Submitted:
10 June 2024
Posted:
11 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Biomonitoring of Carbon in Tropical Forests
1.4. Machine Learning (ML) to Estimate Forest Properties
2. Materials and Methods
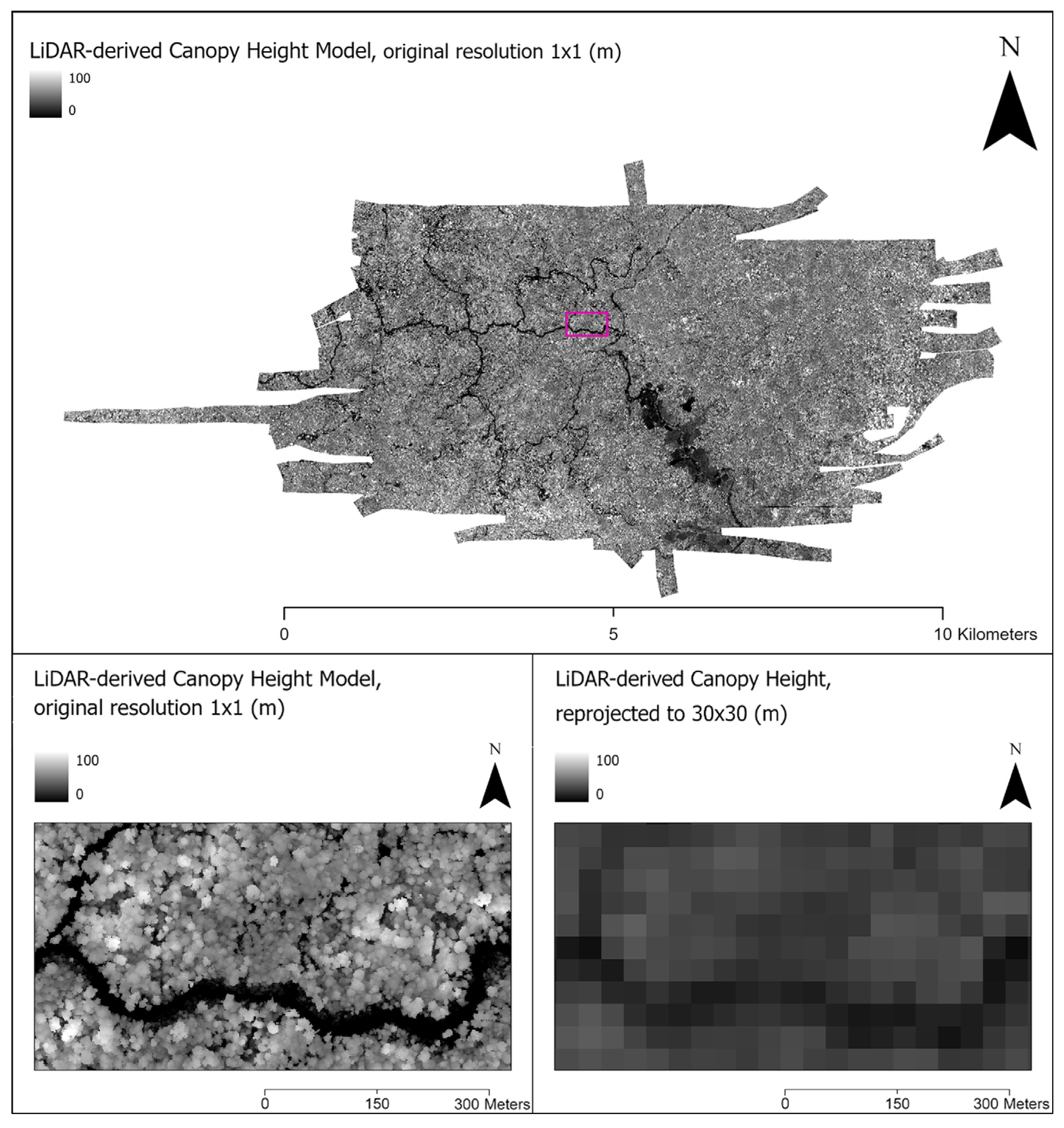
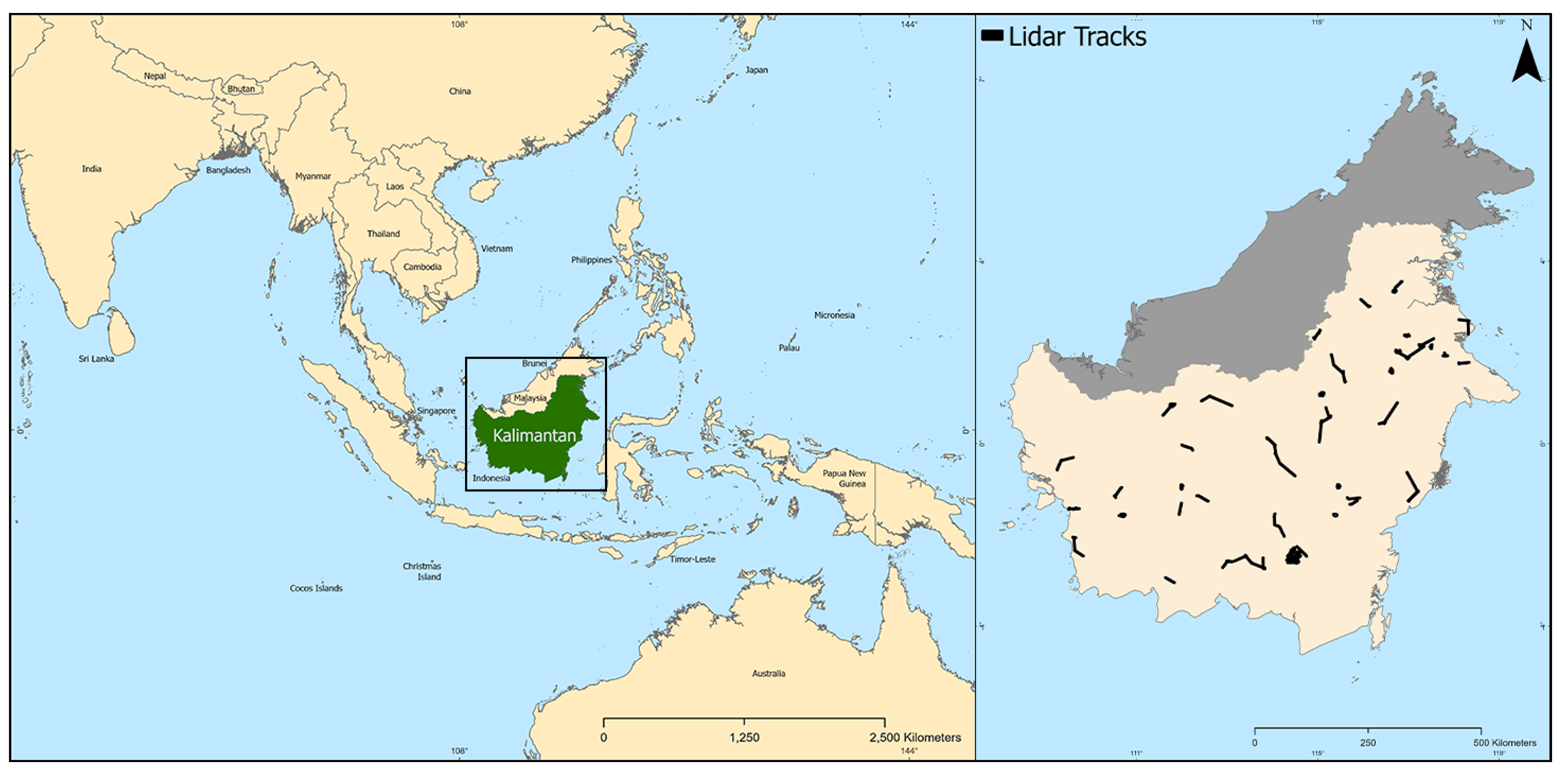
2.1. Study Area and Data Pipeline
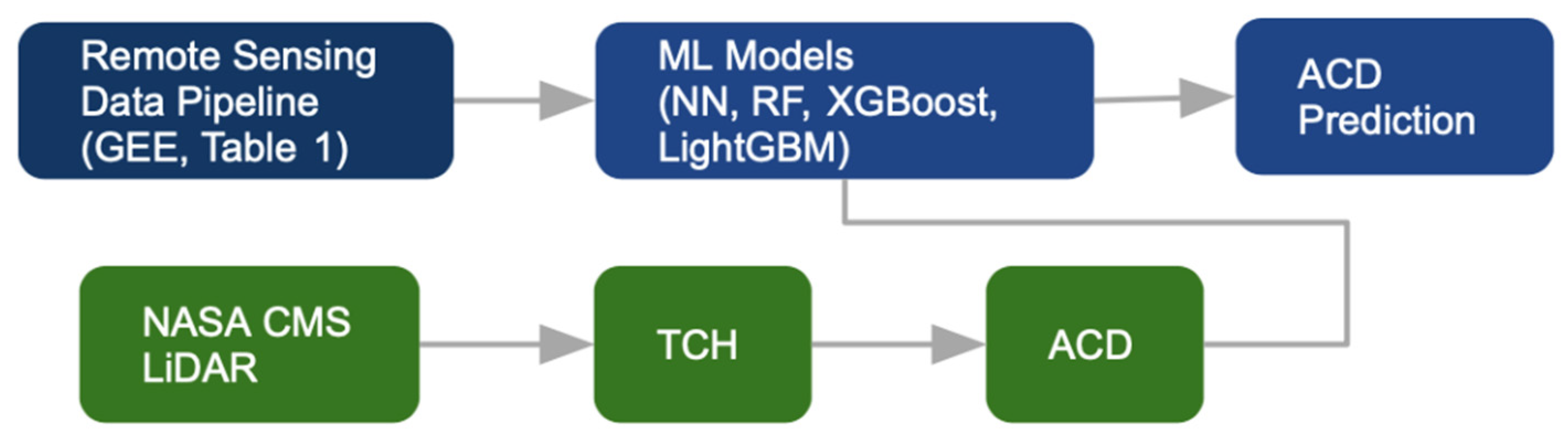
2.2. ACD Estimation
2.3. ML Framework
2.3.1. Tree-Based Models
2.3.2. Neural Network Models
2.3.3. AutoML (Automated Machine Learning)
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baccini, A.; Goetz, S.J.; Walker, W.S.; Laporte, N.T.; Sun, M.; Sulla-Menashe, D.; Hackler, J.; Beck, P.S.A.; Dubayah, R.; Friedl, M.A.; Samanta, S.; Houghton, R.A. Estimated Carbon Dioxide Emissions from Tropical Deforestation Improved by Carbon-Density Maps. Nature Clim Change 2012, 2, 182–185. [Google Scholar] [CrossRef]
- Mitchell, L.E.; Lin, J.C.; Bowling, D.R.; Pataki, D.E.; Strong, C.; Schauer, A.J.; Bares, R.; Bush, S.E.; Stephens, B.B.; Mendoza, D.; Mallia, D.; Holland, L.; Gurney, K.R.; Ehleringer, J.R. Long-Term Urban Carbon Dioxide Observations Reveal Spatial and Temporal Dynamics Related to Urban Characteristics and Growth. Proceedings of the National Academy of Sciences 2018, 115, 2912–2917. [Google Scholar] [CrossRef] [PubMed]
- Chave, J.; Andalo, C.; Brown, S.; Cairns, M.A.; Chambers, J.Q.; Eamus, D.; Fölster, H.; Fromard, F.; Higuchi, N.; Kira, T.; Lescure, J.-P.; Nelson, B.W.; Ogawa, H.; Puig, H.; Riéra, B.; Yamakura, T. Tree Allometry and Improved Estimation of Carbon Stocks and Balance in Tropical Forests. Oecologia 2005, 145, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Brearley, F.Q.; Adinugroho, W.C.; Cámara-Leret, R.; Krisnawati, H.; Ledo, A.; Qie, L.; Smith, T.E.L.; Aini, F.; Garnier, F.; Lestari, N.S.; Mansur, M.; Murdjoko, A.; Oktarita, S.; Soraya, E.; Tata, H.L.; Tiryana, T.; Trethowan, L.A.; Wheeler, C.E.; Abdullah, M.; Aswandi; Buckley, B.J.W.; Cantarello, E.; Dunggio, I.; Gunawan, H.; Heatubun, C.D.; Arini, D.I.D.; Istomo; Komar, T.E.; Kuswandi, R.; Mutaqien, Z.; Pangala, S.R.; Ramadhanil; Prayoto; Puspanti, A.; Qirom, M.A.; Rozak, A.H.; Sadili, A.; Samsoedin, I.; Sulistyawati, E.; Sundari, S.; Sutomo; Tampubolon, A.P.; Webb, C.O. Opportunities and Challenges for an Indonesian Forest Monitoring Network. Annals of Forest Science 2019, 76, 54. [Google Scholar] [CrossRef]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.V.; Goetz, S.J.; Loveland, T.R.; Kommareddy, A.; Egorov, A.; Chini, L.; Justice, C.O.; Townshend, J.R.G. High-Resolution Global Maps of 21st-Century Forest Cover Change. Science 2013, 342, 850–853. [Google Scholar] [CrossRef] [PubMed]
- Mascaro, J.; Asner, G.P.; Knapp, D.E.; Kennedy-Bowdoin, T.; Martin, R.E.; Anderson, C.; Higgins, M.; Chadwick, K.D. A Tale of Two “Forests”: Random Forest Machine Learning Aids Tropical Forest Carbon Mapping. PLOS ONE 2014, 9, e85993. [Google Scholar] [CrossRef] [PubMed]
- Mascaro, J.; Asner, G.P.; Muller-Landau, H.C.; van Breugel, M.; Hall, J.; Dahlin, K. Controls over Aboveground Forest Carbon Density on Barro Colorado Island, Panama. Biogeosciences 2011, 8, 1615–1629. [Google Scholar] [CrossRef]
- Asner, G.P.; Mascaro, J. Mapping Tropical Forest Carbon: Calibrating Plot Estimates to a Simple LiDAR Metric. Remote Sensing of Environment 2014, 140, 614–624. [Google Scholar] [CrossRef]
- Jucker, T.; Bongalov, B.; Burslem, D.F.R.P.; Nilus, R.; Dalponte, M.; Lewis, S.L.; Phillips, O.L.; Qie, L.; Coomes, D.A. Topography Shapes the Structure, Composition and Function of Tropical Forest Landscapes. Ecology Letters 2018, 21, 989–1000. [Google Scholar] [CrossRef]
- Asner, G.P.; Mascaro, J.; Muller-Landau, H.C.; Vieilledent, G.; Vaudry, R.; Rasamoelina, M.; Hall, J.S.; van Breugel, M. A Universal Airborne LiDAR Approach for Tropical Forest Carbon Mapping. Oecologia 2012, 168, 1147–1160. [Google Scholar] [CrossRef]
- Bouvet, A.; Mermoz, S.; Le Toan, T.; Villard, L.; Mathieu, R.; Naidoo, L.; Asner, G.P. An Above-Ground Biomass Map of African Savannahs and Woodlands at 25m Resolution Derived from ALOS PALSAR. Remote Sensing of Environment 2018, 206, 156–173. [Google Scholar] [CrossRef]
- Vaglio Laurin, G.; Chen, Q.; Lindsell, J.; Coomes, D.; Del Frate, F.; Guerriero, L.; Pirotti, F.; Valentini, R. Above Ground Biomass Estimation in an African Tropical Forest with Lidar and Hyperspectral Data. ISPRS Journal of Photogrammetry and Remote Sensing 2014, 89, 49–58. [Google Scholar] [CrossRef]
- Hughes, R.F.; Asner, G.P.; Baldwin, J.A.; Mascaro, J.; Bufil, L.K.K.; Knapp, D.E. Estimating Aboveground Carbon Density across Forest Landscapes of Hawaii: Combining FIA Plot-Derived Estimates and Airborne LiDAR. Forest Ecology and Management 2018, 424, 323–337. [Google Scholar] [CrossRef]
- Asner, G.P.; Clark, J.K.; Mascaro, J.; Galindo García, G.A.; Chadwick, K.D.; Navarrete Encinales, D.A.; Paez-Acosta, G.; Cabrera Montenegro, E.; Kennedy-Bowdoin, T.; Duque, Á.; Balaji, A.; von Hildebrand, P.; Maatoug, L.; Phillips Bernal, J.F.; Yepes Quintero, A.P.; Knapp, D.E.; García Dávila, M.C.; Jacobson, J.; Ordóñez, M.F. High-Resolution Mapping of Forest Carbon Stocks in the Colombian Amazon. Biogeosciences 2012, 9, 2683–2696. [Google Scholar] [CrossRef]
- Saatchi, S.S.; Harris, N.L.; Brown, S.; Lefsky, M.; Mitchard, E.T.A.; Salas, W.; Zutta, B.R.; Buermann, W.; Lewis, S.L.; Hagen, S.; Petrova, S.; White, L.; Silman, M.; Morel, A. Benchmark Map of Forest Carbon Stocks in Tropical Regions across Three Continents. Proceedings of the National Academy of Sciences 2011, 108, 9899–9904. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Saatchi, S.S.; Xu, L.; Yu, Y.; Choi, S.; Phillips, N.; Kennedy, R.; Keller, M.; Knyazikhin, Y.; Myneni, R.B. Post-Drought Decline of the Amazon Carbon Sink. Nat Commun 2018, 9, 3172. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Rudel, T.K.; Aide, T.M.; Defries, R.; Emerson, R. A Contemporary Assessment of Change in Humid Tropical Forests. Conserv Biol 2009, 23, 1386–1395. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Brodrick, P.G.; Philipson, C.; Vaughn, N.R.; Martin, R.E.; Knapp, D.E.; Heckler, J.; Evans, L.J.; Jucker, T.; Goossens, B.; Stark, D.J.; Reynolds, G.; Ong, R.; Renneboog, N.; Kugan, F.; Coomes, D.A. Mapped Aboveground Carbon Stocks to Advance Forest Conservation and Recovery in Malaysian Borneo. Biological Conservation 2018, 217, 289–310. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Martin, R.E.; Tupayachi, R.; Anderson, C.B.; Mascaro, J.; Sinca, F.; Chadwick, K.D.; Higgins, M.; Farfan, W.; Llactayo, W.; Silman, M.R. Targeted Carbon Conservation at National Scales with High-Resolution Monitoring. Proceedings of the National Academy of Sciences 2014, 111, E5016–E5022. [Google Scholar] [CrossRef]
- Baccini, A.; Asner, G.P. Improving Pantropical Forest Carbon Maps with Airborne LiDAR Sampling. Carbon Management 2013, 4, 591–600. [Google Scholar] [CrossRef]
- Ali, A.M.; Darvishzadeh, R.; Skidmore, A.K.; van Duren, I. Effects of Canopy Structural Variables on Retrieval of Leaf Dry Matter Content and Specific Leaf Area From Remotely Sensed Data. IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing 2016, 9, 898–909. [Google Scholar] [CrossRef]
- Baccini, A.; Walker, W.; Carvalho, L.; Farina, M.; Sulla-Menashe, D.; Houghton, R.A. Tropical Forests Are a Net Carbon Source Based on Aboveground Measurements of Gain and Loss. Science 2017, 358, 230–234. [Google Scholar] [CrossRef] [PubMed]
- De’ath, G.; Fabricius, K.E. Classification and Regression Trees: A Powerful yet Simple Technique for Ecological Data Analysis. Ecology 2000, 81, 3178–3192. [Google Scholar] [CrossRef]
- Gleason, C.J.; Im, J. Forest Biomass Estimation from Airborne LiDAR Data Using Machine Learning Approaches. Remote Sensing of Environment 2012, 125, 80–91. [Google Scholar] [CrossRef]
- Prasad, A.M.; Iverson, L.R.; Liaw, A. Newer Classification and Regression Tree Techniques: Bagging and Random Forests for Ecological Prediction. Ecosystems 2006, 9, 181–199. [Google Scholar] [CrossRef]
- Evans, J.S.; Murphy, M.A.; Holden, Z.A.; Cushman, S.A. Modeling Species Distribution and Change Using Random Forest [Chapter 8]. In Predictive Species and Habitat Modeling in Landscape Ecology; Drew, A.C., Wiersma, Y., Huettmann, F., Eds.; Springer: New York, NY, USA, 2011; pp. 139–159. [Google Scholar] [CrossRef]
- Couteron, P.; Barbier, N.; Gautier, D. Textural Ordination Based on Fourier Spectral Decomposition: A Method to Analyze and Compare Landscape Patterns. Landscape Ecology 2006, 21, 555–567. [Google Scholar] [CrossRef]
- Ploton, P.; Pélissier, R.; Proisy, C.; Flavenot, T.; Barbier, N.; Rai, S.N.; Couteron, P. Assessing Aboveground Tropical Forest Biomass Using Google Earth Canopy Images. Ecological Applications 2012, 22, 993–1003. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Transactions on Systems, Man, and Cybernetics 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Meng, S.; Pang, Y.; Zhang, Z.; Jia, W.; Li, Z. Mapping Aboveground Biomass Using Texture Indices from Aerial Photos in a Temperate Forest of Northeastern China. Remote Sensing 2016, 8, 230. [Google Scholar] [CrossRef]
- Wood, E.M.; Pidgeon, A.M.; Radeloff, V.C.; Keuler, N.S. Image Texture as a Remotely Sensed Measure of Vegetation Structure. Remote Sensing of Environment 2012, 121, 516–526. [Google Scholar] [CrossRef]
- Wen, D.; Huang, X.; Bovolo, F.; Li, J.; Ke, X.; Zhang, A.; Benediktsson, J.A. Change Detection From Very-High-Spatial-Resolution Optical Remote Sensing Images: Methods, Applications, and Future Directions. IEEE Geoscience and Remote Sensing Magazine 2021, 9, 68–101. [Google Scholar] [CrossRef]
- Pargal, S.; Fararoda, R.; Rajashekar, G.; Balachandran, N.; Réjou-Méchain, M.; Barbier, N.; Jha, C.S.; Pélissier, R.; Dadhwal, V.K.; Couteron, P. Inverting Aboveground Biomass–Canopy Texture Relationships in a Landscape of Forest Mosaic in the Western Ghats of India Using Very High Resolution Cartosat Imagery. Remote Sensing 2017, 9, 228. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Q.; Liu, J.; Du, X.; Dong, T.; McNairn, H.; Champagne, C.; Liu, M.; Shang, J. Object-Based Crop Classification Using Multi-Temporal SPOT-5 Imagery and Textural Features with a Random Forest Classifier. Geocarto International 2018, 33, 1017–1035. [Google Scholar] [CrossRef]
- Kayitakire, F.; Hamel, C.; Defourny, P. Retrieving Forest Structure Variables Based on Image Texture Analysis and IKONOS-2 Imagery. Remote Sensing of Environment 2006, 102, 390–401. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; KDD’16; Association for Computing Machinery: New York, NY, USA, 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. LightGBM: A Highly Efficient Gradient Boosting Decision Tree. In Advances in Neural Information Processing Systems; Curran Associates, Inc., 2017; Vol. 30. [Google Scholar]
- Gaveau, D.L.A.; Sloan, S.; Molidena, E.; Yaen, H.; Sheil, D.; Abram, N.K.; Ancrenaz, M.; Nasi, R.; Quinones, M.; Wielaard, N.; Meijaard, E. Four Decades of Forest Persistence, Clearance and Logging on Borneo. PLOS ONE 2014, 9, e101654. [Google Scholar] [CrossRef] [PubMed]
- Melendy, L.; Hagen, S.; Sullivan, F.B.; Pearson, T.; Walker, S.M.; Ellis, P.; KUSTIYO; Sambodo, K.A.; Roswintiarti, O.; Hanson, M.; Klassen, A.W.; Palace, M.W.; Braswell, B.H.; Delgado, G.M.; Saatchi, S.S.; Ferraz, A. CMS: LiDAR-Derived Canopy Height, Elevation for Sites in Kalimantan, Indonesia, 2014. ORNL DAAC 2017. [Google Scholar] [CrossRef]
- Vafaei, S.; Soosani, J.; Adeli, K.; Fadaei, H.; Naghavi, H.; Pham, T.D.; Tien Bui, D. Improving Accuracy Estimation of Forest Aboveground Biomass Based on Incorporation of ALOS-2 PALSAR-2 and Sentinel-2A Imagery and Machine Learning: A Case Study of the Hyrcanian Forest Area (Iran). Remote Sensing 2018, 10, 172. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-Km Spatial Resolution Climate Surfaces for Global Land Areas. International Journal of Climatology 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-Scale Geospatial Analysis for Everyone. Remote Sensing of Environment 2017, 202, 18–27. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Machine Learning 2001, 45, 5–32. [Google Scholar] [CrossRef]
- He, X.; Zhao, K.; Chu, X. AutoML: A Survey of the State-of-the-Art. Knowledge-Based Systems 2021, 212, 106622. [Google Scholar] [CrossRef]
- Karmaker (“Santu”), S.K.; Hassan, Md. M.; Smith, M.J.; Xu, L.; Zhai, C.; Veeramachaneni, K. AutoML to Date and Beyond: Challenges and Opportunities. ACM Comput. Surv. 2021, 54, 175:1–175:36. [Google Scholar] [CrossRef]
- Bhagwat, R.U.; Uma Shankar, B. A Novel Multilabel Classification of Remote Sensing Images Using XGBoost. In Proceedings of the 2019 IEEE 5th International Conference for Convergence in Technology (I2CT); 2019; pp. 1–5. [Google Scholar] [CrossRef]
- Samat, A.; Li, E.; Wang, W.; Liu, S.; Lin, C.; Abuduwaili, J. Meta-XGBoost for Hyperspectral Image Classification Using Extended MSER-Guided Morphological Profiles. Remote Sensing 2020, 12, 1973. [Google Scholar] [CrossRef]
- Łoś, H.; Mendes, G.S.; Cordeiro, D.; Grosso, N.; Costa, H.; Benevides, P.; Caetano, M. Evaluation of Xgboost and Lgbm Performance in Tree Species Classification with Sentinel-2 Data. In Proceedings of the 2021 IEEE International Geoscience and Remote Sensing Symposium IGARSS; 2021; pp. 5803–5806. [Google Scholar] [CrossRef]
- Nielsen, M. Neural Networks and Deep Learning; Determination Press, 2015; Available online: http://neuralnetworksanddeeplearning.com/.
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press, 2016. [Google Scholar]
- Nair, V.; Hinton, G. Rectified Linear Units Improve Restricted Boltzmann Machines. In Proceedings of the 27th International Conference on Machine Learning; 2010; Vol. 27, p. 814. [Google Scholar]
- Jin, H.; Song, Q.; Hu, X. Auto-Keras: An Efficient Neural Architecture Search System. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining; ACM: Anchorage, AK, USA, 2019; pp. 1946–1956. [Google Scholar] [CrossRef]
- Olson, R.S.; Moore, J.H. TPOT: A Tree-Based Pipeline Optimization Tool for Automating Machine Learning. In Automated Machine Learning; Hutter, F., Kotthoff, L., Vanschoren, J., Eds.; The Springer Series on Challenges in Machine Learning; Springer International Publishing: Cham, 2019; pp. 151–160. [Google Scholar] [CrossRef]
- Pham, T.D.; Yoshino, K.; Le, N.N.; Bui, D.T. Estimating Aboveground Biomass of a Mangrove Plantation on the Northern Coast of Vietnam Using Machine Learning Techniques with an Integration of ALOS-2 PALSAR-2 and Sentinel-2A Data. International Journal of Remote Sensing 2018, 39, 7761–7788. [Google Scholar] [CrossRef]
- Ng, D.T.K.; Leung, J.K.L.; Chu, S.K.W.; Qiao, M.S. Conceptualizing AI Literacy: An Exploratory Review. Computers and Education: Artificial Intelligence 2021, 2, 100041. [Google Scholar] [CrossRef]
- Jarrahi, M.H.; Lutz, C.; Newlands, G. Artificial Intelligence, Human Intelligence and Hybrid Intelligence Based on Mutual Augmentation. Big Data & Society 2022, 9, 20539517221142824. [Google Scholar] [CrossRef]
- Whang, S.E.; Roh, Y.; Song, H.; Lee, J.-G. Data Collection and Quality Challenges in Deep Learning: A Data-Centric AI Perspective. The VLDB Journal 2023, 32, 791–813. [Google Scholar] [CrossRef]
- Liu, T.; Yao, L.; Qin, J.; Lu, J.; Lu, N.; Zhou, C. A Deep Neural Network for the Estimation of Tree Density Based on High-Spatial Resolution Image. IEEE Transactions on Geoscience and Remote Sensing 2022, 60, 1–11. [Google Scholar] [CrossRef]
- Csillik, O.; Kumar, P.; Mascaro, J.; O’Shea, T.; Asner, G.P. Monitoring Tropical Forest Carbon Stocks and Emissions Using Planet Satellite Data. Sci Rep 2019, 9, 17831. [Google Scholar] [CrossRef] [PubMed]
- McFEETERS, S.K. The Use of the Normalized Difference Water Index (NDWI) in the Delineation of Open Water Features. International Journal of Remote Sensing 1996, 17, 1425–1432. [Google Scholar] [CrossRef]
- Kriegler, F.; Malila, W.; Nalepka, R.; Richardson, W. Preprocessing transformations and their effect on multispectral recognition. In Proceedings of the 6th International Symposium on Remote Sensing of Environment, Ann Arbor, MI; 1969. [Google Scholar]
- Hunt, E.R.; Rock, B.N. Detection of Changes in Leaf Water Content Using Near- and Middle-Infrared Reflectances. Remote Sensing of Environment 1989, 30, 43–54. [Google Scholar] [CrossRef]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the Radiometric and Biophysical Performance of the MODIS Vegetation Indices. Remote Sensing of Environment 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very High Resolution Interpolated Climate Surfaces for Global Land Areas. International Journal of Climatology 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Farr, T.G.; Rosen, P.A.; Caro, E.; Crippen, R.; Duren, R.; Hensley, S.; Kobrick, M.; Paller, M.; Rodriguez, E.; Roth, L.; Seal, D.; Shaffer, S.; Shimada, J.; Umland, J.; Werner, M.; Oskin, M.; Burbank, D.; Alsdorf, D. The Shuttle Radar Topography Mission. Reviews of Geophysics 2007, 45. [Google Scholar] [CrossRef]
- Copernicus Global Land Service: Land Cover 100m: Collection 3: Epoch 2015: Globe. [CrossRef]
- Bisong, E. Google Colaboratory. In Building Machine Learning and Deep Learning Models on Google Cloud Platform: A Comprehensive Guide for Beginners; Bisong, E., Ed.; Apress: Berkeley, CA, USA, 2019; pp. 59–64. [Google Scholar] [CrossRef]
- Ferraz, A.; Saatchi, S.S.; Xu, L.; Hagen, S.; Chave, J.; Yu, Y.; Meyer, V.; Garcia, M.; Silva, C.; Roswintiarti, O.; Samboko, A.; Sist, P.; Walker, S.M.; Pearson, T.; Wijaya, A.; Sullivan, F.B.; Rutishauser, E.; Hoekman, D.; Ganguly, S. Aboveground Biomass, Landcover, and Degradation, Kalimantan Forests, Indonesia, 2014. ORNL DAAC 2019. [Google Scholar] [CrossRef]
- Clay Content in % (Kg / Kg) at 6 Standard Depths (0, 10, 30, 60, 100 and 200 Cm) at 250 m Resolution. [CrossRef]
- Sand Content in % (Kg / Kg) at 6 Standard Depths (0, 10, 30, 60, 100 and 200 Cm) at 250 m Resolution. [CrossRef]
- Soil Water Content (Volumetric %) for 33kPa and 1500kPa Suctions Predicted at 6 Standard Depths (0, 10, 30, 60, 100 and 200 Cm) at 250 m Resolution. [CrossRef]
- Soil Organic Carbon Content in x 5 g / Kg at 6 Standard Depths (0, 10, 30, 60, 100 and 200 Cm) at 250 m Resolution. [CrossRef]
- Soil pH in H2O at 6 Standard Depths (0, 10, 30, 60, 100 and 200 Cm) at 250 m Resolution. [CrossRef]
- Soil Bulk Density (Fine Earth) 10 x Kg / m-Cubic at 6 Standard Depths (0, 10, 30, 60, 100 and 200 Cm) at 250 m Resolution. [CrossRef]
- Yamazaki, D.; Ikeshima, D.; Sosa, J.; Bates, P.D.; Allen, G.H.; Pavelsky, T.M. MERIT Hydro: A High-Resolution Global Hydrography Map Based on Latest Topography Dataset. Water Resources Research 2019, 55, 5053–5073. [Google Scholar] [CrossRef]
- Amatulli, G.; McInerney, D.; Sethi, T.; Strobl, P.; Domisch, S. Geomorpho90m, Empirical Evaluation and Accuracy Assessment of Global High-Resolution Geomorphometric Layers. Sci Data 2020, 7, 162. [Google Scholar] [CrossRef]


| Data | Type | Temporal Range | Spatial Resolution | Source |
|---|---|---|---|---|
| LANDSAT-8 | OLI/TIRS sensors | Aug. 2014 – Jan. 2015 | 30 m | USGS |
| Vegetation Indices - NDVI, NDWI, NDII, EVI, calculated for Landsat-8 | Various measures associated with vegetation properties | Same as input | Same as input | Same as input |
| Sentinel-1 | Synthetic Aperture Radar (SAR) instrument | Aug. 2014 – May 2015 | 10 m | ESA |
| Gray-Level Co-Occurrence Matrix (GLCM), derived from Landsat-8, Sentinel-1, and Landsat-8 vegetation indices | Textural image features derived from pixel spatial relationships | Same as input | Same as input | Same as input |
| Canopy Height Model (CHM) | Plane-mounted LiDAR | 2014 | 1 m | NASA |
| NASA SRTM V3 | Digital Elevation Model (elevation, slope, aspect) | 2000 | 30 m | NASA |
| Bioclim | Climate | 1970-2000 | 927.67 m | WorldClim |
| CopCover | Land Cover - Copernicus | 2015 | 100 m | ESA |
| Land Use/Land Cover | Land Cover classification of Kalimantan - includes types of forests, other natural habitats, and developed lands | 2011 | 30 m | Indonesian Ministry of Forestry |
| OpenLandMap soil variables - soil bulk density (kg/m3), clay content (%), sand content (%), soil organic carbon (g/kg), soil pH in H20, soil water content (volumetric %), all at 0 cm depth. | Modeled soil properties from various global datasets of soil samples | 1950-2018 | 250 m | OpenLandMap |
| Upstream drainage area (km2) and height above nearest drainage (m) | Hydrological flow dataset | 1987-2017 | 90 m | MERIT Hydro |
| Geomorphic layers - compound topographic index, terrain roughness index, vector ruggedness measure, roughness, topographic position index, and stream power index | Topographical relief characterization derived from MERIT Digital Elevation Model | 1987-2017 | 90 m | Geomorpho90m Geomorphometric Layers |
| Model | Band Name/Feature | Importance Rank |
|---|---|---|
| Random Forest | Sentinel-1 VH Cluster Shade | 1 |
| Random Forest | Landsat-8 Thermal Infrared Information Measure of Correlation 2 | 2 |
| Random Forest | Landsat-8 Thermal Infrared Inertia | 3 |
| XGBoost | Sentinel-1 VH Cluster Shade | 1 |
| XGBoost | Landsat-8 Red Difference Entropy | 2 |
| XGBoost | Landsat-8 Red Information Measure of Correlation 2 | 3 |
| LightGBM | Sentinel-1 VH Cluster Shade | 1 |
| LightGBM | Landsat-8 Shortwave Infrared 1 Sum Entropy | 2 |
| LightGBM | Landsat-8 Blue Difference Entropy | 3 |
| Layer (type) | Output shape | Param # |
|---|---|---|
| Input Layer | (None, 336) | 0 |
| Multi-category Encoding | (None, 336) | 0 |
| Normalization | (None, 336) | 705 |
| Dense | (None, 32) | 11,296 |
| ReLu | (None, 32) | 0 |
| Dense | (None, 32) | 1,056 |
| ReLu | (None, 32) | 0 |
| Dense (regression head) | (None, 1) | 33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





