Submitted:
01 June 2024
Posted:
03 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Sampling Collection and SNP Calling
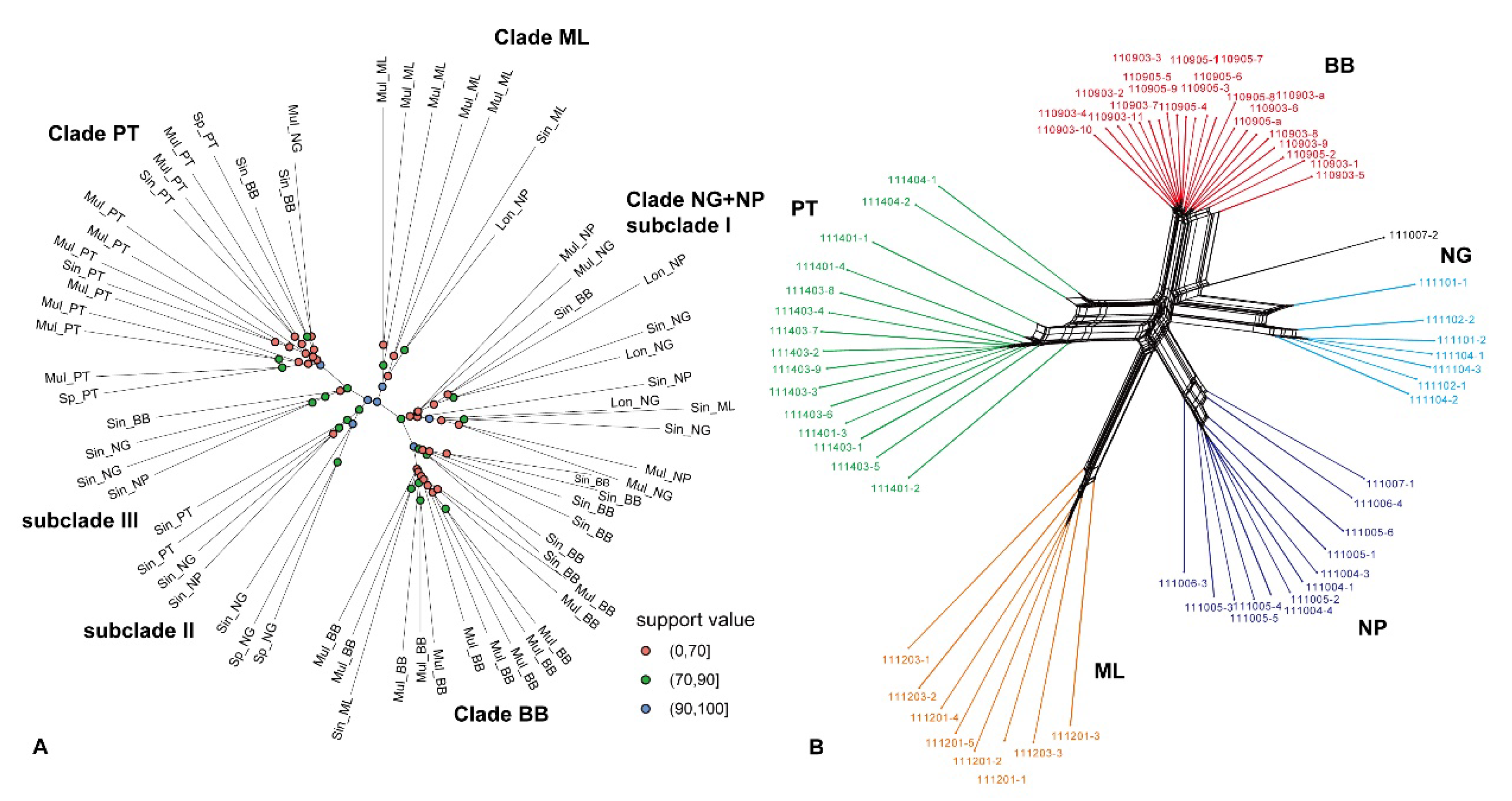
2.2. Phylogenetic Trees and Neighbor Net
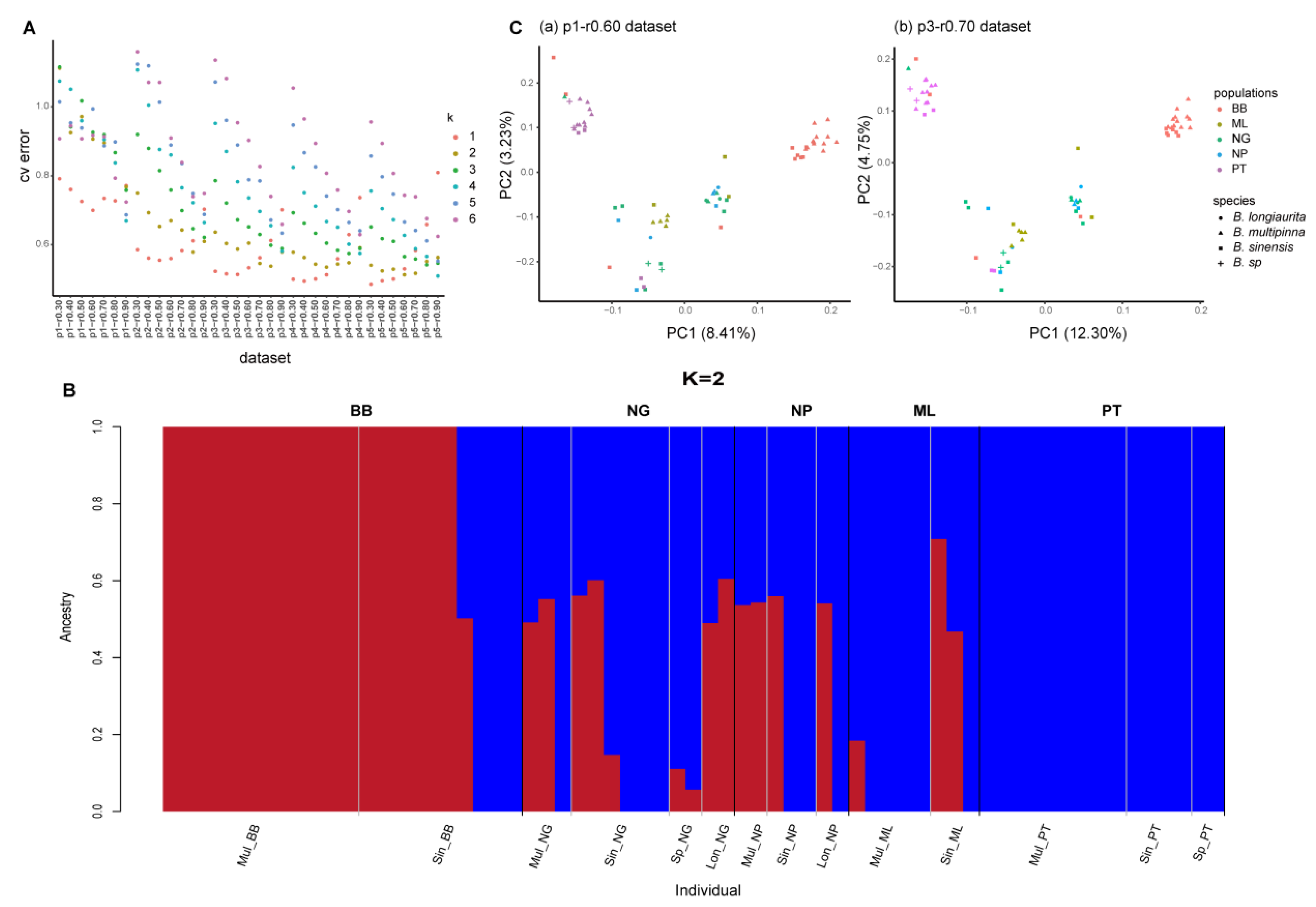
2.3. Genetic Structure Among B. sinensis Species Complex
2.4. Genetic Diversity and Differentiation Among Five Populations
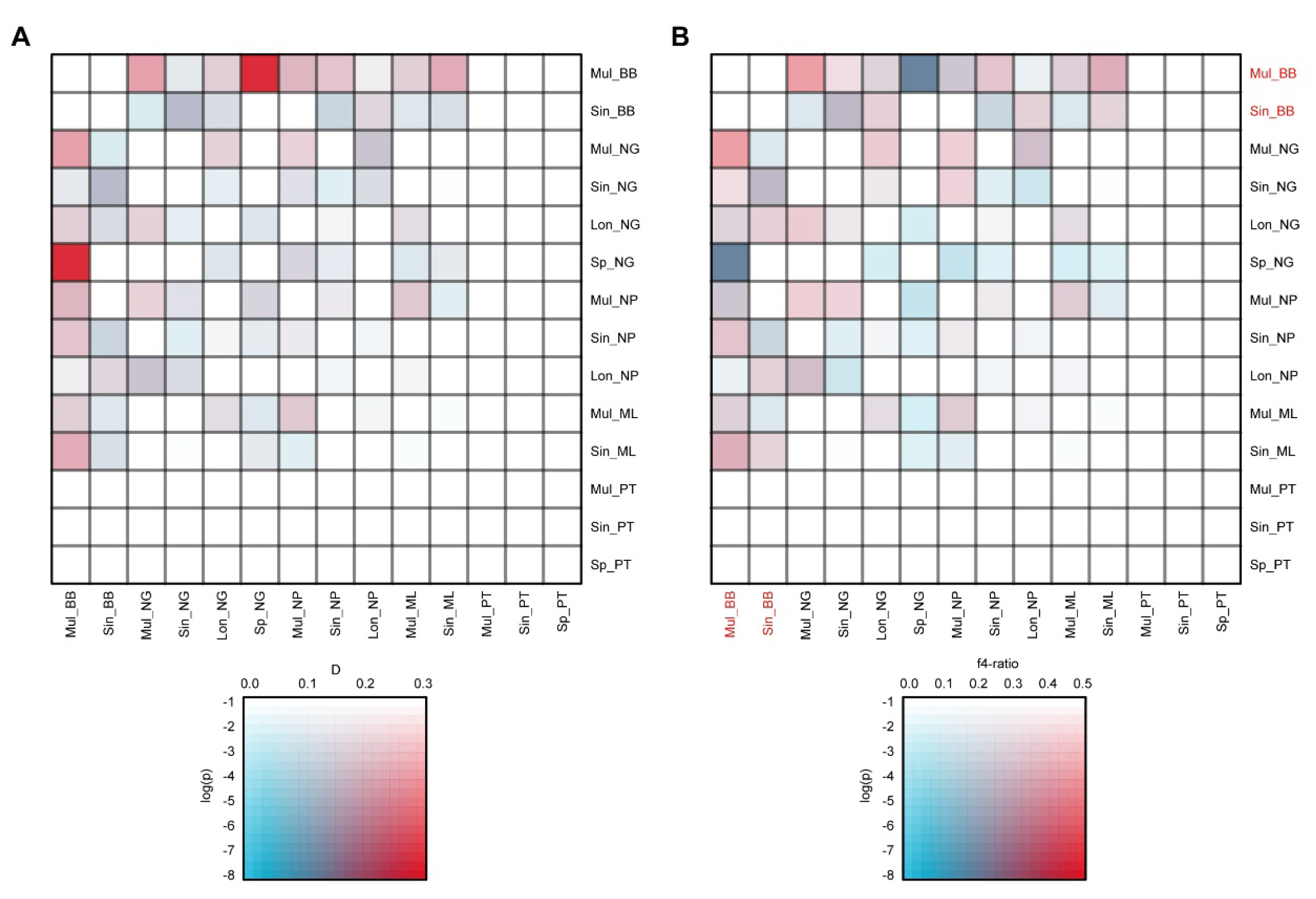
2.5. Potential Gene Flow Among Five Populations
3. Discussion
3.1. Reclassification of Bolbitis sinensis Species Complex
3.2. Genetic Diversity and Differentiation Among Five Populations
4. Materials and Methods
4.1. Taxon Sampling
4.2. RAD-seq Libraries Preparation and Sequencing
4.3. Data Processing
4.4. Phylogenetic Inference
4.5. Genetic Diversity and Structure Analysis
4.6. Introgression Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pinheiro, F.; Dantas-Queiroz, M.V.; Palma-Silva, C. Plant Species Complexes as Models to Understand Speciation and Evolution: A Review of South American Studies. Critical Reviews in Plant Sciences 2018, 37, 54–80. [Google Scholar] [CrossRef]
- Reydon, T.A.; Kunz, W. Species as natural entities, instrumental units and ranked taxa: new perspectives on the grouping and ranking problems. Biological Journal of the Linnean Society 2019, 126, 623–636. [Google Scholar] [CrossRef]
- Rannala, B.; Yang, Z. Species delimitation. In Phylogenetics in the Genomic Era, Scornavacca, C., Delsuc, F., and Galtier, N., Eds.; Self Published: 2020, pp. 1–18.
- Gage, J.L.; Jarquin, D.; Romay, C.; Lorenz, A.; Buckler, E.S.; Kaeppler, S.; Alkhalifah, N.; Bohn, M.; Campbell, D.A.; Edwards, J.; et al. The effect of artificial selection on phenotypic plasticity in maize. Nature Communications 2017, 8, 1348. [Google Scholar] [CrossRef] [PubMed]
- Abdelkrim, J.; Aznar-Cormano, L.; Buge, B.; Fedosov, A.; Kantor, Y.; Zaharias, P.; Puillandre, N. Delimiting species of marine gastropods (Turridae, Conoidea) using RAD sequencing in an integrative taxonomy framework. Molecular Ecology 2018, 27, 4591–4611. [Google Scholar] [CrossRef] [PubMed]
- Ru, D.; Sun, Y.; Wang, D.; Chen, Y.; Wang, T.; Hu, Q.; Abbott, R.J.; Liu, J. Population genomic analysis reveals that homoploid hybrid speciation can be a lengthy process. Molecular Ecology 2018, 27, 4875–4887. [Google Scholar] [CrossRef] [PubMed]
- Aguillon, S.M.; Dodge, T.O.; Preising, G.A.; Schumer, M. Introgression. Current Biology 2022, 32, R865–R868. [Google Scholar] [CrossRef] [PubMed]
- Moran, Robbin C. ; Labiak, Paulo H.; Sundue, M. Phylogeny and Character Evolution of the Bolbitidoid Ferns (Dryopteridaceae). International Journal of Plant Sciences 2010, 171, 547–559. [Google Scholar] [CrossRef]
- Zhang, X.; Wei, R.; Liu, H.; He, L.; Wang, L.; Zhang, G. Phylogeny and Classification of the Extant Lycophytes and Ferns from China. Chinese Bulletin of Botany 2013, 48, 119–137. [Google Scholar] [CrossRef]
- Wu, Z.H.a.W., Z.H. In Flora reipublicae popularis sinicae; Beijing, 1999; Volume 6, pp. 104–105.
- Ching, R.-C.; Wang, C.-H. Materiae ad floram filicum Sinensium. Journal of Systematics and Evolution 1983, 21, 211. [Google Scholar]
- Wang, F.; Kunio, I.; Xing, F. A new name of Bolbitis from China. American Fern Journal 2008, 98, 96–97. [Google Scholar]
- Hennipman, E. A monograph of the fern genus Bolbitis (Lomariopsidaceae). Leiden Botanical Series 1977, 2, 1–329. [Google Scholar]
- Dong, S.Y.; Zhang, X.C. A taxonomic revision of the fern genus Bolbitis (Bolbitidaceae) from China. Journal of Systematics and Evolution 2005, 43, 97. [Google Scholar] [CrossRef]
- Wang, F.; Xing, F. A new name in Chinese Bolbitidaceae. Novon: A Journal for Botanical Nomenclature 2006, 16, 434–435. [Google Scholar]
- Nie, L.Y.; Zhang, L.; Liang, Z.L.; Pollawatn, R.; Yan, Y.H.; Thi Lu, N.; Knapp, R.; Wan, X.; Cicuzza, D.; Cheng, X.X.; et al. Phylogeny, character evolution, and biogeography of the fern genus Bolbitis (Dryopteridaceae). Molecular Phylogenetics and Evolution 2023, 178, 107633. [Google Scholar] [CrossRef]
- Yu, X.; Yang, D.; Guo, C.; Gao, L. Plant phylogenomics based on genome-partitioning strategies: Progress and prospects. Plant Diversity 2018, 40, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.R.; Dunham, J.P.; Amores, A.; Cresko, W.A.; Johnson, E.A. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome research 2007, 17, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP Discovery and Genetic Mapping Using Sequenced RAD Markers. Plos One 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Andrews, K.R.; Good, J.M.; Miller, M.R.; Luikart, G.; Hohenlohe, P.A. Harnessing the power of RADseq for ecological and evolutionary genomics. Nature Reviews Genetics 2016, 17, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Ledent, A.; Gauthier, J.; Pereira, M.; Overson, R.; Laenen, B.; Mardulyn, P.; Gradstein, S.R.; de Haan, M.; Ballings, P.; Van der Beeten, I.; et al. What do tropical cryptogams reveal? Strong genetic structure in Amazonian bryophytes. New Phytologist 2020, 228, 640–650. [Google Scholar] [CrossRef]
- Hipp, A.L.; Manos, P.S.; Hahn, M.; Avishai, M.; Bodenes, C.; Cavender-Bares, J.; Crowl, A.A.; Deng, M.; Denk, T.; Fitz-Gibbon, S.; et al. Genomic landscape of the global oak phylogeny. New Phytologist 2020, 226, 1198–1212. [Google Scholar] [CrossRef]
- Hipp, A.L.; Manos, P.S.; Gonzalez-Rodriguez, A.; Hahn, M.; Kaproth, M.; McVay, J.D.; Avalos, S.V.; Cavender-Bares, J. Sympatric parallel diversification of major oak clades in the Americas and the origins of Mexican species diversity. New Phytologist 2018, 217, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ye, X.; Zhao, L.; Li, D.; Guo, Z.; Zhuang, H. Genome-wide RAD sequencing data provide unprecedented resolution of the phylogeny of temperate bamboos (Poaceae: Bambusoideae). Scientific Reports 2017, 7. [Google Scholar] [CrossRef]
- Zhang, Y.-X.; Guo, C.; Li, D.-Z. A new subtribal classification of Arundinarieae (Poaceae, Bambusoideae) with the description of a new genus. Plant Diversity 2020, 42, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.Y.; Wen, J.; Tian, J.P.; Gui, L.L.; Liu, X.Q. Testing morphological trait evolution and assessing species delimitations in the grape genus using a phylogenomic framework. Molecular Phylogenetics and Evolution 2020, 148, 106809. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Bian, L.; Zhang, X.; Zhao, B.; Zheng, R.; Su, S.; Ye, D.; Zheng, X.; El-Kassaby, Y.A.; Shi, J. Genetic diversity and structure of the 4(th) cycle breeding population of Chinese fir (Cunninghamia lanceolata (lamb.) hook). Frontiers in Plant Science 2023, 14, 1106615. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dong, S.; Yang, L.; Harris, A.; Schneider, H.; Kang, M. Allopolyploid Speciation Accompanied by Gene Flow in a Tree Fern. Molecular Biology and Evolution 2020, 37, 2487–2502. [Google Scholar] [CrossRef] [PubMed]
- Lovell, J.T.; Williamson, R.J.; Wright, S.I.; McKay, J.K.; Sharbel, T.F. Mutation Accumulation in an Asexual Relative of Arabidopsis. PLoS Genetics 2017, 13, e1006550. [Google Scholar] [CrossRef] [PubMed]
- Hojsgaard, D.; Horandl, E. A little bit of sex matters for genome evolution in asexual plants. Frontiers in Plant Science 2015, 6, 82. [Google Scholar] [CrossRef] [PubMed]
- Horandl, E. Apomixis and the paradox of sex in plants. Annals of Botany 2024, mcae044. [Google Scholar] [CrossRef]
- Rodriguez-Gonzalez, P.M.; Garcia, C.; Albuquerque, A.; Monteiro-Henriques, T.; Faria, C.; Guimaraes, J.B.; Mendonca, D.; Simoes, F.; Ferreira, M.T.; Mendes, A.; et al. A spatial stream-network approach assists in managing the remnant genetic diversity of riparian forests. Scientific Reports 2019, 9, 6741. [Google Scholar] [CrossRef]
- Sexton, J.P.; Hangartner, S.B.; Hoffmann, A.A. Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution 2014, 68, 1–15. [Google Scholar] [CrossRef]
- Wang, I.J.; Glor, R.E.; Losos, J.B. Quantifying the roles of ecology and geography in spatial genetic divergence. Ecology letters 2013, 16, 175–182. [Google Scholar] [CrossRef]
- Garot, E.; Joet, T.; Combes, M.C.; Lashermes, P. Genetic diversity and population divergences of an indigenous tree (Coffea mauritiana) in Reunion Island: role of climatic and geographical factors. Heredity 2019, 122, 833–847. [Google Scholar] [CrossRef] [PubMed]
- Govindaraju, D.R. Variation in gene flow levels among predominantly self-pollinated plants. Journal of Evolutionary Biology 1989, 2, 173–181. [Google Scholar] [CrossRef]
- Twyford, A.D.; Wong, E.L.Y.; Friedman, J. Multi-level patterns of genetic structure and isolation by distance in the widespread plant Mimulus guttatus. Heredity 2020, 125, 227–239. [Google Scholar] [CrossRef]
- Wang, S.Q. Genetic diversity and population structure of the endangered species Paeonia decomposita endemic to China and implications for its conservation. BMC Plant Biology 2020, 20, 510. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Wang, I.J.; Comes, H.P.; Peng, H.; Qiu, Y.X. Contributions of historical and contemporary geographic and environmental factors to phylogeographic structure in a Tertiary relict species, Emmenopterys henryi (Rubiaceae). Scientific Reports 2016, 6, 24041. [Google Scholar] [CrossRef]
- Casteleyn, G.; Leliaert, F.; Backeljau, T.; Debeer, A.E.; Kotaki, Y.; Rhodes, L.; Lundholm, N.; Sabbe, K.; Vyverman, W. Limits to gene flow in a cosmopolitan marine planktonic diatom. Proceedings of the National Academy of Sciences 2010, 107, 12952–12957. [Google Scholar] [CrossRef] [PubMed]
- Abdelaziz, M.; Munoz-Pajares, A.J.; Berbel, M.; Garcia-Munoz, A.; Gomez, J.M.; Perfectti, F. Asymmetric Reproductive Barriers and Gene Flow Promote the Rise of a Stable Hybrid Zone in the Mediterranean High Mountain. Frontiers in Plant Science 2021, 12, 687094. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, Y.; Bi, H.; Lu, Z.; Ma, Y.; Yang, X.; Chen, N.; Tian, B.; Liu, B.; Mao, X.; et al. Hybrid speciation via inheritance of alternate alleles of parental isolating genes. Molecular Plant 2021, 14, 208–222. [Google Scholar] [CrossRef]
- Pickup, M.; Brandvain, Y.; Fraisse, C.; Yakimowski, S.; Barton, N.H.; Dixit, T.; Lexer, C.; Cereghetti, E.; Field, D.L. Mating system variation in hybrid zones: facilitation, barriers and asymmetries to gene flow. New Phytologist 2019, 224, 1035–1047. [Google Scholar] [CrossRef] [PubMed]
- Kling, M.M.; Ackerly, D.D. Global wind patterns shape genetic differentiation, asymmetric gene flow, and genetic diversity in trees. Proceedings of the National Academy of Sciences 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.W.; Fridman, E.; Mascher, M.; Himmelbach, A.; Schmid, K. Physical geography, isolation by distance and environmental variables shape genomic variation of wild barley (Hordeum vulgare L. ssp. spontaneum) in the Southern Levant. Heredity 2022, 128, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J. DNA protocols for plants. In Molecular techniques in taxonomy; Springer, 1991; pp. 283–293. [Google Scholar]
- Ali, O.A.; O’Rourke, S.M.; Amish, S.J.; Meek, M.H.; Luikart, G.; Jeffres, C.; Miller, M.R. RAD capture (Rapture): flexible and efficient sequence-based genotyping. Genetics 2016, 202, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Rochette, N.C.; Catchen, J.M. Deriving genotypes from RAD-seq short-read data using Stacks. Nature Protocols 2017, 12, 2640–2659. [Google Scholar] [CrossRef] [PubMed]
- Rochette, N.C.; Rivera-Colon, A.G.; Catchen, J.M. Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. Molecular Ecology 2019, 28, 4737–4754. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, E.M. vcf2phylip v2. 0: convert a VCF matrix into several matrix formats for phylogenetic analysis. URL https://doi org/105281/zenodo 2019, 2540861. [Google Scholar]
- Kalyaanamoorthy, S.; Bui Quang, M.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 2017, 14, 587. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Molecular Biology and Evolution 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Systematic Biology 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Revell, L.J. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Robinson, D.F.; Foulds, L.R. Comparison of phylogenetic trees. Mathematical Biosciences 1981, 53, 131–147. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome research 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Patterson, N.; Moorjani, P.; Luo, Y.; Mallick, S.; Rohland, N.; Zhan, Y.; Genschoreck, T.; Webster, T.; Reich, D. Ancient Admixture in Human History. Genetics 2012, 192, 1065. [Google Scholar] [CrossRef]
- Green, R.E.; Krause, J.; Briggs, A.W.; Maricic, T.; Stenzel, U.; Kircher, M.; Patterson, N.; Li, H.; Zhai, W.; Fritz, M.H.; et al. A draft sequence of the Neandertal genome. Science 2010, 328, 710–722. [Google Scholar] [CrossRef]
- Malinsky, M.; Matschiner, M.; Svardal, H. Dsuite-Fast D-statistics and related admixture evidence from VCF files. Molecular Ecology Resources 2021, 21, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. A direct approach to false discovery rates. Journal of the Royal Statistical Society Series B: Statistical Methodology 1995, 57, 289–300. [Google Scholar] [CrossRef]

| Taxon | Population code | Locality | Population size |
|---|---|---|---|
| B. sinensis | Sin_BB | Bubeng, Mengla, Yunnan, China | 10 |
| Sin_NG | Nangongshan, Mengla, Yunnan, China | 6 | |
| Sin_NP | Nanping, Mengla, Yunnan, China | 3 | |
| Sin_ML | Menglun, Mengla, Yunnan, China | 3 | |
| Sin_PT | Puwen, Jinghong, Yunnan, China | 4 | |
| B. × multipinna | Mul_BB | Bubeng, Mengla, Yunnan, China | 12 |
| Mul_NG | Nangongshan, Mengla, Yunnan, China | 3 | |
| Mul_NP | Nanping, Mengla, Yunnan, China | 2 | |
| Mul_ML | Menglun, Mengla, Yunnan, China | 5 | |
| Mul_PT | Puwen, Jinghong, Yunnan, China | 9 | |
| B. longiaurita | Lon_NG | Nangongshan, Mengla, Yunnan, China | 2 |
| Lon_NP | Nanping, Mengla, Yunnan, China | 2 | |
| B. sp | Sp_NG | Nangongshan, Mengla, Yunnan, China | 2 |
| B. sp | Sp_PT | Puwen, Jinghong, Yunnan, China | 2 |
| Pop ID | Private | Num_Indv | Obs_Het | Obs_Hom | Exp_Het | Exp_Hom | Pi | Fis |
|---|---|---|---|---|---|---|---|---|
| BB | 381 | 17.17512 | 0.1168 | 0.8832 | 0.11173 | 0.88827 | 0.11527 | 0.03708 |
| NG | 573 | 9.8632 | 0.15594 | 0.84406 | 0.14675 | 0.85325 | 0.15519 | 0.03164 |
| NP | 221 | 5.71069 | 0.16309 | 0.83691 | 0.14162 | 0.85838 | 0.15601 | 0.00498 |
| ML | 390 | 6.13749 | 0.22184 | 0.77816 | 0.18258 | 0.81742 | 0.20043 | -0.01523 |
| PT | 616 | 11.23925 | 0.16946 | 0.83054 | 0.15015 | 0.84985 | 0.15775 | 0.01291 |
| weighted Fst | BB | NG | NP | ML | PT |
|---|---|---|---|---|---|
| BB | / | ||||
| NG | 0.0689 | / | |||
| NP | 0.071 | -0.0033 | / | ||
| ML | 0.0834 | 0.0258 | 0.0198 | / | |
| PT | 0.1574 | 0.0678 | 0.078 | 0.0817 | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





