Submitted:
05 June 2024
Posted:
07 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
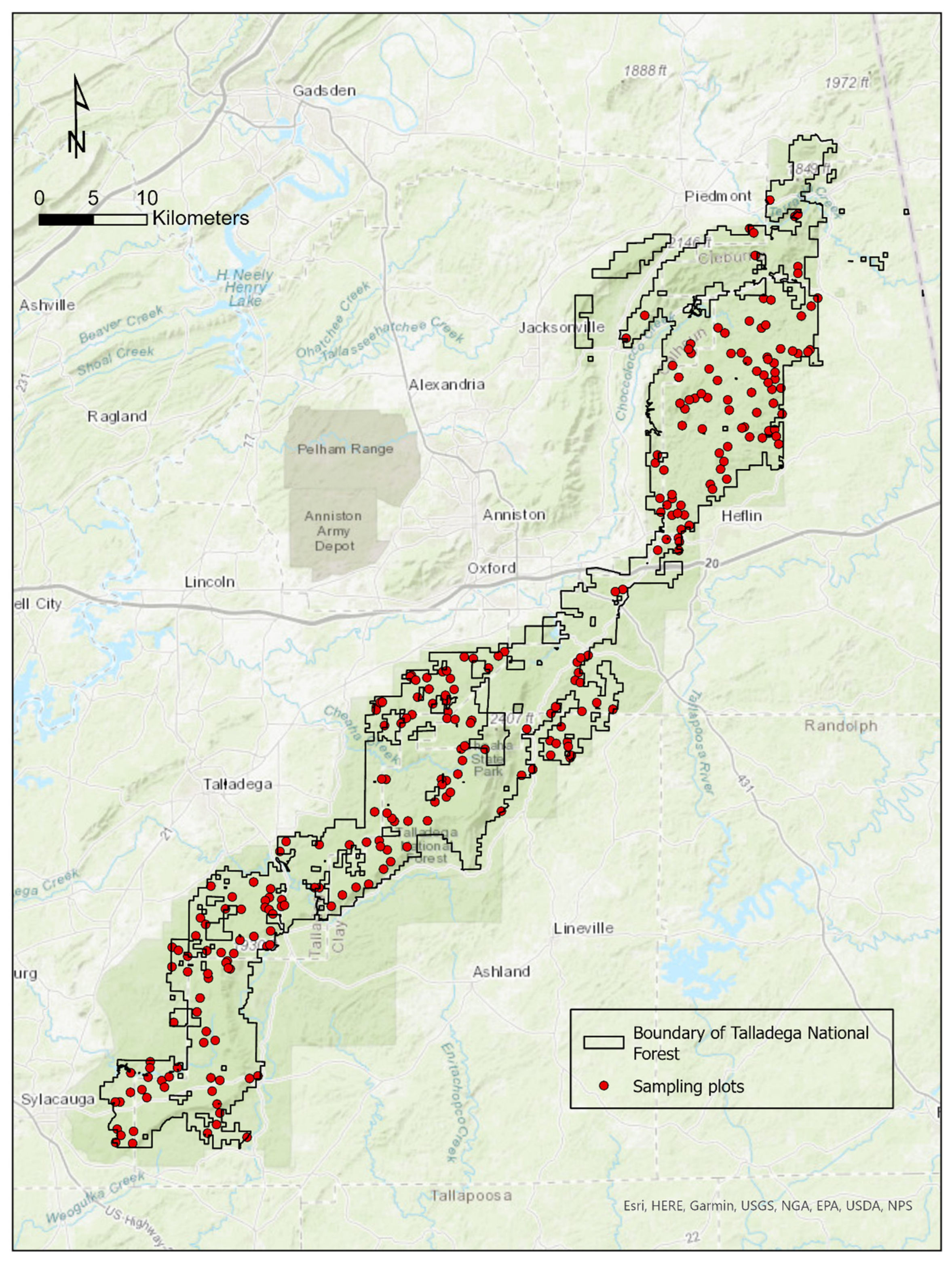
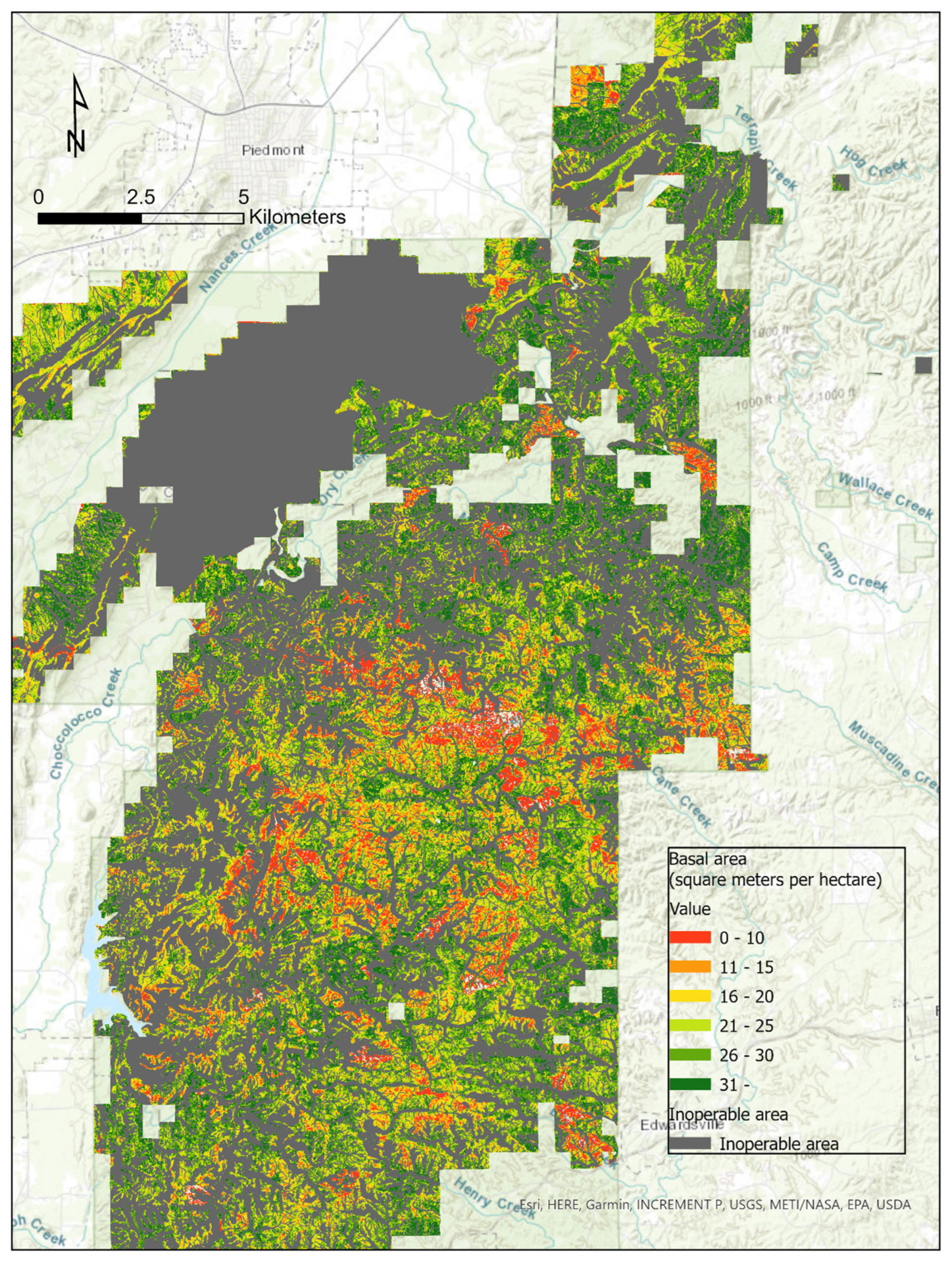
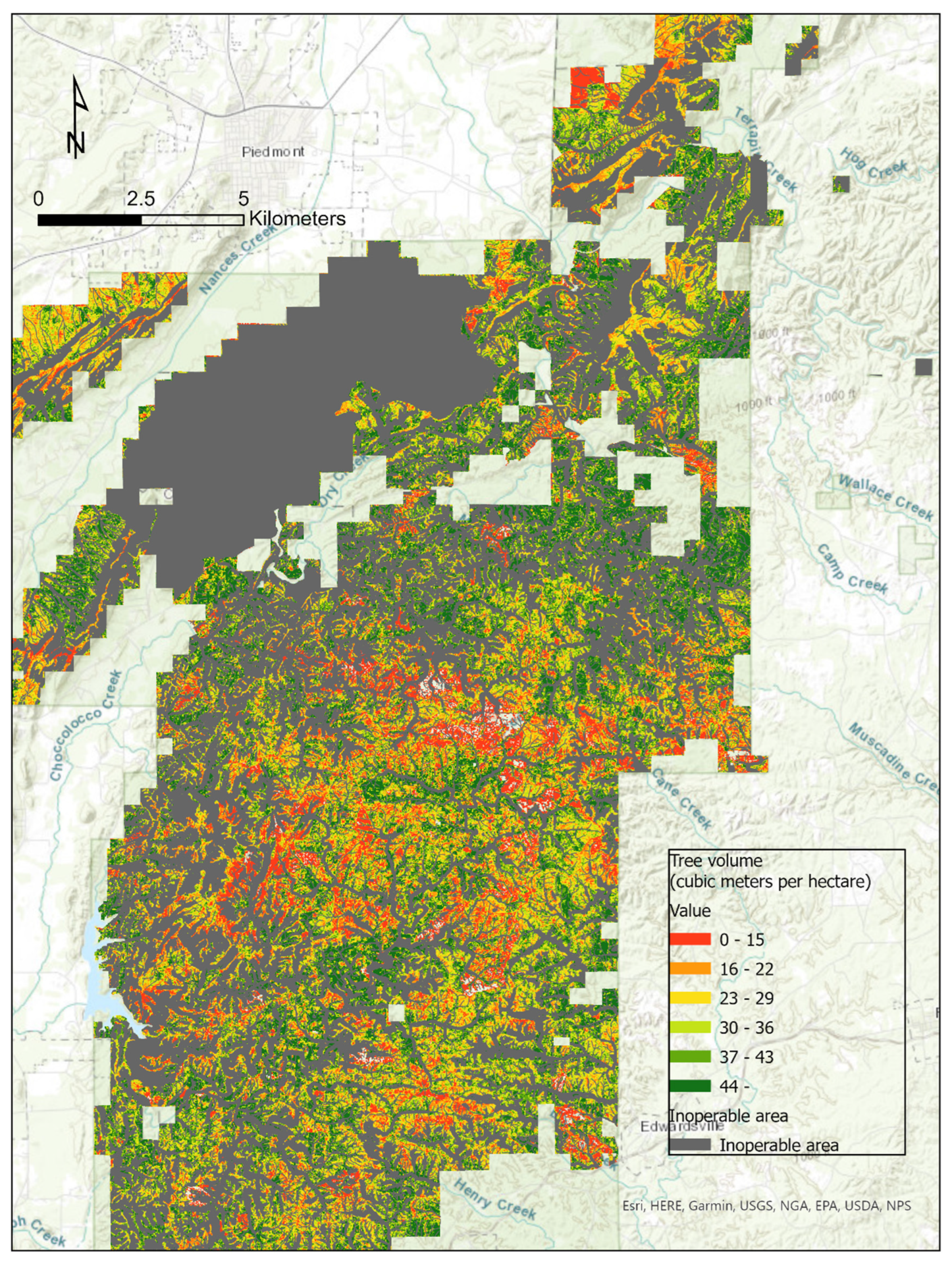
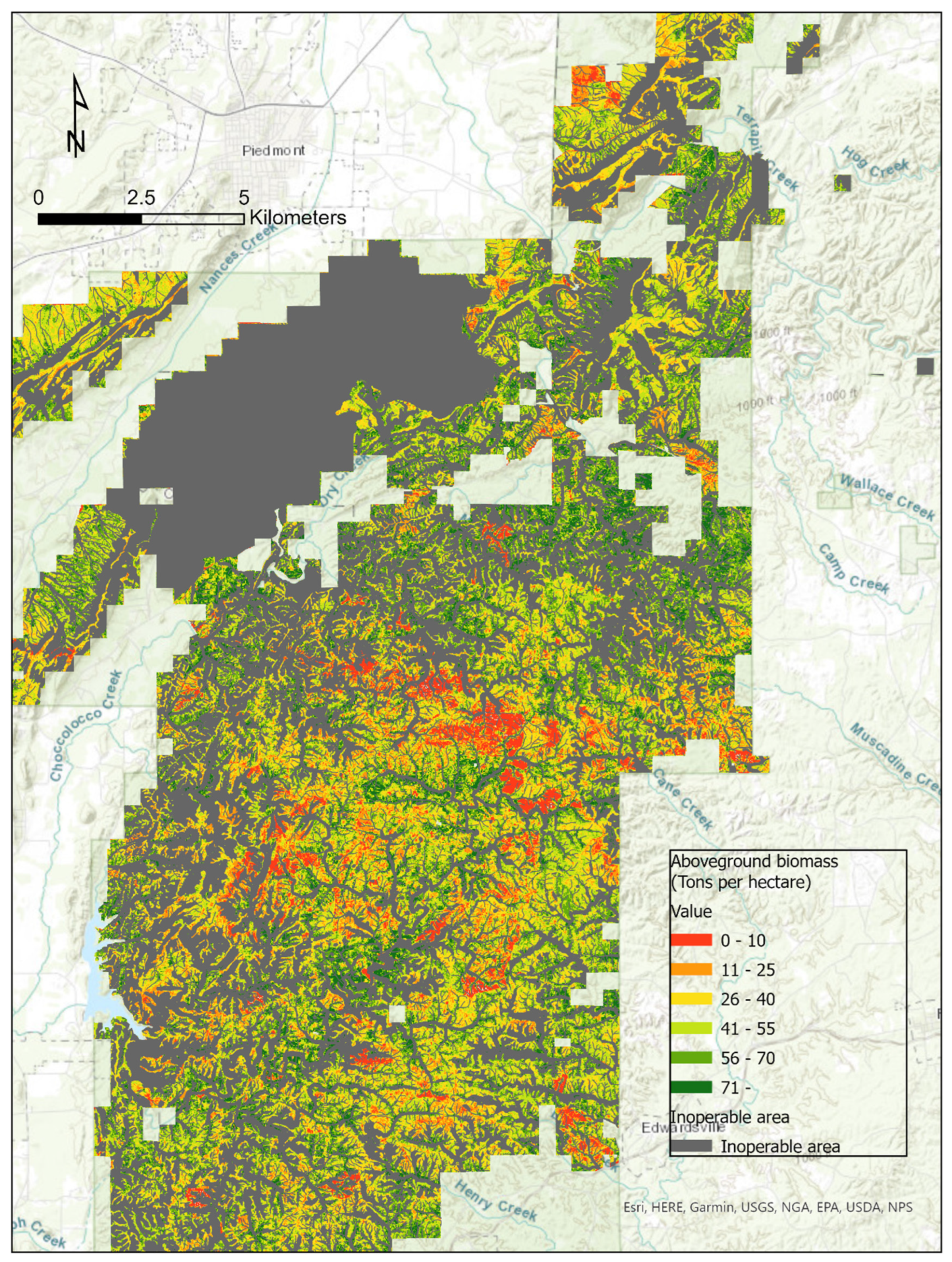
2.1. Study Area
2.2. Data Collection
2.2.1. Field Data Collection
2.2.2. Remote Data Collection
2.3. Data Processing
2.3.1. Field Data Processing
2.3.2. Remote Data Processing
2.4. Modeling
3. Results
3.1. Field-Based Forest Inventory
3.2. Regression Models
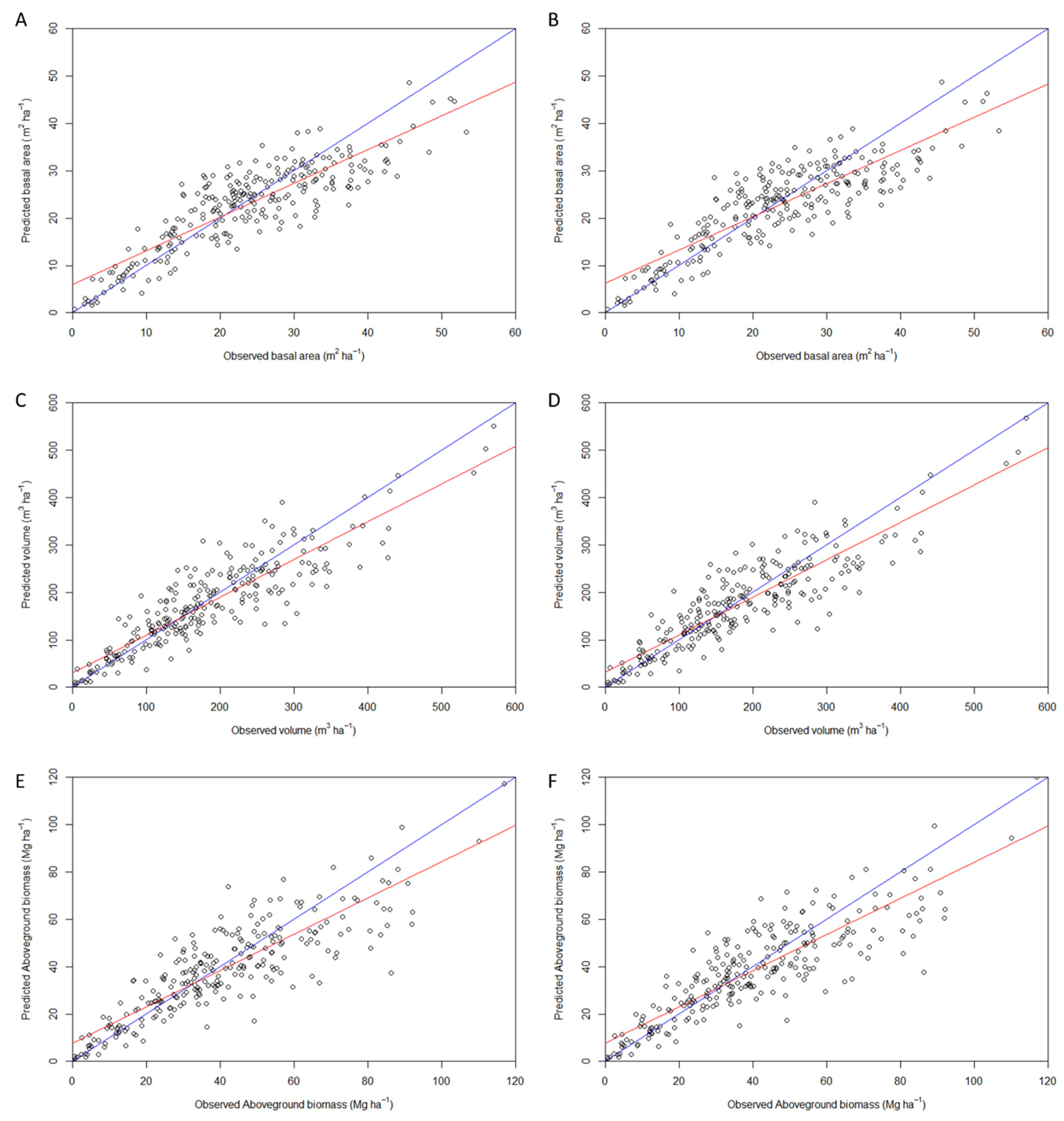
3.2.1. Estimation of Forest Attributes Based on General Models
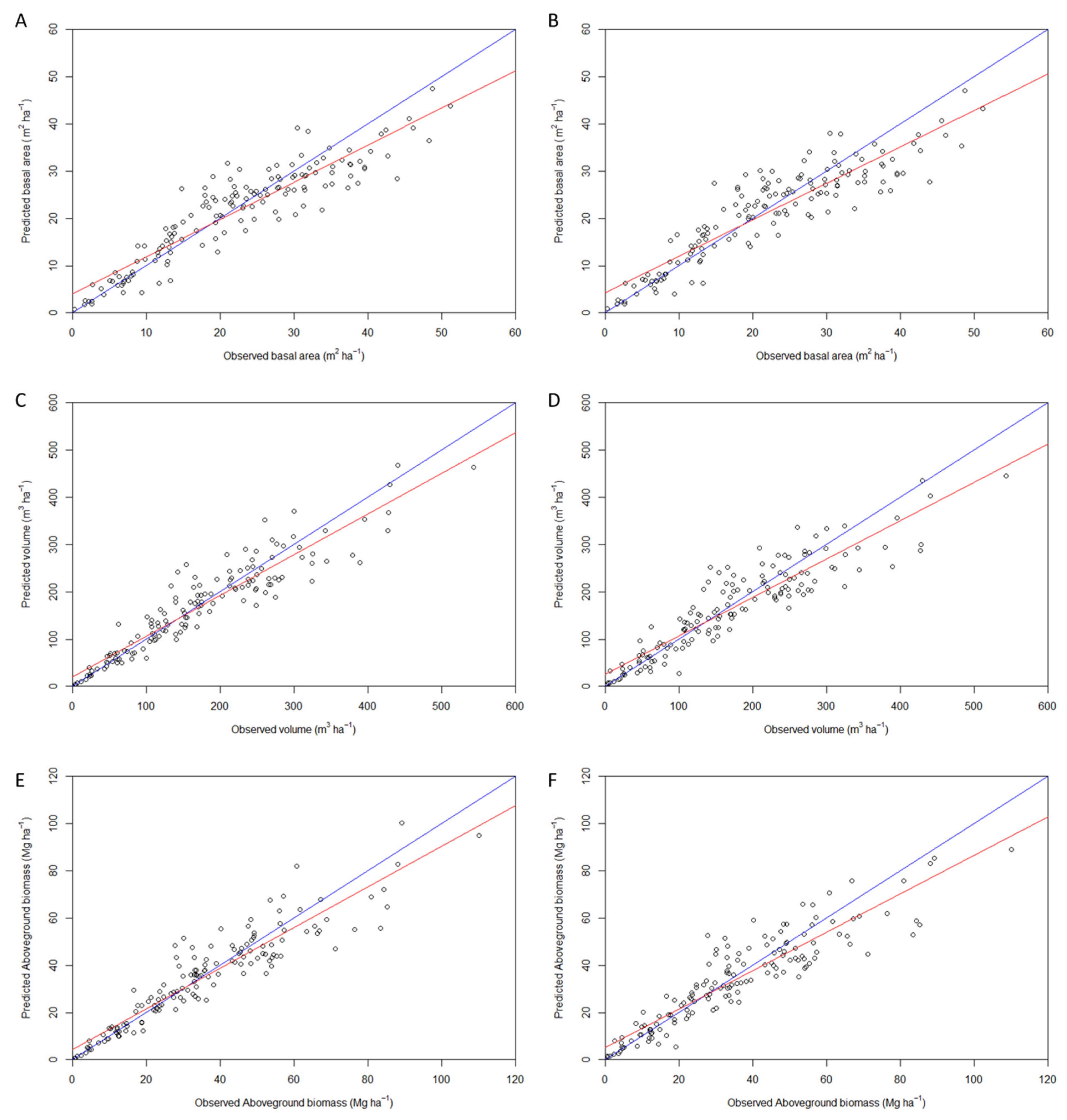
3.2.2. Estimation of Forest Attributes Based on Pine Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bettinger, P.; Boston, K.; Siry, J.P.; Grebner, D.L. Forest management and planning. Academic Press, London. 2017.
- Jarron, L.R.; Coops, N.C.; MacKenzie, W.H.; Tompalski, P.; Dykstra, P. Detection of sub-canopy forest structure using airborne LiDAR. Remote Sens. Environ. 2020, 244, 111770. [CrossRef]
- Bouvier, M.; Durrieu, S.; Fournier, R.A.; Renaud, J.P. Generalizing predictive models of forest inventory attributes using an area-based approach with airborne LiDAR data. Remote Sens. Environ. 2015, 156, 322-334. [CrossRef]
- Chen, Y.; Kershaw, J.A.; Hsu, Y.H.; Yang, T.R. Carbon estimation using sampling to correct LiDAR-assisted enhanced forest inventory estimates. Forestry Chron. 2020, 96 (1), 9-19. [CrossRef]
- Du, L.; Pang, Y.; Wang, Q.; Huang, C.; Bai, Y.; Chen, D.; Lu, W.; Kong, D. A LiDAR biomass index-based approach for tree-and plot-level biomass mapping over forest farms using 3D point clouds. Remote Sens. Environ. 2023, 290, 113543. [CrossRef]
- Tian, L.; Wu, X.; Tao, Y.; Li, M.; Qian, C.; Liao, L.; Fu, W. Review of remote sensing-based methods for forest aboveground biomass estimation: Progress, challenges, and prospects. Forests. 2023, 14 (6), 1086. [CrossRef]
- Vatandaşlar, C.; Seki, M.; Zeybek, M. Assessing the potential of mobile laser scanning for stand-level forest inventories in near-natural forests. Forestry. 2023, 96 (4), 448-464. [CrossRef]
- Nowak, J.T.; Meeker, J.R.; Coyle, D.R.; Steiner, C.A.; Brownie, C. Southern pine beetle infestations in relation to forest stand conditions, previous thinning, and prescribed burning: Evaluation of the southern pine beetle prevention program. J. Forest. 2015, 113(5), 454-462. [CrossRef]
- Ager, A.A.; Vaillant, N.M.; Finney, M.A. Integrating fire behavior models and geospatial analysis for wildland fire risk assessment and fuel management planning. J. Combust. 2011. [CrossRef]
- Adhikari, A.; Peduzzi, A.; Montes, C.R.; Osborne, N.; Mishra, D.R. Assessment of understory vegetation in a plantation forest of the southeastern United States using terrestrial laser scanning. Ecol. Inform. 2023, 77, 102254. [CrossRef]
- Seki, M.; Sakici, O.E. Ecoregion-based height-diameter models for Crimean pine. J. For. Res. 2022, 27, 36-44. [CrossRef]
- García, M.; Riaño, D.; Chuvieco, E.; Danson, F.M. Estimating biomass carbon stocks for a Mediterranean forest in central Spain using LiDAR height and intensity data. Remote Sens. Environ. 2010, 114 (4), 816-830. [CrossRef]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote sensing technologies for enhancing forest inventories: A review. Can. J. Remote Sens. 2016, 42 (5): 619-641. [CrossRef]
- White, J.C.; Tompalski, P.; Vastaranta, M.; Wulder, M.A.; Saarinen, N.; Stepper, C.; Coops, N.C. A model development and application guide for generating an enhanced forest inventory using airborne laser scanning data and an area based approach. Canadian Wood Fibre Centre: Victoria, BC, Canada. Information Report FI-X-018. 2017, 38. ISBN 978-0-660-09738-1.
- Ferraz, A.; Saatchi, S.; Mallet, C.; Meyer, V. Lidar detection of individual tree size in tropical forests. Remote Sens. Environ. 2016, 183, 318-333. [CrossRef]
- Lim, K.; Treitz, P.; Wulder, M.A.; St-Onge, B.; Flood, M.; LiDAR remote sensing of forest structure. Prog. Phys. Geog. 2003, 27 (1), 88-106. https://doi.org/10.1191/0309133303pp360.
- McRoberts, R.E.; Næsset, E.; Sannier, C.; Stehman, S.V.; Tomppo, E.O. Remote sensing support for the gain-loss approach for greenhouse gas inventories. Remote Sens. 2020, 12 (11), 1891. [CrossRef]
- Bater, C.W.; Coops, N.C. Evaluating error associated with Lidar-derived DEM interpolation. Comput. Geosci. 2009, 35, 289-300. [CrossRef]
- García-Gutiérrez, J.; Martínez-Álvarez, F.; Troncoso, A.; Riquelme, J.C. A comparison of machine learning regression techniques for LiDAR-derived estimation of forest variables. Neurocomputing. 2015, 167, 24-31. [CrossRef]
- Potapov, P.; Li, X.; Hernandez-Serna, A.; Tyukavina, A.; Hansen, M.C.; Kommareddy, A.; Pickens, A.; Turubanova, S.; Tang, H.; Silva, C.E.; Armston, J.; Dubayah, R.; Blair, J.B.; Hofton, M. Mapping global forest canopy height through integration of GEDI and Landsat data. Remote Sens. Environ. 2021, 253, 112165. https:// doi. org/10. 3390/ rs120 30505.
- Dubayah, R.; Armston, J.; Healey, S.P.; Bruening, J.M.; Patterson, P.L.; Kellner, J.R.; Duncanson, L.; Saarela, S.; Ståhl, G.; Yang, Z.; Tang, H. GEDI launches a new era of biomass inference from space. Environ. Res. Lett. 2022, 17(9), 095001. [CrossRef]
- Liang, X.; Kankare, V.; Hyyppä, J.; Wang, Y.; Kukko, A.; Haggrén, H.; Yu, X.; Kaartinen, H.; Jaakkola, A.; Guan, F.; Holopainen M.; Vastaranta, M. Terrestrial laser scanning in forest inventories. ISPRS J. Photogramm. 2016, 115, 63-77. [CrossRef]
- Liang, X.; Kukko, A.; Balenović, I.; Saarinen, N.; Junttila, S.; Kankare, V.; Holopainen, M.; Mokroš, M.; Surový, P.; Kaartinen, H.; Jurjević, L.; Honkavaara, E.; Näsi, R.; Liu, J.; Hollaus, M.; Tian, J.; Yu, X.; Pan, J.; Cai, S.; Virtanen, J.P.; Wang, Y.; Hyyppä, J.; Close-range remote sensing of forests: The state of the art, challenges, and opportunities for systems and data acquisitions. IEEE Geosci. Remote Sens. Mag. 2022, 10 (3), 32-71. [CrossRef]
- Arseniou, G.; MacFarlane, D.W.; Calders, K.; Baker, M. Accuracy differences in aboveground woody biomass estimation with terrestrial laser scanning for trees in urban and rural forests and different leaf conditions. Trees. 2023, 37, 761-779. [CrossRef]
- Mathes, T.; Seidel, D.; Häberle, K.H.; Pretzsch, H.; Annighöfer, P.; What are we missing? Occlusion in laser scanning point clouds and its impact on the detection of single-tree morphologies and stand structural variables. Remote Sens. 2023, 15 (2), 450. [CrossRef]
- Chen, S.; Liu, H.; Feng, Z.; Shen, C.; Chen, P. Applicability of personal laser scanning in forestry inventory. PLoS One. 2019, 14 (2), e0211392. [CrossRef]
- Ryding, J.; Williams, E.; Smith, M.; Eichhorn, M. Assessing handheld mobile laser scanners for forest surveys. Remote Sens. 2015, 7 (1), 1095–1111. [CrossRef]
- Gollob, C.; Ritter, T.; Nothdurft, A. Forest inventory with long range and high-speed personal laser scanning (PLS) and simultaneous localization and mapping (SLAM) technology. Remote Sens. 2020, 12 (9), 1509. [CrossRef]
- Mokroš, M.; Mikita T.; Singh A.; Tomastík J.; Chudá J.; Wezyk P.; Kuželka, K.; Surový, P.; Klimánek, M.; Zięba-Kulawik, K.; Bobrowski, R.; Liang, X. Novel low-cost mobile mapping systems for forest inventories as terrestrial laser scanning alternatives. Int. J. Appl. Earth Obs. Geoinf. 2021, 104, 102512. [CrossRef]
- Li, C.; Yu, Z.; Dai, H.; Zhou X.; Zhou M. Effect of sample size on the estimation of forest inventory attributes using airborne LiDAR data in large-scale subtropical areas. Ann. Forest Sci. 2023, 80, 40. [CrossRef]
- Stober, J.; Merry, K.; Bettinger, P. Analysis of fire frequency on the Talladega National Forest, USA, 1998–2018. Int. J. Wildland Fire. 2020, 29 (10), 919–925. [CrossRef]
- Yao, H.; Qin, R.; Chen, X. Unmanned aerial vehicle for remote sensing applications-A review. Remote Sens. 2019, 11 (12), 1443. [CrossRef]
- Penner, M.; Woods, M.; Bilyk, A. Assessing site productivity via remote sensing—age-independent site index estimation in even-aged forests. Forests. 2023, 14 (8), 1541. [CrossRef]
- Ozkan, U.Y.; Demirel, T.; Ozdemir, I.; Saglam, S.; Mert, A. Predicting forest stand attributes using the integration of airborne laser scanning and Worldview-3 data in a mixed forest in Turkey. Adv. Space Res. 2022, 69 (2), 1146-1158. [CrossRef]
- Adhikari, A.; Montes, C.R.; Peduzzi, A. A comparison of modeling methods for predicting forest attributes using Lidar metrics. Remote Sens. 2023, 15 (5), 1284. [CrossRef]
- Chen, X.; Xie, D.; Zhang, Z.; Sharma, R.P.; Chen, Q.; Liu, Q.; Fu, L. Compatible biomass model with measurement error using airborne LiDAR data. Remote Sens. 2023, 15 (14), 3546. [CrossRef]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B (Methodol.). 1996, 73 (1), 267–288.
- Laes, D.; Reutebuch, S.E.; McGaughey, R.J.; Mitchell, B. Guidelines to estimate forest inventory parameters from lidar and field plot data. 2011, Available at: https://fsapps.nwcg.gov/gtac/CourseDownloads/Reimbursables/FY21/Lidar_Material/GTAC_Guidelines%20to%20estimate%20forest%20inventory%20parameters%20from%20lidar%20and%20field%20plot%20data.pdf.
- U.S. Department of Agriculture. National Agriculture Imagery Program (NAIP). U.S. Department of Agriculture, Washington, D.C. 2023. https://naip-usdaonline.hub.arcgis.com/ (Accessed May 2, 2024).
- U.S. Geologic Survey. Topographic data quality levels (QLs). U.S. Geologic Survey, Reston, VA. 2024. https://www.usgs.gov/3d-elevation-program/topographic-data-quality-levels-qls (Accessed April 22, 2024).
- McCullagh, M.J. Terrain and surface modelling systems: Theory and practice. Photogramm. Rec. 1988, 12 (72), 747-779. [CrossRef]
- Hengl, T. Finding the right pixel size. Comput. Geosci. 2006, 32 (9), 1283-1298. [CrossRef]
- Henn, K.A.; Peduzzi, A. Biomass estimation of urban forests using LiDAR and high-resolution aerial imagery in Athens–Clarke County, GA. Forests. 2023, 14 (5), 1064. [CrossRef]
- Leboeuf, A.; Riopel, M.; Munger, D.; Fradette, M.S.; Bégin, J. Modeling merchantable wood volume using Airborne LiDAR metrics and historical forest inventory plots at a provincial scale. Forests. 2022, 13 (7), 985. [CrossRef]
- Brown, S.; Narine, L.L.; Gilbert, J. Using airborne lidar, multispectral imagery, and field inventory data to estimate basal area, volume, and aboveground biomass in heterogeneous mixed species forests: A case study in southern Alabama. Remote Sens. 2022, 14 (11), 2708. [CrossRef]
- Hawbaker, T.J.; Keuler, N.S.; Lesak, A.A.; Gobakken, T.; Contrucci, K.; Radeloff, V.C. Improved estimates of forest vegetation structure and biomass with a LiDAR-optimized sampling design. J. Geophys. Res. Biogeo. 2009, 114, G00E04. [CrossRef]
- Silva, C.A.; Hudak, A.T.; Vierling, L.A.; Loudermilk, E.L.; O’Brien, J.J.; Hiers, J.K.; Jack, S.B.; Gonzalez-Benecke, C.; Lee, H.; Falkowski, M.J.; Khosravipour, A. Imputation of individual longleaf pine (Pinus palustris Mill.) tree attributes from field and LiDAR data. Can. J. Remote Sens. 2016, 42 (5), 554-573. ttps://doi.org/10.1080/07038992.2016.1196582.
- Sumnall, M.J.; Hill, R.A.; Hinsley, S.A. Towards forest condition assessment: evaluating small-footprint full-waveform airborne laser scanning data for deriving forest structural and compositional metrics. Remote Sens. 2022, 14 (20), 5081. [CrossRef]
- Yan, W.Y.; Van Ewijk, K.; Treitz, P.; Shaker, A. Effects of radiometric correction on cover type and spatial resolution for modeling plot level forest attributes using multispectral airborne LiDAR data. ISPRS J. Photogramm. 2020, 169, 152-165. [CrossRef]
| Vegetation Index | Equation | Calculated statistics and its abbreviation |
| Greenness | Minimum of greenness (GMIN) | |
| Maximum of greenness (GMAX) | ||
| Range of greenness (GRANGE) | ||
| Mean of greenness (GMEAN) | ||
| Standard deviation of greenness (GSTD) | ||
| Sum of greenness (GSUM) | ||
| Median of greenness (GMEDIAN) | ||
| 90 percentage of greenness (GPCT90) | ||
| Normalized Difference Vegetation Index, NDVI | Minimum of NDVI (NDVIMIN) | |
| Maximum of NDVI (NDVIMAX) | ||
| Range of NDVI (NDVIRANGE) | ||
| Mean of NDVI (NDVIMEAN) | ||
| Standard deviation of NDVI (NDVISTD) | ||
| Sum of NDVI (NDVISUM) | ||
| Median of NDVI (NDVIMEDIAN) | ||
| 90 percentage of NDVI (NDVIPCT90) | ||
| Enhanced Vegetation Index, EVI | Minimum of EVI (EVIMIN) | |
| Maximum of EVI (EVIMAX) | ||
| Range of EVI (EVIRANGE) | ||
| Mean of EVI (EVIMEAN) | ||
| Standard deviation of EVI (EVISTD) | ||
| Sum of EVI (EVISUM) | ||
| Median of EVI (EVIMEDIAN) | ||
| 90 percentage of EVI (EVIPCT90) |
| Metrics | Descriptions | Metrics | Descriptions |
| zmean | Mean height | zpcum x (from 1st to 9th) |
Cumulative percentage of return in the ith layer |
| zsd |
Standard deviation of height distribution | isd | standard deviation of intensity |
| zskew |
Skewness of height distribution | iskew | skewness of intensity distribution |
| zkurt | Kurtosis of height distribution | ikurt | kurtosis of intensity distribution |
| zentropy | Entropy of height distribution | ipground | percentage of intensity returned by points classified as "ground" |
| pzabovezmean | Percentage of returns above z mean | ipcumzq x (10th, 30th, 50th, 70th, and 90th) |
Percentage of intensity returned below the xth percentile of height |
| Pzabove2 |
Percentage of returns above 2 m | P xth (1, 2, 3, 4, and 5) |
Percentage xth returns |
| zq x (From 5th to 95th) |
xth percentile (quantile) of height distribution | pground | Percentage of returns classified as "ground" |
| Diameter at breast height (cm) | Basal area (m2 ha-1) | Volume (m3 ha-1) | Aboveground biomass (Mg ha-1) | |
| All plots (n =254) | ||||
| Average | 22.39 | 23.43 | 180.85 | 40.13 |
| Standard deviation | 7.39 | 10.95 | 105.48 | 23.07 |
| Minimum | 8.65 | 0.33 | 0.77 | 0.13 |
| Maximum | 54.36 | 53.29 | 569.93 | 119.03 |
| Pine plots (n =149) | ||||
| Average | 22.42 | 22.28 | 168.97 | 34.62 |
| Standard deviation | 8.40 | 16.92 | 108.26 | 21.94 |
| Minimum | 8.66 | 0.33 | 0.77 | 0.13 |
| Maximum | 54.36 | 51.19 | 543.27 | 110.02 |
| Forest variables | Data sources | eequation |
| General models | ||
| Basal area | LiDAR+ NAIP | -1.828 + 0.017 * pzabove2 + 0.015 * zq25 + 0.017 * zq95 - 7.83 * e-4 * zpcum5 - 0.002 * zpcum6 + 2 * e-5 * isd + 0.24 * iskew - 0.091 * ikurt + 0.024 * ipcumzq90 + 0.043 * p2th + 0.116 * GMIN + 0.52 * NDVIMIN + 0.428 * NDVIMEDIAN + 3.58 * e-5 * EVIMAX - 0.012 * EVIPCT90 |
| LiDAR | -1.197 + 0.018 * pzabove2 + 0.017 * zq25 + 1.13 * e-4 * zq30 + 0.015 * zq95 - 3.79 * e-4 * zpcum5 - 0.003 * zpcum6 + 1.83 * e-5 * isd + 0.196 * iskew - 0.105 * ikurt + 0.017 * ipcumzq90 + 0.046 * p2th | |
| Volume | LiDAR+ NAIP | -0.148 + 0.018 * pzabove2 + 0.004 * zq25 + 0.056 * zq95 - 7.91 * e-4 * zpcum5 - 0.00566 * zpcum6 + 2.51 * e-5 * isd + 0.237 * iskew - 0.128 * ikurt - 0.018 * ipcumzq10 - 0.002 * ipcumzq30 + 0.029 * ipcumzq90 - 0.002 * p1th + 0.029 * p2th + 0.091 * GMIN - 0.165 * GRANGE + 0.522 * NDVIMIN + 0.229 * NDVIMEAN + 1.45 * e-5 * NDVISUM + 0.158 * NDVIMEDIAN + 1.96 * e-4 * EVIMAX - 0.010 * EVIPCT90 |
| LiDAR | 0.380 + 0.020 * pzabove2 + 0.007 * zq25 + 0.053 * zq95 - 0.006 * zpcum6 + 3.088 * e-05 * isd + 0.209 * iskew - 0.121 * ikurt - 0.020 * ipcumzq10 - 0.004 * ipcumzq30 + 0.020 * ipcumzq90 - 4.642 * e-5 * p1th + 0.037 * p2th | |
| Aboveground biomass | LiDAR+ NAIP | 0.033 + 0.021 * pzabove2 + 0.062 * zq95 - 0.004 * zpcum6 + 3.93 * e-5 * isd + 0.132 * iskew - 0.184 * ikurt - 0.011 * ipcumzq10 + 0.083 * ipcumzq90 - 0.006 * p1th + 0.031 * p2th - 1.51 * e-4 * pground + 0.385 * GMIN + 0.505 * NDVIMIN + 0.218 * NDVIMEAN + 9.36 * e-6 * NDVISUM - 0.005 * EVIPCT90 |
| LiDAR | 0.516 + 0.022 * pzabove2 + 0.238 * zq5 + 0.002 * zq25 + 0.059 * zq95 - 0.005 * zpcum6 + 3.83 * e-5 * isd + 0.103 * iskew - 0.198 * ikurt - 0.012 * ipcumzq10 + 0.078 * ipcumzq90 - 0.006 * p1th + 0.034 * p2th | |
| Pine models | ||
| Basal area | LiDAR+ NAIP | 0.657 + 0.009 * pzabove2 + 3.600 * zq5 + 0.021 * zq25 + 0.001 * zq40 + 0.007 * zq95 - 0.009 * zpcum5 + 0.173 * iskew - 0.067 * ikurt + 0.057 * p2th + 0.426 * NDVIMIN + 0.985 * NDVIMEDIAN - 8.6 * e-4 * EVIPCT90 |
| LiDAR | 5.195 + 0.011 * pzabove2 + 2.873 * zq5 + 0.021 * zq25 + 1.681 * e-4 * zq40 + 0.003 * zq95 - 0.008 * zpcum5 - 1.96 * e-4 * zpcum6 + 0.168 * iskew - 0.061 * ikurt - 0.004 * ipcumzq30 - 0.0475 * ipcumzq90 + 0.057 * p2th | |
| Volume | LiDAR+ NAIP | 1.593 - 0.002 * zkurt + 0.021 * pzabove2 + 8.41 * zq5 - 1.91 * zq15 + 0.019 * zq25 + 0.006 * zq40 - 0.008 * zq65 + 0.016 * zq75 - 0.071 * zq80 + 0.096 * zq95 + 0.010 * zpcum1 - 0.010 * zpcum5 - 0.005 * zpcum6 + 1.29 * e-5 * zpcum8 + 0.002 * zpcum9 + 3.04 * e-5 * isd + 0.241 * iskew - 0.16 * ikurt + 0.016 * ipcumzq10 - 0.008 * ipcumzq30 + 0.042 * p2th - 0.039 * p5th + 0.486 * GMIN - 0.132 * GRANGE + 0.475 * NDVIMIN + 1.28 * NDVIMEDIAN + 2.7 * e-4 * EVIMAX - 0.004 * EVISTD - 0.019 * EVIMEDIAN - 0.015 * EVIPCT90 |
| LiDAR | 3.592 + 0.008 * pzabove2 + 3.401 * zq5 + 0.004 * zq25 + 0.043 * zq95 - 0.013 * zpcum5 + 0.222 * iskew - 0.094 * ikurt - 2.769 *e-4 * ipcumzq10 - 0.033 * ipcumzq30 + 0.047 * p2th + 0.013 * p3th | |
| Aboveground biomass | LiDAR+ NAIP | 6.466 - 0.005 * zkurt + 0.41 * zentropy + 0.025 * pzabove2 + 8.66 * zq5 - 0.037 * zq10 - 2.18 * zq15 + 0.017 * zq25 - 7.48 *e-4 * zq30 + 0.004 * zq40 - 0.001 * zq65 + 0.010 * zq75 - 0.079 * zq80 + 0.11 * zq95 + 0.013 * zpcum1 - 0.009 * zpcum5 - 0.005 * zpcum6 + 0.003 * zpcum9 + 4.75 * e-05 * isd + 0.078 * iskew - 0.175 * ikurt + 0.020 * ipcumzq10 - 0.008 * ipcumzq90 + 0.042 * p2th - 0.016 * p5th + 0.673 * GMIN - 0.094 * GRANGE + 0.171 * GSTD - 0.362 * GMEDIAN + 0.364 * NDVIMIN - 0.327 * NDVISTD + 1.51 * NDVIMEDIAN + 1.52 *e-4 * EVIMAX - 0.002 * EVISTD - 0.033 * EVIPCT90 |
| LiDAR | 10.699 + 0.013 * pzabove2 + 2.062 * zq5 + 0.052 * zq95 - 0.011 * zpcum5 + 0.010 * iskew - 0.129 * ikurt - 0.010 * ipcumzq30 - 0.024 * ipcumzq90 - 0.009 * p1th + 0.045 * p2th | |
| Quality metrics | Basal area (m2 ha-1) | Total volume (m3 ha-1) | Total aboveground biomass (Mg ha-1) | |||
| LiDAR + NAIP | LiDAR | LiDAR + NAIP | LiDAR | LiDAR + NAIP | LiDAR | |
| R2adj. | 0.72 | 0.71 | 0.77 | 0.77 | 0.73 | 0.72 |
| # of variables | 15 | 11 | 21 | 12 | 16 | 12 |
| RMSE | 5.58 | 5.73 | 48.44 | 49.34 | 11.68 | 11.84 |
| R2 | 0.74 | 0.72 | 0.79 | 0.78 | 0.74 | 0.74 |
| Bias | - 0.78 | -0.80 | -6.27 | -6.54 | -1.62 | -1.64 |
| Bias (%) | -3.33 | -3.40 | -3.45 | -3.61 | -4.03 | -4.08 |
| AIC | 0.08 | -76.05 | -118.05 | -137.76 | -137.43 | -145.17 |
| BIC | -68.19 | -38.28 | -47.95 | -96.66 | -83.13 | -104.02 |
| CP | -17.21 | 0.08 | 0.11 | 0.12 | 0.12 | 0.13 |
| Quality metrics | Basal area (m2 ha-1) | Total volume (m3 ha-1) | Total aboveground biomass (Mg ha-1) | |||
| LiDAR + NAIP | LiDAR | LiDAR + NAIP | LiDAR | LiDAR + NAIP | LiDAR | |
| R2adj. | 0.69 | 0.71 | 0.67 | 0.73 | 0.64 | 0.65 |
| R2 | 0.72 | 0.69 | 0.72 | 0.75 | 0.69 | 0.68 |
| RMSE | 5.90 | 5.91 | 55.87 | 53.10 | 13.05 | 13.09 |
| Bias | -0.76 | -0.75 | -4.06 | -7.56 | -1.12 | -1.21 |
| Bias (%) | -3.20 | -3.20 | -2.26 | -4.12 | -2.63 | -2.85 |
| LiDAR metrics | NAIP metrics | |
| General & pine model | pzabove2, zq95, iskew, ikurt, p2th | NDVIMIN, EVIPCT90 |
| General model | zpcum6, isd, ipcumzq90 | GMIN |
| Pine model | zq5, zpcum5 | NDVIMEDIAN |
| Quality metrics | Basal area (m2 ha-1) | Total volume (m3 ha-1) | Total aboveground biomass (Mg ha-1) | |||
| LiDAR + NAIP | LiDAR | LiDAR + NAIP | LiDAR | LiDAR + NAIP | LiDAR | |
| R2adj. | 0.81 | 0.80 | 0.84 | 0.82 | 0.83 | 0.82 |
| # of variables | 12 | 12 | 30 | 11 | 34 | 10 |
| RMSE | 4.80 | 5.10 | 37.86 | 43.45 | 7.89 | 8.93 |
| R2 | 0.83 | 0.81 | 0.87 | 0.84 | 0.87 | 0.83 |
| Bias | -0.72 | -0.75 | -3.65 | -6.71 | -0.68 | -1.36 |
| Bias (%) | -3.23 | -3.38 | -2.12 | -3.92 | -1.93 | -3.89 |
| AIC | -5.84 | -54.77 | -56.60 | -108.83 | -49.23 | -113.69 |
| BIC | -22.09 | -21.01 | 17.08 | -77.80 | 31.20 | -85.33 |
| CP | 0.07 | 0.08 | 0.06 | 0.11 | 0.06 | 0.10 |
| Quality metrics | Basal area (m2 ha-1) | Total volume (m3 ha-1) | Total aboveground biomass (Mg ha-1) | |||
| LiDAR + NAIP | LiDAR | LiDAR + NAIP | LiDAR | LiDAR + NAIP | LiDAR | |
| R2adj. | 0.79 | 0.75 | 0.78 | 0.79 | 0.80 | 0.78 |
| R2 | 0.82 | 0.78 | 0.82 | 0.81 | 0.84 | 0.81 |
| RMSE | 5.21 | 5.71 | 47.44 | 48.94 | 9.42 | 10.24 |
| Bias | -0.72 | -0.99 | -4.41 | -8.43 | -0.97 | -0.92 |
| Bias (%) | -3.17 | -4.33 | -2.48 | -4.92 | -2.21 | -2.71 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





