Since 2022, there have been increasing reports of deadly outbreaks among mammals also caused by influenza A(H5) – including influenza A(H5N1) – viruses. There are likely to be more outbreaks that have not been detected or reported. Both land and sea mammals have been affected, including outbreaks in farmed fur animals, seals, sea lions, and detections in other wild and domestic animals such as foxes, bears, otters, raccoons, cats, dogs, cows, goats and others. " [1]
"We show that H5Nx viruses emerged during the successful suppression of H5N1 virus populations in poultry [in China], providing an opportunity for antigenically distinct H5Nx viruses to propagate. Avian influenza vaccination programs would benefit from universal vaccines targeting a wider diversity of influenza viruses to prevent the emergence of novel subtypes."[6].
"The HPAI H5N1 viruses that were detected in Newfoundland in November and December 2021 originated from Northwest Europe and belonged to HPAI clade 2.3.4.4b. Most likely, these viruses emerged in Northwest Europe in winter 2020/2021, dispersed from Europe in late winter or early spring 2021, and arrived in Newfoundland in autumn 2021. The viruses may have been carried across the Atlantic by migratory birds using different routes, including Icelandic, Greenland/Arctic, or pelagic routes. The unusually high presence of the viruses in European wild bird populations in late winter and spring 2021, as well as the greater involvement of barnacle and greylag geese in the epidemiology of HPAI in Europe since October 2020, may explain why spread to Newfoundland happened this winter (2021/2022), and not in the previous winters." [10].
"In the laboratory, I could make it [H5N1] more infectious to humans in months … it’s been published the four amino acids that I need to change … That’s the real biosecurity threat, that these University labs are doing these bio experiments … Bird Flu, I think, is gonna be the cause of the great pandemic, where they are teaching these viruses how to be more infectious for humans." [17]
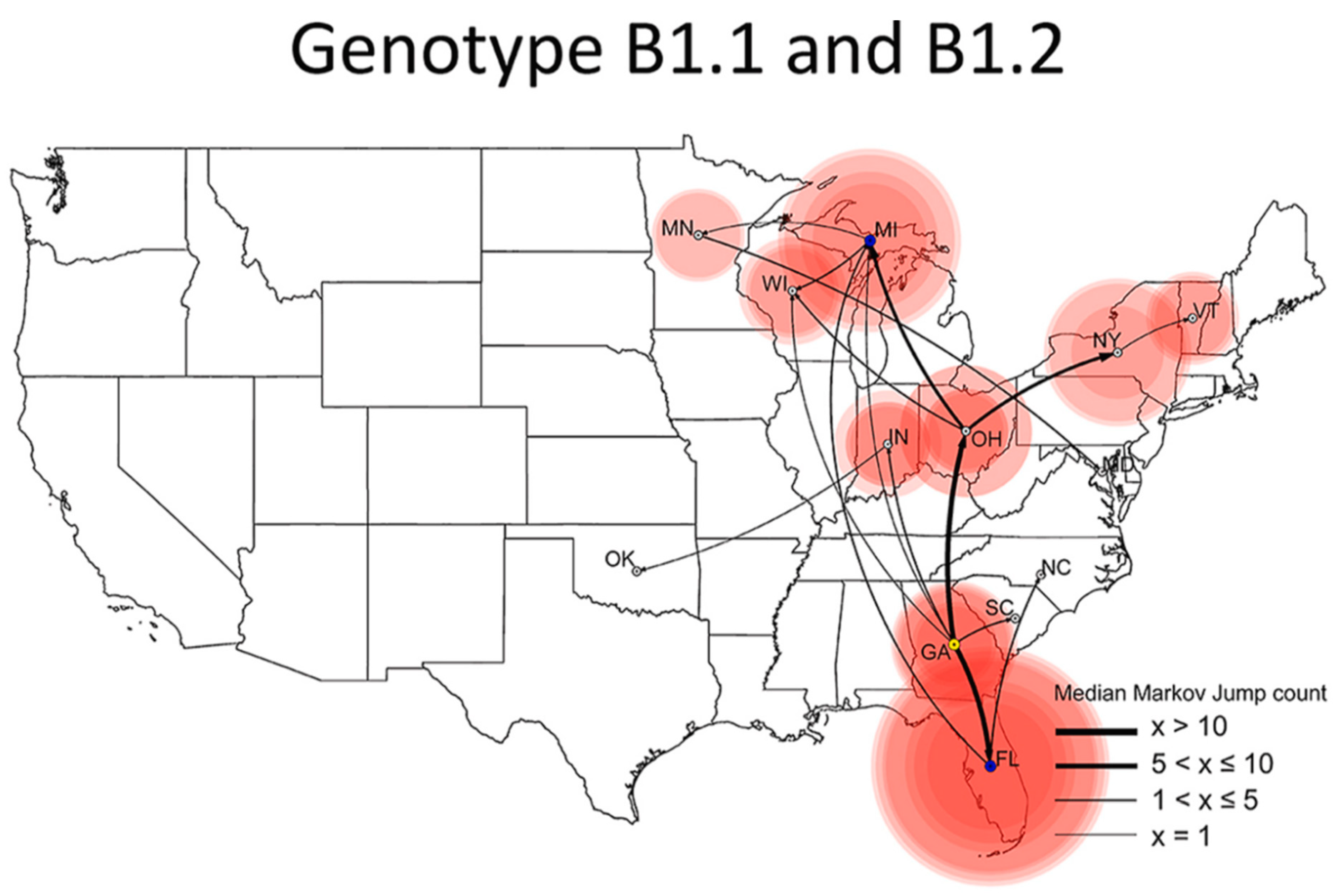
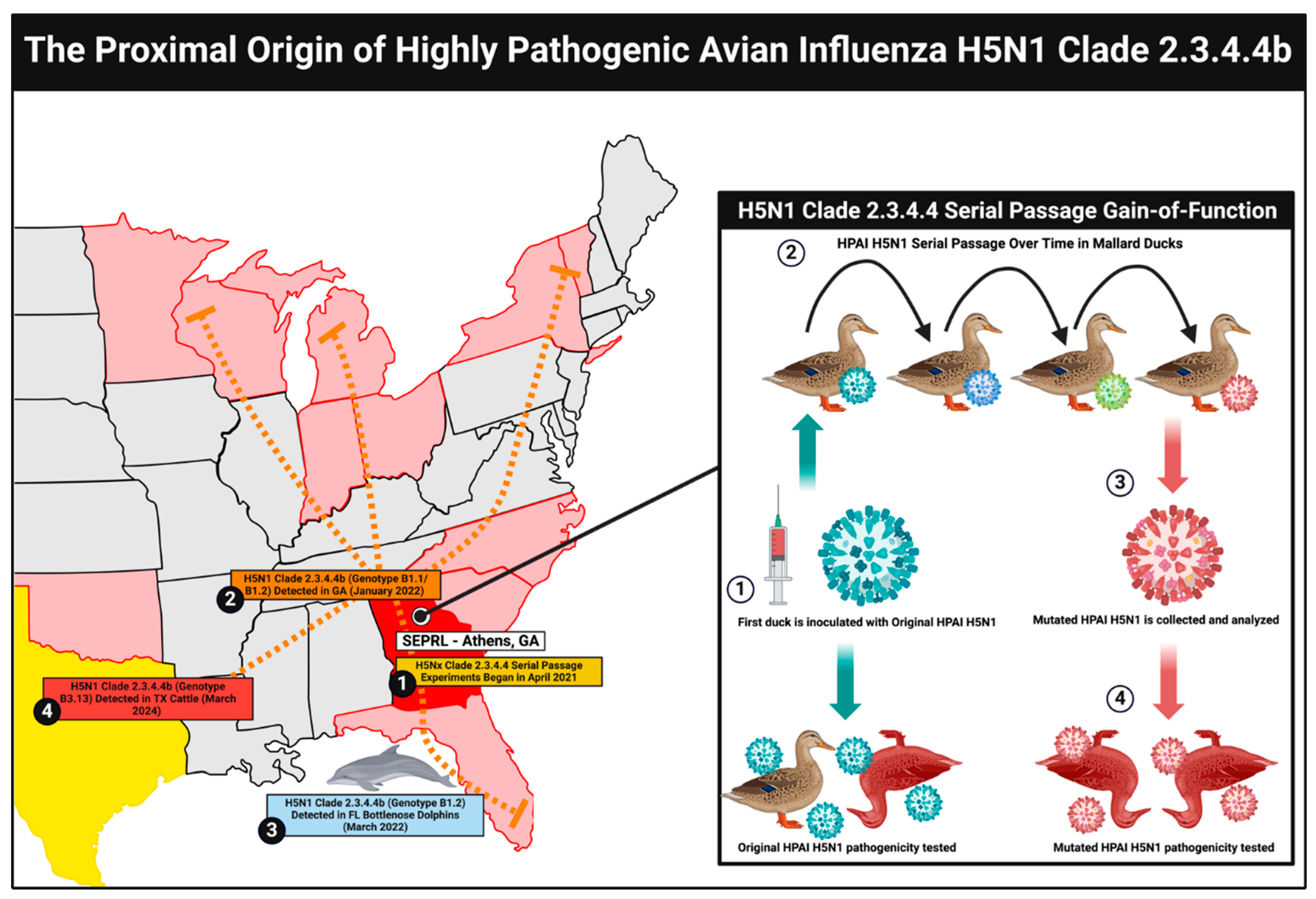
H5N1 Gain-of-Function Research at SEPRL
Genetic Mutations Raise Suspicion of H5N1 Laboratory Leaks
H5N1 Gain-of-Function Risks and Biosecurity Concerns
Conclusions
Ethics statement
Funding
Acknowledgments
Conflicts of Interest
References
- Influenza: A(H5N1). World Health Organization. Published May 16, 2024. Available online: https://www.who.int/news-room/questions-and-answers/item/influenza-h5n1 (accessed on 29 May 2024).
- Graziosi, G.; Lupini, C.; Catelli, E.; Carnaccini, S. Highly Pathogenic Avian Influenza (HPAI) H5 Clade 2.3.4.4b Virus Infection in Birds and Mammals. Animals 2024, 14, 1372. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; de Vries, E.; McBride, R.; Dekkers, J.; Peng, W.; Bouwman, K.M.; Nycholat, C.; Verheije, M.H.; Paulson, J.C.; van Kuppeveld, F.J.; et al. Highly Pathogenic Influenza A(H5Nx) Viruses with Altered H5 Receptor-Binding Specificity. Emerg. Infect. Dis. 2017, 23, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Imai, M.; Kawaoka, Y. The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr. Opin. Virol. 2012, 2, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Chutinimitkul, S.; Van Riel, D.; Munster, V.J.; van den Brand, J.M.A.; Rimmelzwaan, G.F.; Kuiken, T.; Osterhaus, A.D.M.E.; Fouchier, R.A.M.; De Wit, E. In Vitro Assessment of Attachment Pattern and Replication Efficiency of H5N1 Influenza A Viruses with Altered Receptor Specificity. J. Virol. 2010, 84, 6825–6833. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-T.; Su, Y.C.F.; Smith, G.J.D. H5Nx Viruses Emerged during the Suppression of H5N1 Virus Populations in Poultry. Microbiol. Spectr. 2021, 9, e0130921. [Google Scholar] [CrossRef] [PubMed]
- Vanden Bossche G. (Open Letter to WHO) Mass infection prevention and mass vaccination with leaky Covid-19 vaccines in the midst of the pandemic can only breed highly infectious variants. Published March 6, 2021. Accessed June 2, 2024. https://37b32f5a-6ed9-4d6d-b3e1-5ec648ad9ed9.filesusr.com/ugd/28d8fe_266039aeb27a4465988c37adec9cd1dc.pdf.
- Lewis, N.S.; Banyard, A.C.; Whittard, E.; Karibayev, T.; Al Kafagi, T.; Chvala, I.; Byrne, A.; (Akberovna), S.M.; King, J.; Harder, T.; et al. Emergence and spread of novel H5N8, H5N5 and H5N1 clade 2.3.4.4 highly pathogenic avian influenza in 2020. Emerg. Microbes Infect. 2021, 10, 148–151. [Google Scholar] [CrossRef] [PubMed]
- CDC. Emergence and Evolution of H5N1 Bird Flu | Avian Influenza (Flu). www.cdc.gov. Published June 6, 2023. Accessed May 29, 2024. https://www.cdc.gov/flu/avianflu/communication-resources/bird-flu-origin-infographic.html.
- Caliendo, V.; Lewis, N.S.; Pohlmann, A.; Baillie, S.R.; Banyard, A.C.; Beer, M.; Brown, I.H.; Fouchier, R.A.M.; Hansen, R.D.E.; Lameris, T.K.; et al. Transatlantic spread of highly pathogenic avian influenza H5N1 by wild birds from Europe to North America in 2021. Sci. Rep. 2022, 12, 11729. [Google Scholar] [CrossRef]
- BirdLife International. East Atlantic Flyway. Accessed May 30, 2024. http://datazone.birdlife.org/userfiles/file/sowb/flyways/4_East_Atlantic_Factsheet.pdf.
- Bevins, S.N.; Shriner, S.A.; Cumbee, J.C.; Dilione, K.E.; Douglass, K.E.; Ellis, J.W.; Killian, M.L.; Torchetti, M.K.; Lenoch, J.B. Intercontinental Movement of Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4 Virus to the United States, 2021. Emerg. Infect. Dis. 2022, 28, 1006–1011. [Google Scholar] [CrossRef]
- Research Project #440252: US-UK-China Collab: Predictive Phylogenetics For Evolutionary and Transmission Dynamics of Newly Emerging Avian Influenza Viruses. USDA ARS. Accessed May 26, 2024. https://www.ars.usda.gov/research/project/?accnNo=440252.
- Research Project #43962: US-UK-China Collab: Predictive Phylogenetics For Evolutionary and Transmission Dynamics of Newly Emerging Avian Influenza Viruses. USDA ARS. Accessed May 26, 2024. https://www.ars.usda.gov/research/project/?accnNo=439621.
- Hu X, Saxena A, Magstadt DR; et al. Highly Pathogenic Avian Influenza A (H5N1) clade 2.3.4.4b Virus detected in dairy cattle. bioRxiv. 2024. [CrossRef]
- Lipsitch, M. Why Do Exceptionally Dangerous Gain-of-Function Experiments in Influenza?. Methods Mol Biol. 2018;1836:589-608. [CrossRef]
- Declassify the COVID documents: Former CDC director | Vargas Reports. NewsNation. Published May 8, 2024. Accessed May 27, 2024. https://www.newsnationnow.com/video/declassify-the-covid-documents-former-cdc-director-vargas-reports/9678111/.
- Youk, S.; Torchetti, M.K.; Lantz, K.; Lenoch, J.B.; Killian, M.L.; Leyson, C.; Bevins, S.N.; Dilione, K.; Ip, H.S.; Stallknecht, D.E.; et al. H5N1 highly pathogenic avian influenza clade 2.3.4.4b in wild and domestic birds: Introductions into the United States and reassortments, December 2021–April 2022. Virology 2023, 587, 109860. [Google Scholar] [CrossRef]
- Wasilenko, J.L.; Lee, C.W.; Sarmento, L.; Spackman, E.; Kapczynski, D.R.; Suarez, D.L.; Pantin-Jackwood, M.J. NP, PB1, and PB2 Viral Genes Contribute to Altered Replication of H5N1 Avian Influenza Viruses in Chickens. J. Virol. 2008, 82, 4544–4553. [Google Scholar] [CrossRef]
- Xu, G.; Wang, F.; Li, Q.; Bing, G.; Xie, S.; Sun, S.; Bian, Z.; Sun, H.; Feng, Y.; Peng, X.; et al. Mutations in PB2 and HA enhanced pathogenicity of H4N6 avian influenza virus in mice. J. Gen. Virol. 2020, 101, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Danzy, S.; Studdard, L.R.; Manicassamy, B.; Solorzano, A.; Marshall, N.; García-Sastre, A.; Steel, J.; Lowen, A.C. Mutations to PB2 and NP Proteins of an Avian Influenza Virus Combine To Confer Efficient Growth in Primary Human Respiratory Cells. J. Virol. 2014, 88, 13436–13446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, G.; Wang, C.; Jiang, M.; Gao, W.; Wang, M.; Sun, H.; Sun, Y.; Chang, K.-C.; Liu, J.; et al. Enhanced pathogenicity and neurotropism of mouse-adapted H10N7 influenza virus are mediated by novel PB2 and NA mutations. J. Gen. Virol. 2017, 98, 1185–1195. [Google Scholar] [CrossRef] [PubMed]
- CDC. Summary Analysis of Genetic Sequences of HPAI A(H5N1) Viruses in Texas. Centers for Disease Control and Prevention. Published April 2, 2024. Accessed May 26, 2024. https://www.cdc.gov/flu/avianflu/spotlights/2023-2024/h5n1-analysis-texas.htm.
- CDC. Technical Update: Summary Analysis of the Genetic Sequence of a Highly. Centers for Disease Control and Prevention. Published May 24, 2024. Accessed May 26, 2024. https://www.cdc.gov/flu/avianflu/spotlights/2023-2024/h5n1-technical-update-may-24-2024.html#_edn1.
- Elsmo, E.J.; Wunschmann, A.; Beckmen, K.B.; Broughton-Neiswanger, L.E.; Buckles, E.L.; Ellis, J.; Fitzgerald, S.D.; Gerlach, R.; Hawkins, S.; Ip, H.S.; et al. Highly Pathogenic Avian Influenza A(H5N1) Virus Clade 2.3.4.4b Infections in Wild Terrestrial Mammals, United States, 2022. Emerg. Infect. Dis. 2023, 29, 2451–2460. [Google Scholar] [CrossRef] [PubMed]
- Murawski, A.; Fabrizio, T.; Ossiboff, R.; Kackos, C.; Jeevan, T.; Jones, J.C.; Kandeil, A.; Walker, D.; Turner, J.C.M.; Patton, C.; et al. Highly pathogenic avian influenza A(H5N1) virus in a common bottlenose dolphin (Tursiops truncatus) in Florida. Commun. Biol. 2024, 7, 476. [Google Scholar] [CrossRef] [PubMed]
- Herfst, S.; Schrauwen, E.J.A.; Linster, M.; Chutinimitkul, S.; de Wit, E.; Munster, V.J.; Sorrell, E.M.; Bestebroer, T.M.; Burke, D.F.; Smith, D.J.; et al. Airborne Transmission of Influenza A/H5N1 Virus Between Ferrets. Science 2012, 336, 1534–1541. [Google Scholar] [CrossRef]
- Restori, K.H.; Septer, K.M.; Field, C.J.; Patel, D.R.; VanInsberghe, D.; Raghunathan, V.; Lowen, A.C.; Sutton, T.C. Risk assessment of a highly pathogenic H5N1 influenza virus from mink. Nat. Commun. 2024, 15, 4112. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Yount, B.L., Jr.; Debbink, K.; Agnihothram, S.; Gralinski, L.E.; Plante, J.A.; Graham, R.L.; Scobey, T.; Ge, X.-Y.; Donaldson, E.F.; et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015, 21, 1508–1513, Erratum in 2020, 16, 1146. [Google Scholar] [CrossRef]
- Menachery, V.D.; Yount, B.L.; Sims, A.C.; Debbink, K.; Agnihothram, S.S.; Gralinski, L.E.; Graham, R.L.; Scobey, T.; Plante, J.A.; Royal, S.R.; et al. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. 2016, 113, 3048–3053. [Google Scholar] [CrossRef]
- DEPARTMENT OF HEALTH & HUMAN SERVICES. Re: Notice of Suspension and Proposed Debarment of Dr. Peter Daszak.; 2024. Accessed June 2, 2024. https://oversight.house.gov/wp-content/uploads/2024/05/HHS-SUSP4D-Notice-of-Dr.-Peter-Daszak_05.21.2024_Redacted.pdf.
- Wang, Y.; Davidson, I.; Fouchier, R.; Spackman, E. Antigenic Cartography of H9 Avian Influenza Virus and Its Application to Vaccine Selection. Avian Dis. 2016, 60, 218–225. [Google Scholar] [CrossRef]
- Davis, C.T.; Chen, L.-M.; Pappas, C.; Stevens, J.; Tumpey, T.M.; Gubareva, L.V.; Katz, J.M.; Villanueva, J.M.; Donis, R.O.; Cox, N.J. Use of Highly Pathogenic Avian Influenza A(H5N1) Gain-Of-Function Studies for Molecular-Based Surveillance and Pandemic Preparedness. mBio 2014, 5, e02431-14. [Google Scholar] [CrossRef] [PubMed]
- Berns, K.I.; Casadevall, A.; Cohen, M.L.; Ehrlich, S.A.; Enquist, L.W.; Fitch, J.P.; Franz, D.R.; Fraser-Liggett, C.M.; Grant, C.M.; Imperiale, M.J.; et al. Adaptations of Avian Flu Virus Are a Cause for Concern. Science 2012, 335, 660–661. [Google Scholar] [CrossRef] [PubMed]
- Keim, P.S. The NSABB Recommendations: Rationale, Impact, and Implications. mBio 2012, 3, e00021-12. [Google Scholar] [CrossRef] [PubMed]
- Osterholm, M.T.; Henderson, D.A. Life Sciences at a Crossroads: Respiratory Transmissible H5N1. Science 2012, 335, 801–802. [Google Scholar] [CrossRef]
- Osterholm, M.T.; Relman, D.A. Creating a Mammalian-Transmissible A/H5N1 Influenza Virus: Social Contracts, Prudence, and Alternative Perspectives. J. Infect. Dis. 2012, 205, 1636–1638. [Google Scholar] [CrossRef]
- Inglesby, T.V. Engineered H5N1: A Rare Time for Restraint in Science. Ann. Intern. Med. 2012, 156, 460–462. [Google Scholar] [CrossRef]
- Blacksell, S.D.; Dhawan, S.; Kusumoto, M.; Le, K.K.; Summermatter, K.; O’keefe, J.; Kozlovac, J.P.; Almuhairi, S.S.; Sendow, I.; Scheel, C.M.; et al. Laboratory-acquired infections and pathogen escapes worldwide between 2000 and 2021: a scoping review. Lancet Microbe 2024, 5, e194–e202. [Google Scholar] [CrossRef] [PubMed]
- Origins Forum Wrap Up: The Science Points to a Lab Leak. United States House Committee on Oversight and Accountability. Published June 29, 2021. Accessed June 2, 2024. https://oversight.house.gov/release/origins-forum-wrap-up-the-science-points-to-a-lab-leak/.
- INVESTIGATION INTO THE REEDLEY BIOLAB. House Select Committee on the Chinese Communist Party; 2023. Accessed June 2, 2024. https://selectcommitteeontheccp.house.gov/sites/evo-subsites/selectcommitteeontheccp.house.gov/files/evo-media-document/scc-reedley-report-11.15.pdf.
- Kim, J.; Negovetich, N.J.; Forrest, H.L.; Webster, R.G. Ducks: The “Trojan Horses” of H5N1 influenza. Influ. Other Respir. Viruses 2009, 3, 121–128. [Google Scholar] [CrossRef]
- Report on the inadvertent cross-contamination and shipment of a laboratory specimen with influenza virus H5N1. stacks.cdc.gov. Published August 15, 2014. Accessed May 26, 2024. https://stacks.cdc.gov/view/cdc/24766.
- Imai, M.; Watanabe, T.; Hatta, M.; Das, S.C.; Ozawa, M.; Shinya, K.; Zhong, G.; Hanson, A.; Katsura, H.; Watanabe, S.; et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 2012, 486, 420–428. [Google Scholar] [CrossRef]
- Young, A. Lab-created bird flu virus accident shows lax oversight of risky “gain of function” research. USA TODAY. Published April 12, 2023. Accessed May 27, 2024. https://www.usatoday.com/story/opinion/2023/04/11/lab-leak-accident-h-5-n-1-virus-avian-flu-experiment/11354399002/.
- Merler, S.; Ajelli, M.; Fumanelli, L.; Vespignani, A. Containing the accidental laboratory escape of potential pandemic influenza viruses. BMC Med. 2013, 11, 252. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




