Submitted:
25 June 2024
Posted:
26 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Animal Design
2.3. Astaxanthin-Rich Dietary Supplement
2.4. Growth Performance
2.5. Tissue Sample Collection
2.6. Total RNA Extraction and cDNA preparation
2.7. Bioinformatics: Genome Assembly and Gene Primer Design
2.8. Quantitative Real-Time RT-PCR (qPCR)
2.9. Gene Ontology
2.10. Statistical Analysis
3. Results
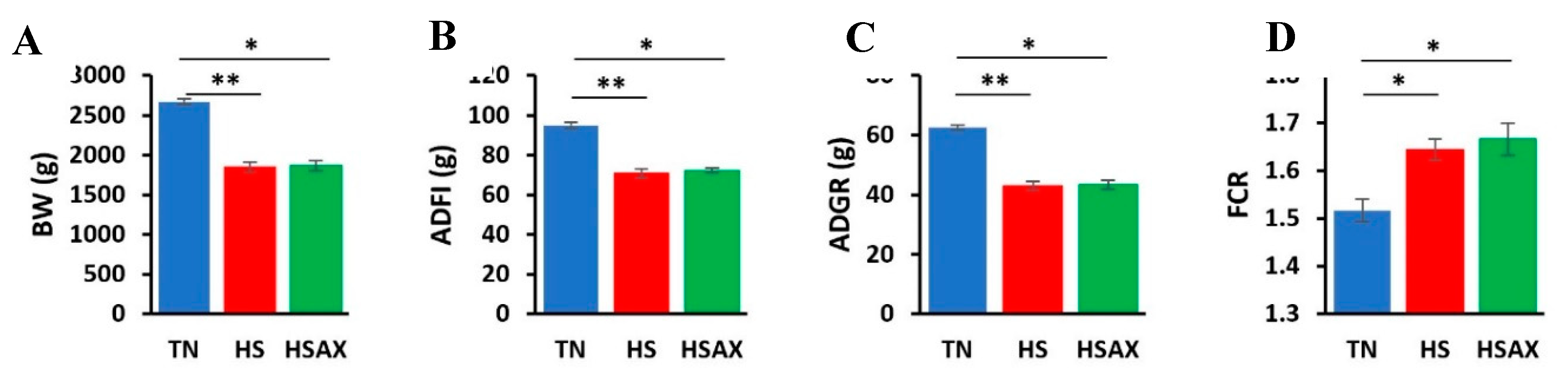
3.1. Growth Performance
3.2. Quantitative Real-time RT-PCR (qPCR) Gene Expression
3.2.1. Statistically Significant Genes Identified in Differential Expression Analysis
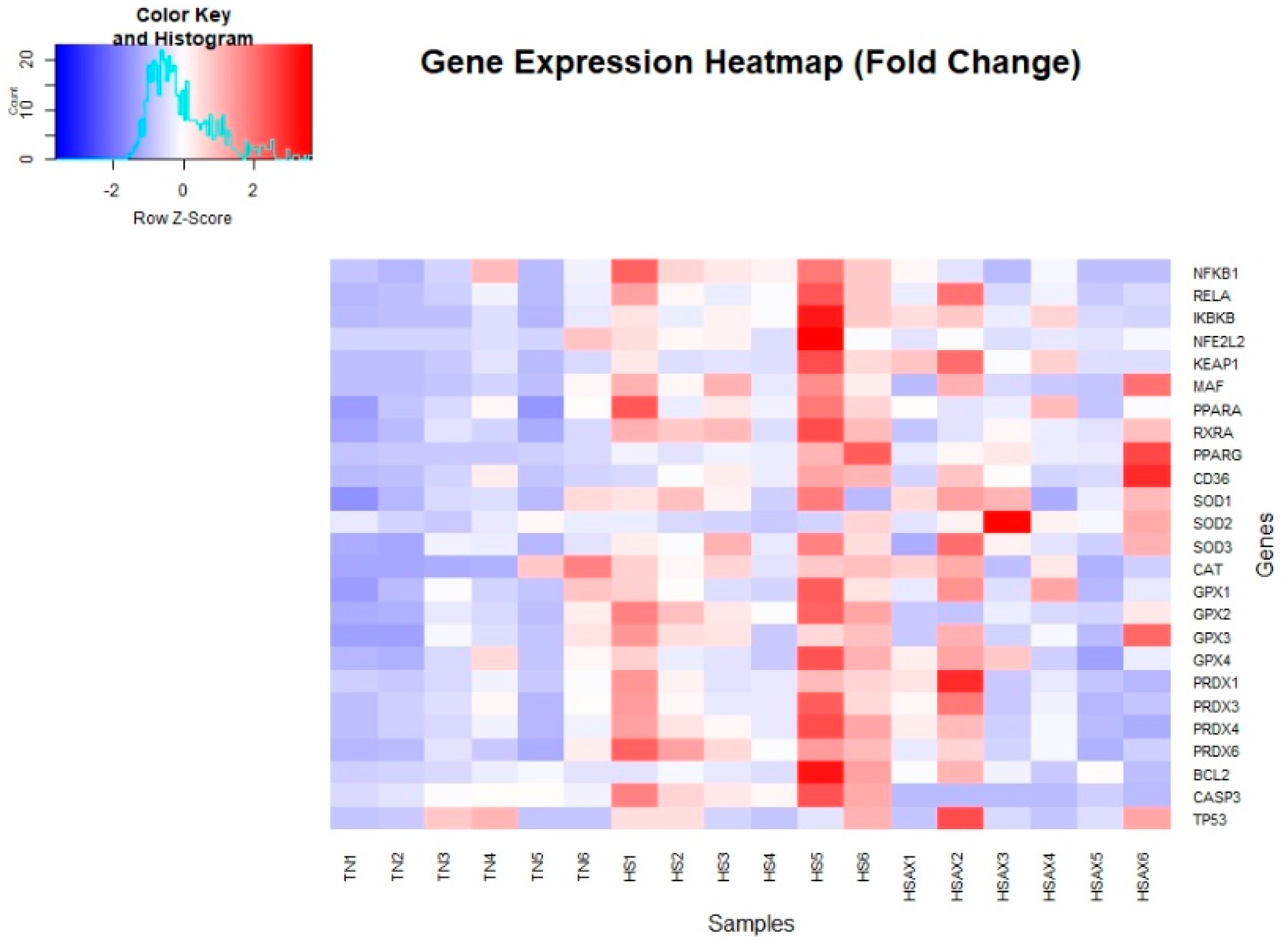
| Gene | TN | HS | HSAX | p-value |
| NFKB1 | 1.263a ± 0.42 |
3.168b ± 0.45 |
1.157a ± 0.25 |
a0.011 |
| RELA |
1.153a ± 0.27 |
3.586b ± 0.72 |
2.261a ±0.74 |
0.012 |
|
IKBKB |
1.257a ± 0.38 |
5.759b ± 1.55 |
3.857a ± 0.74 |
0.009 |
|
NFE2L2 |
2.275a ± 1.55 |
8.637b ± 3.90 |
2.582a ± 0.46 |
0.039 |
|
KEAP1 |
1.177b ± 0.29 |
4.375a ± 1.36 |
4.675a ± 1.28 |
0.011 |
|
MAF |
1.199a ± 0.38 |
4.330b ± 0.72 |
2.824a ± 1.19 |
0.030 |
|
PPARα |
1.192 ± 0.28 |
2.658 ± 0.45 |
1.791 ± 0.25 |
0.064 |
|
RXRA |
1.343a ± 0.37 |
6.121b ± 1.01 |
3.148a ± 0.63 |
0.004 |
|
PPARγ |
1.859b ± 0.81 |
25.560a ± 9.53 |
5.452a ± 9.84 |
0.003 |
|
CD36 |
1.744a ± 0.84 |
5.811b ± 1.37 |
6.030a ± 2.57 |
0.042 |
|
SOD1 |
1.170 ± 0.28 |
2.098 ± 0.41 |
2.228 ± 0.39 |
0.128 |
|
SOD2 |
1.070 ± 0.17 |
1.046 ± 0.29 |
2.638 ± 0.82 |
0.049 |
|
SOD3 |
1.381a ± 0.41 |
3.994b ± 0.64 |
3.272a ± 1.01 |
0.045 |
|
CAT |
6.447 ± 3.92 |
12.313 ± 1.47 |
8.614 ± 2.81 |
0.291 |
|
GPX1 |
1.232 ± 0.33 |
2.156 ± 0.44 |
1.862 ± 0.44 |
0.209 |
|
GPX2 |
1.764a ± 0.68 |
6.997b ± 1.07 |
2.392a ± 0.52 |
0.005 |
|
GPX3 |
1.266 ± 0.34 |
2.599 ± 0.36 |
2.143 ± 0.60 |
0.085 |
|
GPX4 |
1.132 ± 0.25 |
2.029 ± 0.45 |
1.578 ± 0.36 |
0.236 |
|
PRDX1 |
1.123 ± 0.25 |
2.950 ± 0.60 |
2.224 ± 1.22 |
0.117 |
|
PRDX3 |
1.119 ± 0.23 |
2.389 ± 0.49 |
1.540 ± 0.51 |
0.135 |
|
PRDX4 |
1.060a ± 0.16 |
2.601b ± 0.43 |
1.360a ± 0.36 |
0.026 |
|
PRDX6 |
1.100a ± 0.23 |
2.888b ± 0.33 |
1.427a ± 0.25 |
0.006 |
|
BCL2 |
1.039 ± 0.13 |
2.479 ± 0.88 |
1.483 ± 0.41 |
0.312 |
|
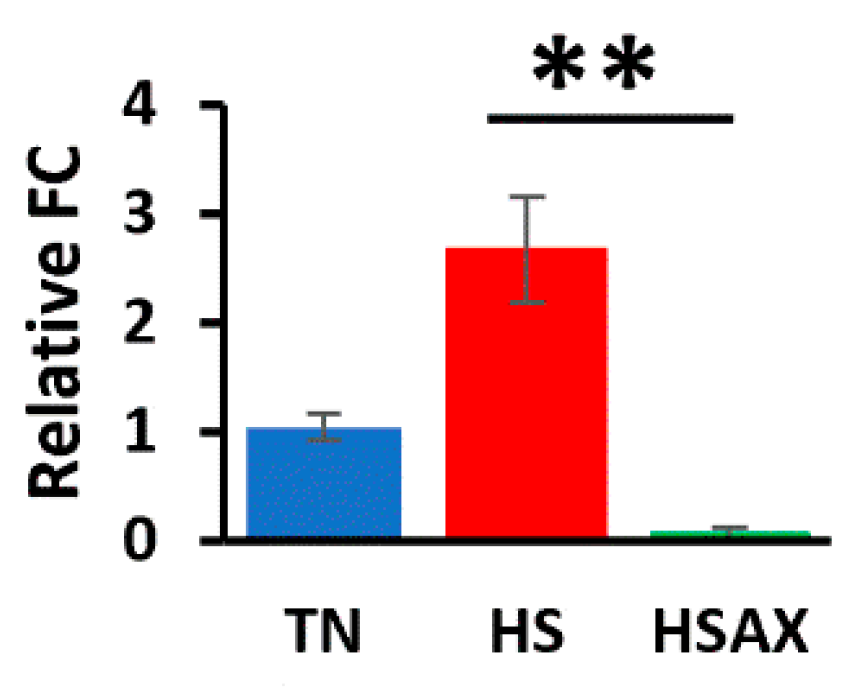
CASP3 |
1.043a ± 0.12 |
2.677b ± 0.48 |
0.067a ± 0.06 |
0.001 |
|
TP53 |
2.976 ± 1.66 |
4.297 ± 1.40 |
4.890 ± 2.55 |
0.567 |
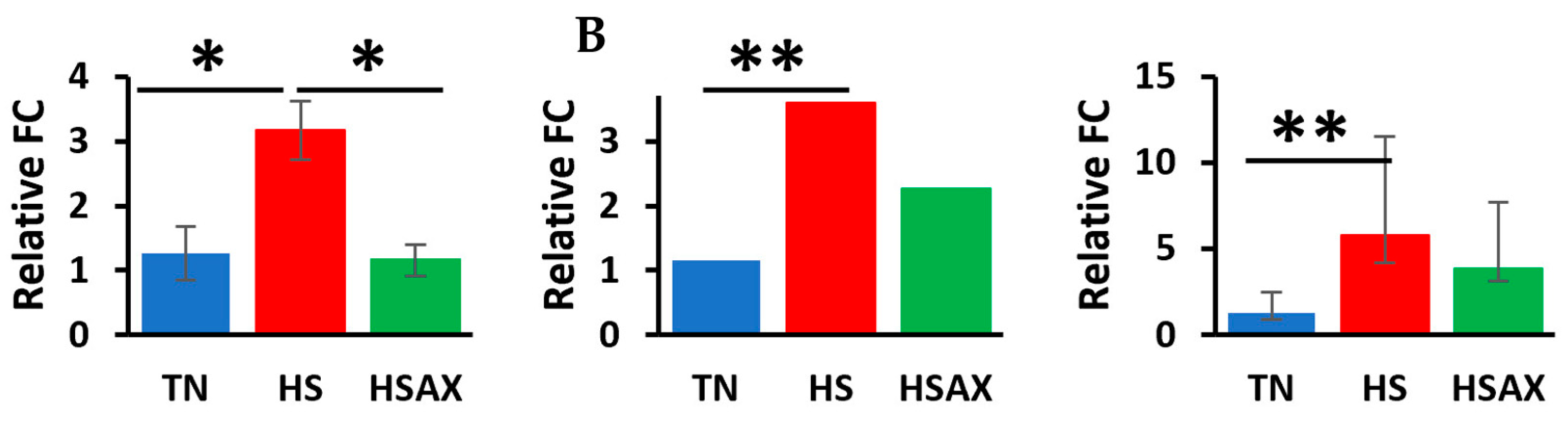
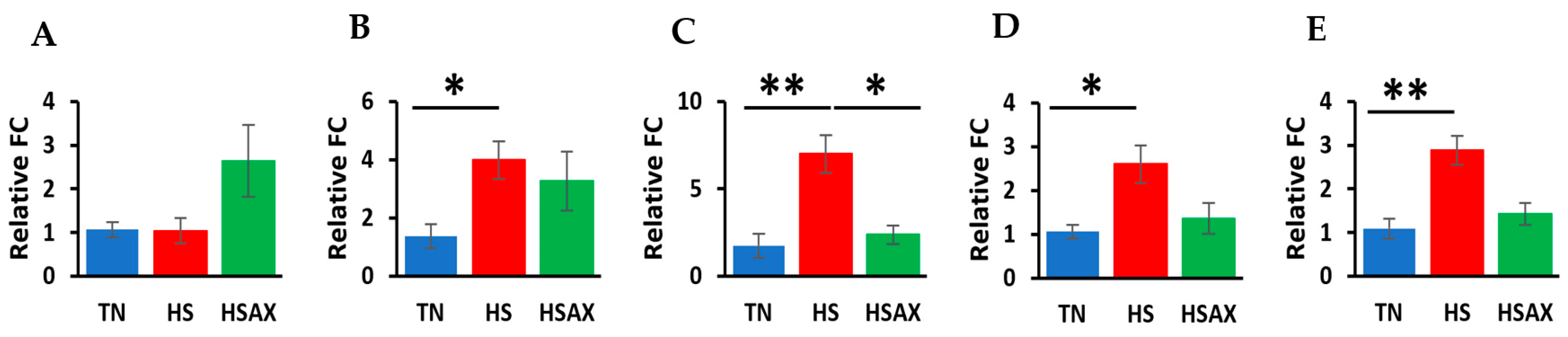
3.2.2. NF-kB Transcription Signaling Pathway Genes
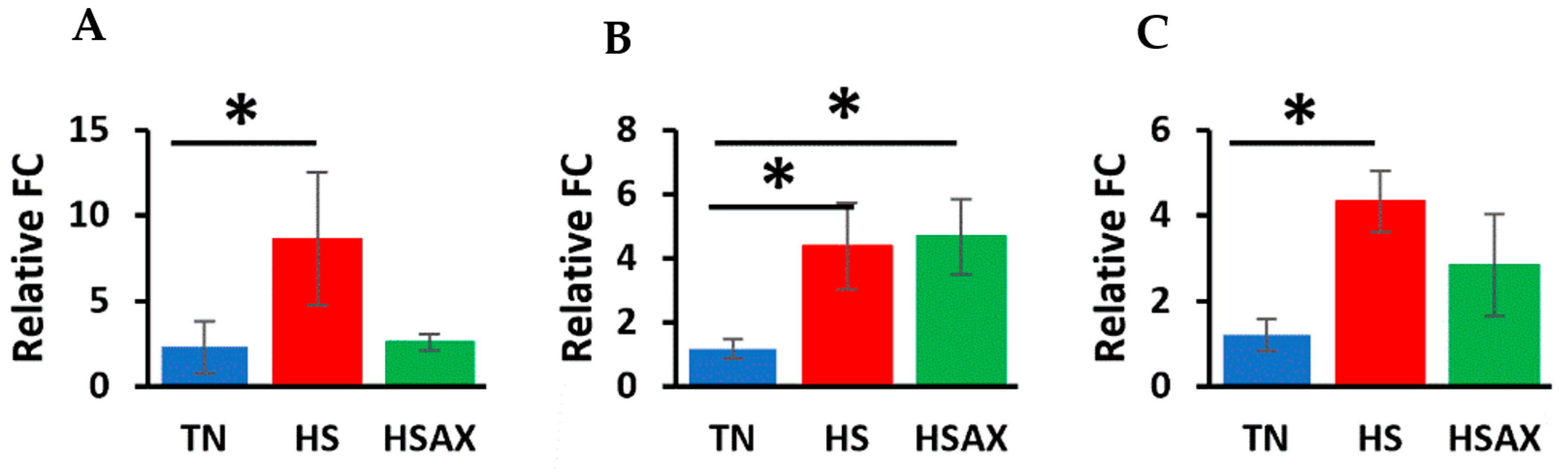
3.2.3. NFE2L2-Mediated Signaling Pathway Genes
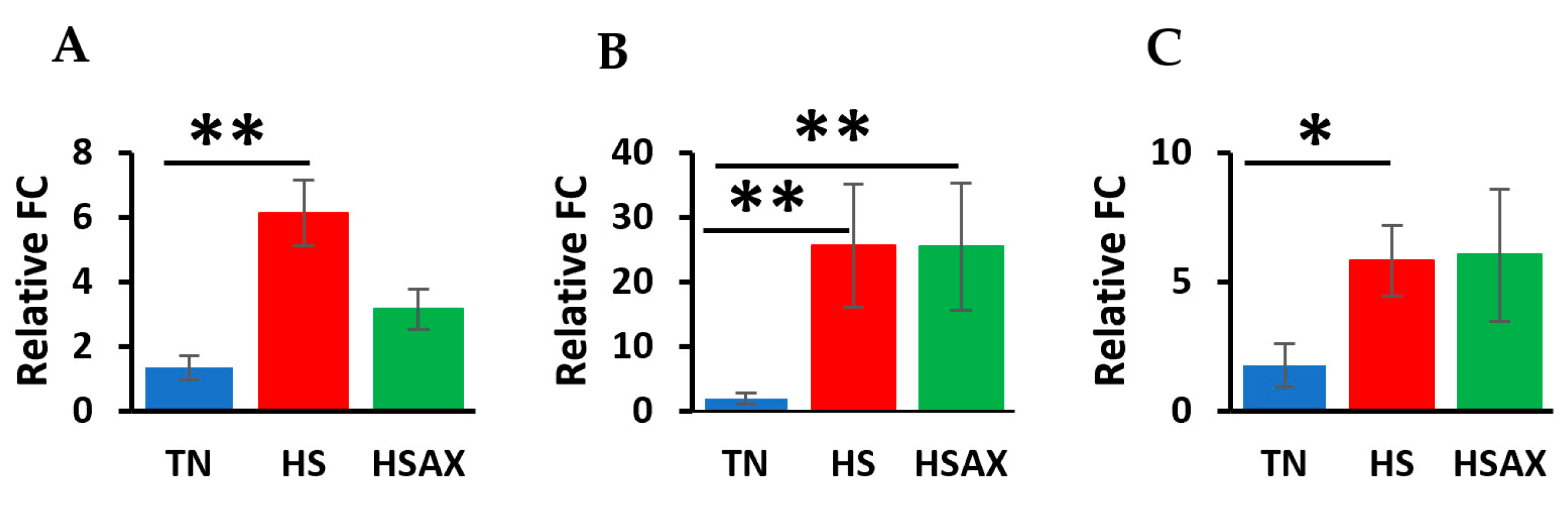
3.2.4. PPARα Signaling Pathway Genes
3.2.5. Cytoprotective Capacity Genes
3.2.6. Apoptotic Gene
3.2.7. Gene Ontology (GO) Gene Enrichment
| Gene | Cellular Component | Molecular Function | Biological Process | Transcript IDs |
|---|---|---|---|---|
| NFKB1 | nucleus, cytoplasm, mitochondrion, chromatin | DNA binding transcription factor activity, RNA polymerase II specific DNA binding, chromatin, protein and actinin binding | regulation of transcription by RNA polymerase II, MAPK cascade, JNK cascade, NIK/NF-kappaB signaling | ENSGAL00010005476 |
| RELA | cytoplasm | DNA-binding transcription factor activity | regulation of DNA-templated transcription | ENSGALG00010001277 |
| IKBKB | cytoplasm, cytosol | protein kinase activity, identical protein binding, protein homodimerization activity, scaffold protein binding, transferrin receptor binding | Protein phosphorylation, I-kappaB kinase/NF-kappaB signaling, cellular response to tumor necrosis factor, regulation of establishment of endothelial barrier, negative regulation of bicellular tight junction assembly | ENSGALG00010017955 |
| NFE2L2 | nucleus, cytoplasm, Golgi apparatus, chromatin, centrosome, plasma membrane, RNA polymerase II transcription regulator complex | DNA binding transcription factor activity, RNA polymerase II specific DNA-binding transcription factor binding, ubiquitin protein ligase binding | response to oxidative stress, inflammatory response, regulation of gene expression, protein ubiquitination, cell redox homeostasis, positive regulation of glutathione biosynthetic process, regulation of removal of superoxide radicals | ENSGAL00010024107 |
| KEAP1 | nucleus, cytoplasm, endoplasmic reticulum, Cul3-RING ubiquitin ligase complex, centriolar satellite | RNA polymerase II-specific DNA-binding transcription factor binding, ubiquitin ligase-substrate adaptor activity, disordered domain specific binding, identical protein binding | cellular response to oxidative stress, ubiquitin-dependent protein catabolic process, regulation of DNA-templated transcription, regulation of autophagy | ENSG00000079999 |
| MAF | nucleus, cytoplasm, RNA polymerase II transcription regulator complex | DNA-binding transcription factor activity, RNA polymerase II sequence-specific DNA binding | regulation of DNA templated transcription, regulation of transcription by RNA polymerase II, cell development | ENSGALG00010007800 |
| PPARα | nucleus, nucleoplasm | RNA polymerase II-specific DNA-binding transcription factor, signaling receptor activity, metal ion binding, lipid binding, ubiquitin conjugating enzyme binding | Negative regulation of transcription by RNA polymerase II, regulation of DNA-templated transcription, negative regulation of reactive oxygen species biosynthesis process, negative regulation of cytokine production ninvolved in inflammatory response, positive regulation of fatty acid oxidation, positive regulation for gluconeogenesis | ENSGALG00010023058 |
| RXRA | nucleus, RNA polymerase II transcription regulator complex, receptor complex | RNA polymerase II transcription regulatory region sequence-specific DNA binding, enzyme, peptide, metal ion binding, retinoic acid-responsive element binding, Vitamin D response element binding | peroxisome proliferator activated receptor signaling pathway, retinoic acid receptor signaling pathway, positive regulation of Vitamin D receptor signaling pathway, cell differentiation | ENSGALG00010028422 |
| PPARγ | nucleus, cytoplasm, intracellular membrane-bounded organelle | DNA-binding transcription factor activity, nuclear receptor activity, zinc and metal ion binding | Regulation of DNA-templated transcription, transcription by RNA polymerase II, intracellular receptor signaling pathway | ENSGALG00010027917 |
| CD36 | external side of plasma membrane, receptor complex, membrane raft | low-density lipoprotein particle receptor activity, high-density lipoprotein particle binding, Toll-like receptor binding, scavenger receptor activity | MAPK cascade, cell surface receptor signaling pathway, positive regulation of cytosolic calcium ion concentration, nitric oxide mediated signal transduction, intestinal cholesterol absorption, positive regulation of reactive oxygen species metabolic process, lipid transport across blood-brain barrier | ENSGALG000010008392 |
| SOD2 | mitochondrion | superoxide dismutase activity, oxidoreductase activity, manganese ion binding, metal ion binding, identical protein binding | response to oxidative stress, oxidation-reduction process, negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway, response to hydrogen peroxide, removal of superoxide radicals |
ENSGALT00000019062 |
| SOD3 | extracellular space, collagen- containing extracellular matrix | superoxide dismutase activity, copper and metal ion binding | superoxide metabolic process, response to hypoxia | ENSGALG00010009833 |
| GPX2 | cytosol, intercellular bridge, mitotic spindle | glutathione peroxidase activity, phospholipid-hydroperoxide glutathione peroxidase activity | response to oxidative stress | ENSGALG00010021537 |
| PRDX4 | cytoplasm, endoplasmic reticulum | antioxidant activity, thioredoxin peroxidase activity, oxidoreductase activity, peroxiredoxin activity, molecular sequestering activity | response to oxidative stress, cell redox homeostasis, hydrogen peroxide catabolic process, reactive oxygen species metabolic process, cellular oxidant detoxification | ENSGALG00010003214 |
| PRDX6 | nucleus, cytoplasm | antioxidant activity, glutathione peroxidase activity, oxidoreductase activity, peroxiredoxin activity, ubiquitin protein ligase binding | response to oxidative stress, cellular oxidant detoxification, glycerophospholipid catabolic process | ENSGALG00010013528 |
| CASP3 | nucleus, cytoplasm | endopeptidase activity, hydrolase activity, cyclin-dependent protein serine/threonine kinase inhibitor activity, cysteine-type endopeptidase activity involved in execution phase of apoptosis | T and B cell homeostasis, negative regulation of cytokine production, intrinsic apoptotic signaling pathway, negative regulation of cell cycle, neuron apoptotic process, epithelial cell apoptotic process | ENSGALG00010007067 |
4. Discussion
4.1. Growth Performance
4.2. Gene Ontology Enrichment and Expression Analysis
4.2.1. NF-kB Transcription Signaling Pathway Genes
4.2.2. NFE2L2-Mediated Signaling Pathway Genes
4.2.3. PPARα Signaling Pathway Genes
4.2.4. Cytoprotective Capacity Genes
4.2.5. Apoptotic Pathway Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vos, J.G.; Moore, J.A. Immune suppression as related to toxicology. CRC Crit. Rev. Toxicol. 1977, 5, 1, 67–101. [CrossRef]
- Murray, J.M.; Kaufmann, G.R.; Hodgkin, P.D.; Lewin, S.R.; Kelleher, A.D.; Davenport, M. P.; Zaunders, J. J. Naive T cells are maintained by thymic output in early ages but by proliferation without phenotypic change after age twenty. Immunol. Cell Biol. 2003, 81, 6, 487–495. [CrossRef]
- Gameiro, J.; Nagib, P.; Verinaud, L. The thymus microenvironment in regulating thymocyte differentiation. Cell Adhes. Migr. 2010, 4, 3, 382–390. [CrossRef]
- Savino, W. The thymus is a common target organ in infectious diseases. PLoS Pathog. 2006, 2, 5, e62. [CrossRef]
- Thapa, P.; Farber, D.L. The role of the thymus in the immune response. Thorac. Surg. Clin. 2019, 29, 2, 123–131. [CrossRef]
- Raviola, E.; Karnovsky, M.J. Evidence for a blood-thymus barrier using electronopaque tracers. J Exp Med. 1972, 136, 3, 466–498. [CrossRef]
- Lynch, H.E.; Goldberg, G.L.; Chidgey, A.; Van den Brink, M. R. M.; Boyd, R.; Sempowski, G. D.. Thymic involution and immune reconstitution. Trends Immunol. 2009, 30, 7, 366–373. [CrossRef]
- Dai, X.; Zhang, D.; Wang, C.; Wu, Z.; Liang, C. The pivotal role of thymus in atherosclerosis mediated by immune and inflammatory response. Int. J. Med. Sci. 2018, 15, 13, 1555–1563. [CrossRef]
- Chrousos, G. P. Stress and disorders of the stress system. Nat. Rev. Endocrinol., 2009, 5, 7, 374-381. [CrossRef]
- Kumar, B.; Manuja, A.; Aich, P. Stress and its impact on farm animals. Front. Biosci. 2012, E4, 1759-1767.
- Li, W. J.; Nie, S. P.; Peng, X. P.; Liu, X. Z., Li, C.; Chen, Y.; Li, J. E.; Song, W. .; Xie, M. Y. Ganoderma atrum polysaccharide improves age-related oxidative stress and immune impairment in mice. J Agric Food Chem. 2012, 60, 6, 1413–1418. [CrossRef]
- Gostner, J.M.; Becker, K.; Ueberall, F.; Fuchs, D. The good and bad of antioxidant foods: an immunological perspective. Food Chem Toxicol, 2015, 80, 72–79. [CrossRef]
- Lessard, M.; Savard, C.; Deschene, K.; Lauzon, K.; Pinilla, V.A.; Gagnon, C. A.; Lapointe, J.; Guay, F.; Chorfi, Y. Impact of deoxynivalenol (DON) contaminated feed on intestinal integrity and immune response in swine. Food Chem Toxicol. 2015, 80, 7–16. [CrossRef]
- Saeidnia, S.; Abdollahi, M. Toxicological and pharmacological concerns on oxidative stress and related diseases. Toxicol Appl Pharmacol. 2013, 273, 3, 442–55. [CrossRef]
- Zhang, X.Y.; Yao, J.K. Oxidative stress and therapeutic implications in psychiatric disorders. Intl. J. Neuropsychopharm Biol. Psych. 2013; 46, 10, 197–9. [CrossRef]
- Neri, M.; Fineschi, V.; Di Paolo, M.; Pomara, C.; Riezzo, I.; Turillazzi, E.; Cerretani, D. Cardiac Oxidative Stress and Inflammatory Cytokines Response after Myocardial Infarction. Curr Vasc Pharmacol. 2015, 13, 1, 26–36. [CrossRef]
- Nunes, V.A.; Gozzo, A.J.; Cruz-Silva, I.; Juliano, M.A.; Viel, T.A.; Godinho, R. O.; Meirelles, F. V.; Sampaio, M. U.; Sampaio, C. A. M.; Araujo, M. S. Vitamin E prevents cell death induced by mild oxidative stress in chicken skeletal muscle cells. Comp Biochem Physiol C Toxicol Pharmacol. 2005, 141, 3, 225–40. [CrossRef]
- Singhal, A.; Morris, V.B.; Labhasetwar, V.; Ghorpade, A. Nanoparticle-mediated catalase delivery protects human neurons from oxidative stress. Cell Death Dis. 2013, 4, 11, e903. [CrossRef]
- Rao, M.; Yang, W.; Seifalian, A.M.; Winslet, M.C. Role of cyclooxygenase-2 in the angiogenesis of colorectal cancer. Int J Color Dis. 2004, 19, 1, 1–11. [CrossRef]
- Ashton, K. J.; Reichelt, M. E.; Mustafa, S. J.; Teng, B.; Lea, C. L.; Hoffman, P. A.; Morrison, R. R.; Headrick, J. P. Transcriptomic effects of adenosine 2A receptor deletion in healthy and endotoxemic murine myocardium. Purinergic Signal. 2016, 13, 1, 27-49. [CrossRef]
- Goodman, W. A.; Omenetti, S.; Date, D.; Martino, L. D.; De Salvo, C.; Kim, G.-D.; Chowdhry, S.; Bamias, G.; Cominelli, F.; Pizarro, T. T., Mahabeleshwar, G. H. KLF6 contributes tomyeloid cell plasticity in the pathogenesis of intestinal inflammation. Mucosal Immunol. 2016, 9, 5, 1250–1262. Doi: 10.1038/mi.2016.1.
- Hu, X.; Chi, Q.; Liu, Q.; Wang, D.; Zhang, Y. Atmospheric H2S triggers immune damage by activating the TLR-7/MyD88/NF-κB pathway and NLRP3 inflammasome in broiler thymus. Chemosphere, 2019, 237, 124427. [CrossRef]
- Di Prisco, G.; Iannaccone, M.; Ianniello, F.; Ferrara, R.; Caprio, E.; Pennacchio, F; Capparelli, R. The neonicotinoid insecticide Clothianidin adversely affects immune signaling in a human cell line. Sci. Rep. 2017, 7,1, 13446. [CrossRef]
- Bajpai, V.K.; Alam, M.B.; Ju, M.-K.; Kwon, K.-R.; Huh, Y.S.; Han, Y.-K.; Lee, S. H. Antioxidant mechanism of polyphenol-rich Nymphaea nouchali leaf extract protecting DNA damage and attenuating oxidative stress-induced cell death via Nrf2-mediated heme-oxygenase-1 induction coupled with ERK/p38 signaling pathway. Biomed Pharmacother. 2018, 103, 1397–1407. [CrossRef]
- Lam. G.Y.; Huang, J.; Brumell, J.H. The many roles of NOX2 NADPH oxidase derived ROS in immunity. Semin Immunopathol. 2010, 32, 4. 415–30. [CrossRef]
- Kotsias, F.; Hoffmann, E.; Amigorena, S.; Savina, A. Reactive oxygen species production in the phagosome: impact on antigen presentation in dendritic cells. Antioxid Redox Signal. 2013, 18, 6. 714–29. [CrossRef]
- Yang, Y.; Bazhin, A.V.; Werner, J.; Karakhanova, S. Reactive oxygen species in the immune system. Int Rev Immunol. 2013, 32, 3, 249–70. DOI: 10.3109/08830185.2012.755176.
- Moraes, L. A.; Piqueras, L.; Bishop-Bailey, D. Peroxisome proliferator-activated receptors and inflammation. Pharmacol. Ther. 2006, 110, 3, 371–385. [CrossRef]
- Mounier, R.; Theret, M.; Arnold, L.; Cuvellier, S.; Bultot, L.; Goransson, O.; Sanz, N.; Ferry, A.; Sakamoto, K.; Foretz, M.; Viollet, B.; Chazaud, B. AMPK alpha1 regulates macrophage skewing at the time of resolution of inflammation during skeletal muscle regeneration. Cell Metab. 2013, 18, 251–264. [CrossRef]
- Okamoto, H.; Iwamoto, T.; Kotake, S.; Momohara, S.; Yamanaka, H.; Kamatani, N. Inhibition of NF-kappaB signaling by fenofibrate, a peroxisome proliferator-activated receptor-alpha ligand, presents a therapeutic strategy for rheumatoid arthritis. Clin. Exp. Rheumatol. 2005, 23, 3, 323–330.
- Eggersdorfer, M.; Wyss, A. Carotenoids in human nutrition and health. Arch Biochem Biophy. 2018, 652, 18–26. [CrossRef]
- Destefanis, S.; Giretto, D.; Muscolo, M. C.; Di Cerbo, A.; Guidetti, G.; Canello, S.; Giovazzino, A.; Centenaro, S.; Terrazzano, G.. Clinical evaluation of a nutraceutical diet as an adjuvant to pharmacological treatment in dogs affected by Keratoconjunctivitis sicca. BMC Vet. Res. 2016, 12, 1, 214. [CrossRef]
- Guidetti, G.; Di Cerbo, A.; Giovazzino, A.; Rubino, V.; Palatucci, A.T.; Centenaro, S.; Fraccaroli, E.; Cortese, K.; Bonomo, M. G.; Ruggiero, G.; Canello, S.; Terrazzano, G. In Vitro Effects of Some Botanicals with Anti-Inflammatory and Antitoxic Activity. J. Immunol. Res. 2016, 1-11. [CrossRef]
- Di Cerbo, A.; Morales-Medina, J. C.; Palmieri, B.; Pezzuto, F.; Cocco, R.; Flores, G.; Iannitti, T. Functional foods in pet nutrition: Focus on dogs and cats. Res. Vet. Sci. 2017, 112, 161–166. [CrossRef]
- Fakhri, S.; Abbaszadeh, F.; Dargahi, L.; Jorjani, M. Astaxanthin: A mechanistic review on its biological activities and health benefits. Pharmacol. Res. 2018, 136, 1–20. [CrossRef]
- Astaxanthin. (n.d.). Retrieved from https://pubchem.ncbi.nlm.nih.gov/compound/Astaxanthin. Accessed 02 Apr 2024.
- Heaney, R. P. Factors Influencing the Measurement of Bioavailability, Taking Calcium as a Model. J. Nutr. 2001, 131, 4. [CrossRef]
- Mularczyk, M.; Michalak, I.; Marycz, K. Astaxanthin and other Nutrients from Haematococcus pluvialis-Multifunctional Applications. Mar Drugs. 2020, 18. 9, 459. PMID: 32906619; PMCID: PMC7551667. [CrossRef]
- Zhang, Z.-W.; Xu, X.-C.; Liu, T.; Yuan, S. Mitochondrion-Permeable Antioxidants to Treat ROS-Burst-Mediated Acute Diseases. Oxid. Med. Cell. Longev. 2016, 1-10. [CrossRef]
- Shahidi, F., Barrow, C. J. Marine nutraceuticals and functional foods. Boca Raton: CRC Press, 2008.
- Livak, K. J.; Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods, 2001, 25, 402-408. [CrossRef]
- Ensembl.org. https://uswest.ensembl.org/Gallus_gallus/Info/Index Accessed 04 Apr 2024.
- R Core Team (2023) https://www.R-project.org/ Accessed 04 Apr 2024.
- Akbarian, A.; Michiels, J.; Degroote, J.; Majdeddin, M.; Golian, A.; De Smet, S. Association between Heat Stress and Oxidative Stress in Poultry; Mitochondrial Dysfunction and Dietary Interventions with Phytochemicals. J. Anim.l Sci. Biotechnol. 2016, 7, 1. [CrossRef]
- Yang, T.; Liu, B.; Wang, Y.; Huang, X.; Yan, Z.; Jiang, Q.; Chen, Q. Ellagic Acid Improves Antioxidant Capacity and Intestinal Barrier Function of Heat-Stressed Broilers via Regulating Gut Microbiota. Animals 2022, 12, 1180. [CrossRef]
- He, W.; Wang, H.; Tang, C.; Zhao, Q.; Zhang, J. Dietary supplementation with astaxanthin alleviates ovarian aging in aged laying hens by enhancing antioxidant capacity and increasing reproductive hormones. Poult. Sci. 2023, 102, 102258. [CrossRef]
- Tolba, S. A.; Magnuson, A. D.; Sun, T.; Lei, X. G. Dietary supplemental microalgal astaxanthin modulates molecular profiles of stress, inflammation, and lipid metabolism in broilers chickens and laying hens under high ambient temperatures. Poult Sci. 2020, 99, 4853-4860. [CrossRef]
- Kouvedaki, I.; Pappas, A. C.; Surai, P. F.; Zoidis, E. Nutrigenomics of Natural Antioxidants in Broilers. Antioxidants. 2024, 13, 270. [CrossRef]
- Tian, Y.; Che, H.; Yang, J.; Jin, Y.; Yu, H.; Wang, C.; Fu, Y.; Li, N. Astaxanthin Alleviates Aflatoxin B1-Induced Oxidative Stress and Apoptosis in IPEC-J2 Cells via the Nrf2 Signaling Pathway. Toxins. 2023, 15, 232. [CrossRef]
- Lee, J.; Lim, J. W.; Kim, H. Astaxanthin Inhibits Oxidative Stress-Induced Ku Protein Degradation and Apoptosis in Gastric Epithelial Cells. Nutrients. 2022, 14, 19, 3939. [CrossRef]
| Ingredients (%) | Starter | Finisher |
|---|---|---|
| Corn | 54.86 | 63.14 |
| Soybean Meal | 39.49 | 29.59 |
| Soybean oil | 2.00 | 4.50 |
| Limestone | 1.27 | 0.85 |
| Monocalcium phosphate | 0.75 | 0.50 |
| L-lysine (98-99%) | 0.23 | 0.18 |
| DL-methionine (99%) | 0.14 | 0.12 |
| L-threonine (98-99%) | 0.20 | 0.16 |
| Sodium chloride | 0.43 | 0.35 |
| Sodium bicarbonate | 0.12 | 0.10 |
| Vitamin-mineral premix1 | 0.50 | 0.50 |
| Astaxanthin supplement2 | 0.01 | 0.01 |
| Total | ||
| Calculated analysis | ||
| ME (kcal/kg) | 2909 | 3203 |
| Soybean Meal-CP (%) | 22.09 | 18.07 |
| Calcium (%) | 0.75 | 0.52 |
| Total Phosphorus (%) | 0.57 | 0.47 |
| dig Phosphorous (%) | 0.30 | 0.23 |
| L-lysine (%) | 1.39 | 1.10 |
| dig L-lysine (%) | 1.25 | 0.99 |
| DL-methionine (%) | 0.48 | 0.41 |
| dig DL-methionine (%) | 0.45 | 0.39 |
| L-cysteine (%) | 0.43 | 0.38 |
| L-threonine (%) | 1.03 | 0.85 |
| dig L-threonine (%) | 0.85 | 0.69 |
| L-tryptophan (%) | 0.33 | 0.26 |
| DL-methionine + L-cysteine (%) | 0.91 | 0.80 |
| L-arginine (%) | 1.61 | 1.31 |
| L-valine (%) | 1.22 | 1.03 |
| L-isoleucine (%) | 0.93 | 0.76 |
| L-leucine (%) | 1.89 | 1.63 |
| Neutral detergent fiber (% DM) | 9.13 | 8.78 |
| Crude fiber (%) | 3.97 | 3.46 |
| Sodium (mg/kg) | 0.22 | 0.18 |
| Chloride (mg/kg) | 0.30 | 0.25 |
| Choline (mg/kg) | 1419 | 1200 |
| Astaxanthin (mg/kg) | --- | 1.33 |
| Gene | NCBI Accession No. | Primer set (5’-3’) |
|---|---|---|
| ACTB | NM_205518.2 | F:5' - AATTGTGCGTGACATCAAGG |
|
TBP GAPDH |
NM_205103.2 NM_204305.2 |
R:3' -CACAGGACTCCATACCCAAG F:5' -GCGGCAGGCTCTGTT R:3' -ACCGAAAAGGTTTTTGACCC F:5' -AAGTCGGAGTCAACGGATTT R:3' -TCACAAGTTTCCCGTTCTCA |
| NFKB1 RELA IKBKB |
NM_205134.2 NM_001396038.1 NM_001395965.1 |
F:5’ -GGACGGCGAAAGGACTCT R:3’ -CCATTGCAAACATTTGGGGAT F:5’ -CGGTTCCGCTATAAGTGTGA R:3’ -GTAATGGTTTACGCGGATGG F:5’ -TCCCTGGGAGATGAAGGAG R:3’ -TTTGGATGGTTCAGCCTCTT |
| NFE2L2 KEAP1 MAF PPARA RXRA PPARG CD36 |
XM_015287264.4 GenBank: KU321503.1 NM_001044671.1 NM_001001464.1 XM_003642291.6 NM_001001460.2 NM_001030731.1 |
F:5’ –CAGGGGTAGCAAGGTATGAG R:3’ -TGCCTCCAAAGGATGTCAAT F:5’ –GATCGACGGGATGATCTACG R:3’ –GGCGTACAGCAGTATGTTCA F:5’ –CCAGAGTTTTTCATGTACCCG R:3’ -CTTTGTAGCTGTCTTCGTGC F:5’ – AGCCACTTGCTATCACCAAT R:3’ – ACTTAAACTCCTTTATGATTCTGGT F:5’ –CTTCCTGCCACTGGATTTCT R:3’ –CTGATGACGGAGAAGGGTG F:5’ – CTTGACAGCGCCAGAGATTA R:3’ –GATTGCACTTTGGCAATCCT F:5’ –TTTCTTGCAAAGCAGGAGGTT R:3’ –CTGATCTTCGTGAGAGAAGCTGTA |
| SOD1 | NM_205064.2 | F:5' -AAAAGATGCAGATAGGCACG |
| R:3' -TTATCTCCCCCTCTACCCAG | ||
| SOD2 SOD3 |
NM_204211.2 XM_040699307.2 |
F:5' -CCTTCGCAAACTTCAAGGAG R:3' -AGCAATGGAATGAGACCTGT F:5’ –CAACTCGCAAACAACGCT R:3’ –CTGGTGAGTGAGAACCTGC |
| CAT | NM_001031215.2 | F:5' -TTCCACGTTAAGACCGATCA |
| R:3' -CAATCTTGCCCACTGGAATG | ||
| GPX1 | NM_001277853.3 | F:5' -AATTCGGGCACCAGGAGAA |
| R:3' - CTCGAACATGGTGAAGTTGG | ||
| GPX2 | NM_001277854.3 | F:5' -AGTTCGGCTACCAGGAGAA |
| R:3' -CTTCTGGAACAGGGTGAAGT | ||
| GPX3 | NM_001163232.3 | F:5' - AGGTGAAATGCTACGACTCC |
|
GPX4 PRDX1 PRDX3 PRDX4 PRDX6 BCL2 CASP3 TP53 |
NM_204220.3 NM_001271932.2 XM_426543.6 XM_046912353.1 NM_001039329.3 NM_205339.3 NM_204725.1 NM_205264.1 |
R:3' - AGTGCATTCAGTTCGAGGTA F:5’ –AATGTGCGCTCAGGCG R:3’ –AGACGAAGCCCCTGTACT F:5’ –TGCGGGGCTCTTTGTATTAAA R:3’ –ATTGCCCATCTGGCATTACA F:5’ –CGTTGTCAATGGGGAGTTC R:3’ –GGGGCACACAAAGGTGAAAT F:5’ –ATCCCCTTGACTTCACGTTT R:3’ –ATCTTCATTGGTCCGAGTCC F:5’ –GACATCAACGCCTACAATGG R:3’ –GGCCAAATATGAACACCACA F:5’ –GAGGATGGGATGCCTTTGTG R:3’ –CCACGATAAACTGGGTGACT F:5’ – GGTGGAGGTGGAGGAGC R:3’ –TGAGCGTGGTCCATCTTTTA F:5’ –GTTACCACGACGAGCCACCAA R:3’ –TGCAGCGCCTCATTGATCTCCTT |
| Measurements | TN | HS | HSAX | p-value |
|
BW (g) |
2673.68b ± 35.71 |
1848.85a ± 61.56 |
1867.83a ± 60.82 |
0.005 |
|
ADFI (g) |
94.98b ± 1.45 |
70.68a ± 2.38 |
72.24a ± 1.14 |
0.004 |
|
ADG (g) |
62.65b ± 0.86 |
42.99a ± 1.46 |
43.45a ± 1.44 |
0.005 |
|
FCR |
1.52b ± 0.02 |
1.64a ± 0.02 |
1.67a ± 0.03 |
0.005 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





