Submitted:
12 June 2024
Posted:
13 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
... the biosphere does not contain a predictable class of objects or events but constitutes a particular occurrence, compatible with first principles but not deducible from these principles, and therefore essentially unpredictable.
There are always some constraints imposed by stability and thermodynamics. But as complexity increases, additional constraints appear (...). Consequently, there cannot be any general law of evolution.
organisms are under constant scrutiny of natural selection and are also subject to the constraints of the physical and chemical factors that severely limit the action of all inhabitants of the biosphere. Put simply, convergence shows that in the real world, not all things are possible.
...monsters are a good system to study the internal properties of generative rules. They represent forms which lack adaptative function while preserving structural order. There is an internal logic to the genesis and transformation of morphologies and in that logic we may learn about the constraints on the normal.
The chemical and physical properties of the different complex molecules are different, and in biology, the functional properties of these tens of thousands of different molecules in cells are also different. The universe is not ergodic because it will not make all the possible different complex molecules on timescales very much longer than the lifetime of the universe. It is true that most complex things will never “get to exist”.
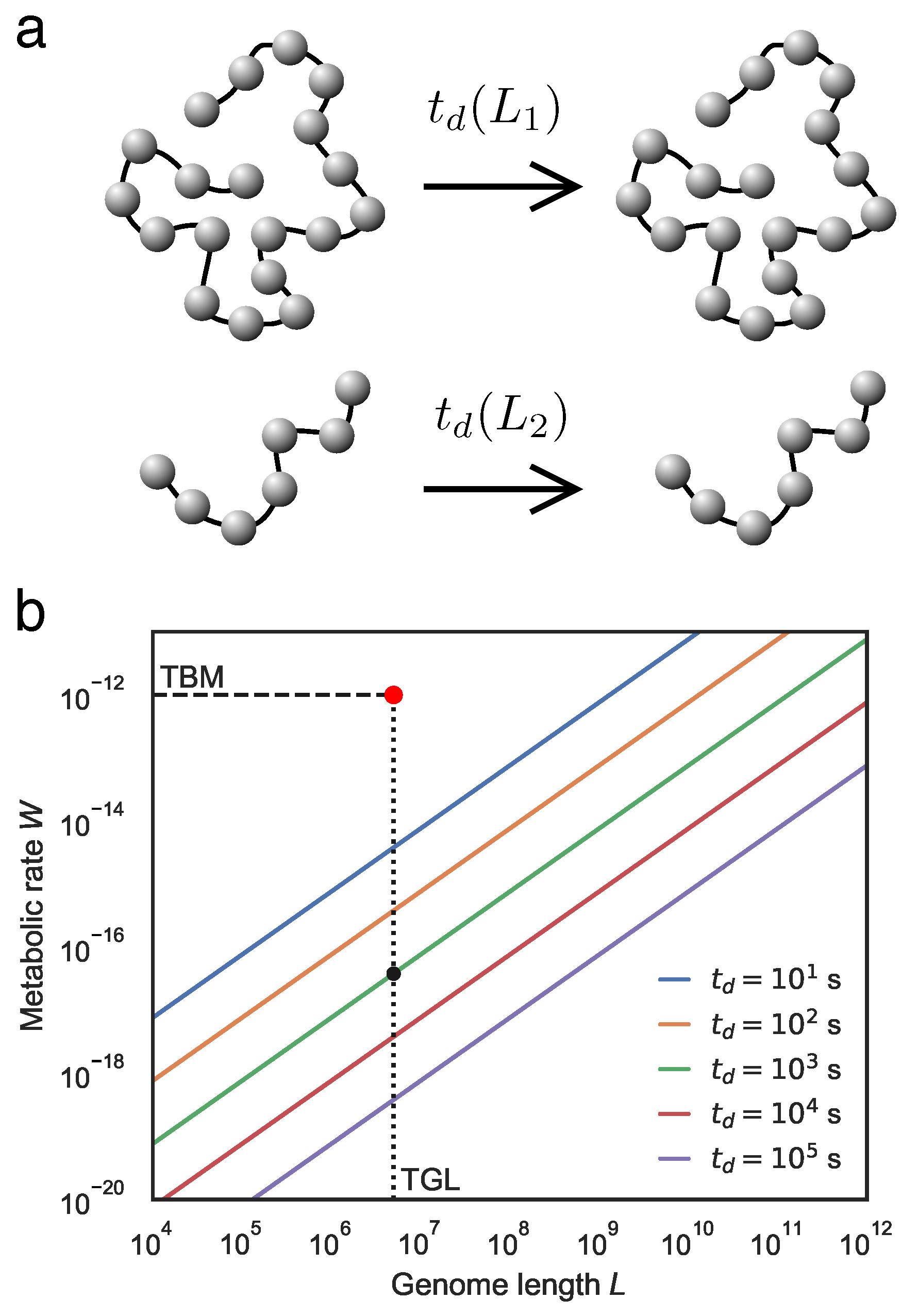
2. Living Systems as Thermodynamic Engines
3. Linear Information Carriers
There is an enormously larger class of natural structures that have nearly equal probabilities of formation because they are one-dimensional and have nearly equivalent energies. They are linear copolymers, like polynucleotides and polypeptides. Life and evolution depend on this class of copolymer that forms an unbounded sequence space, undetermined by laws. (...) This unbounded sequence space is the first component of the freedom from laws necessary for evolution.
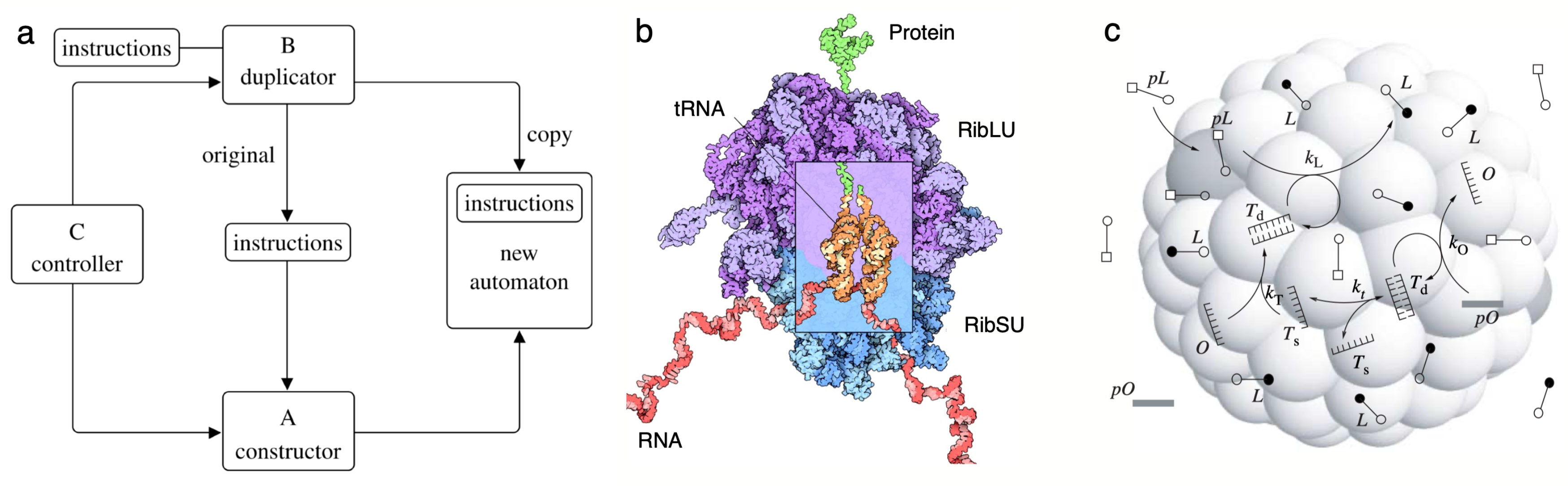
4. Cells as Minimal Units of Life
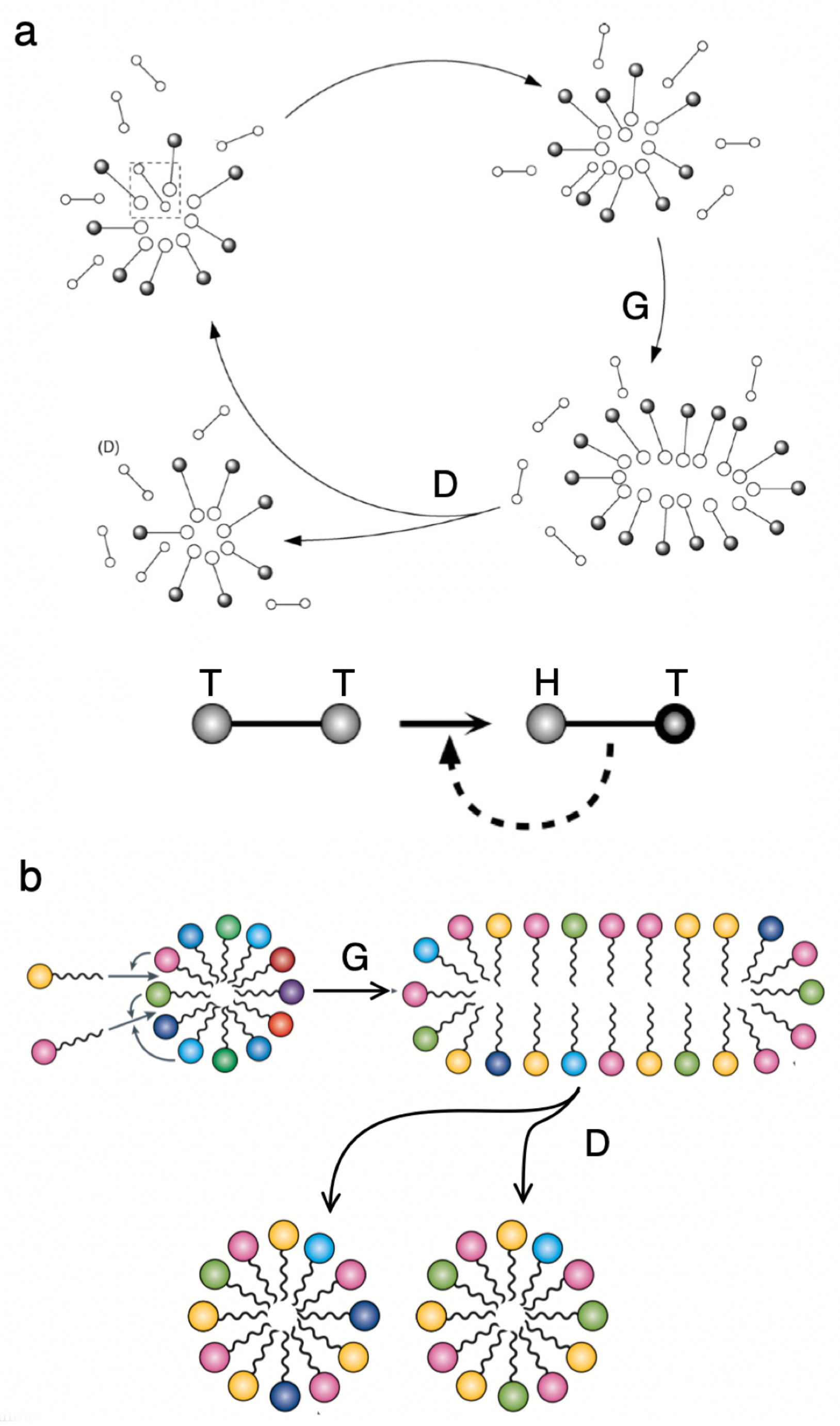
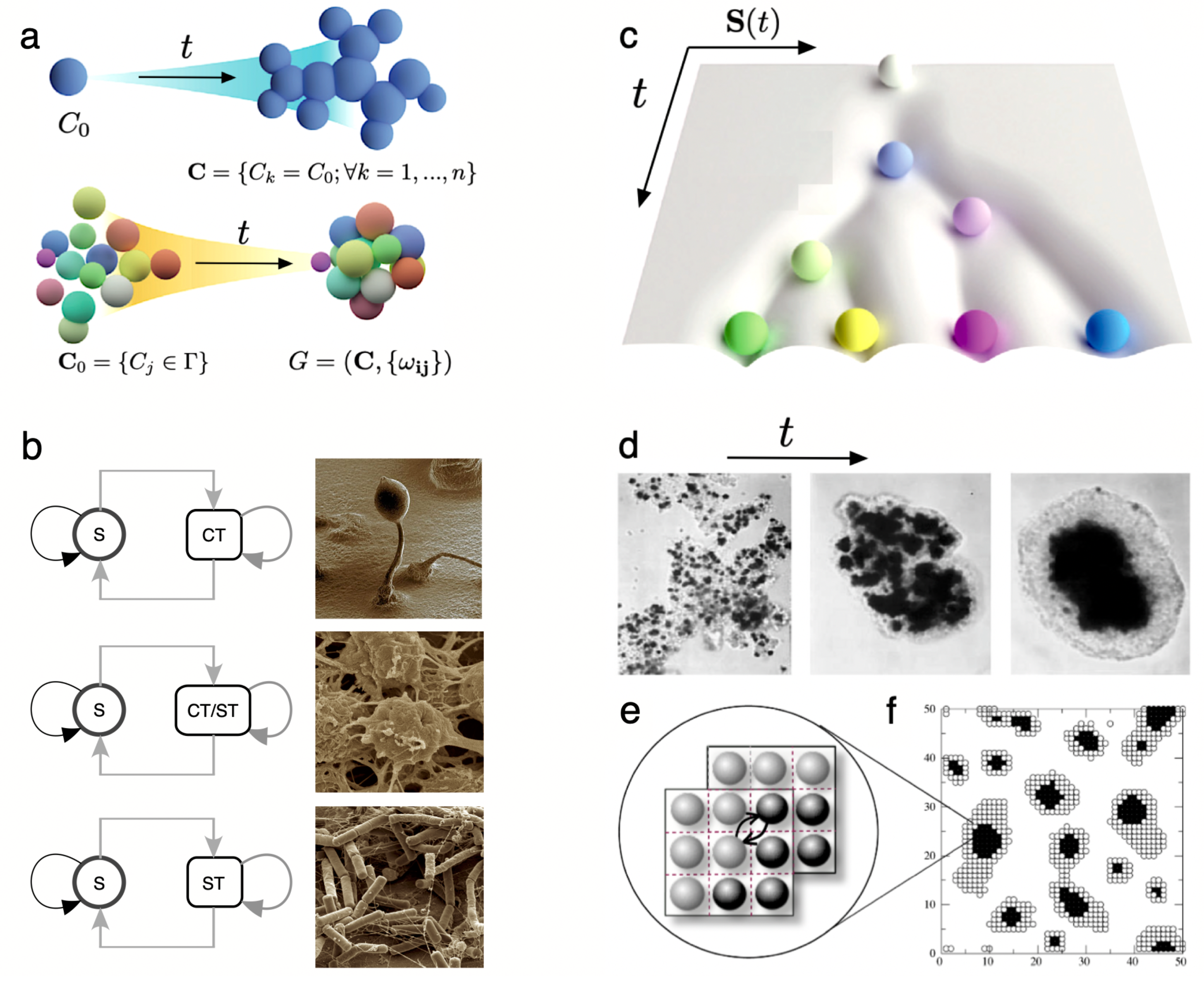
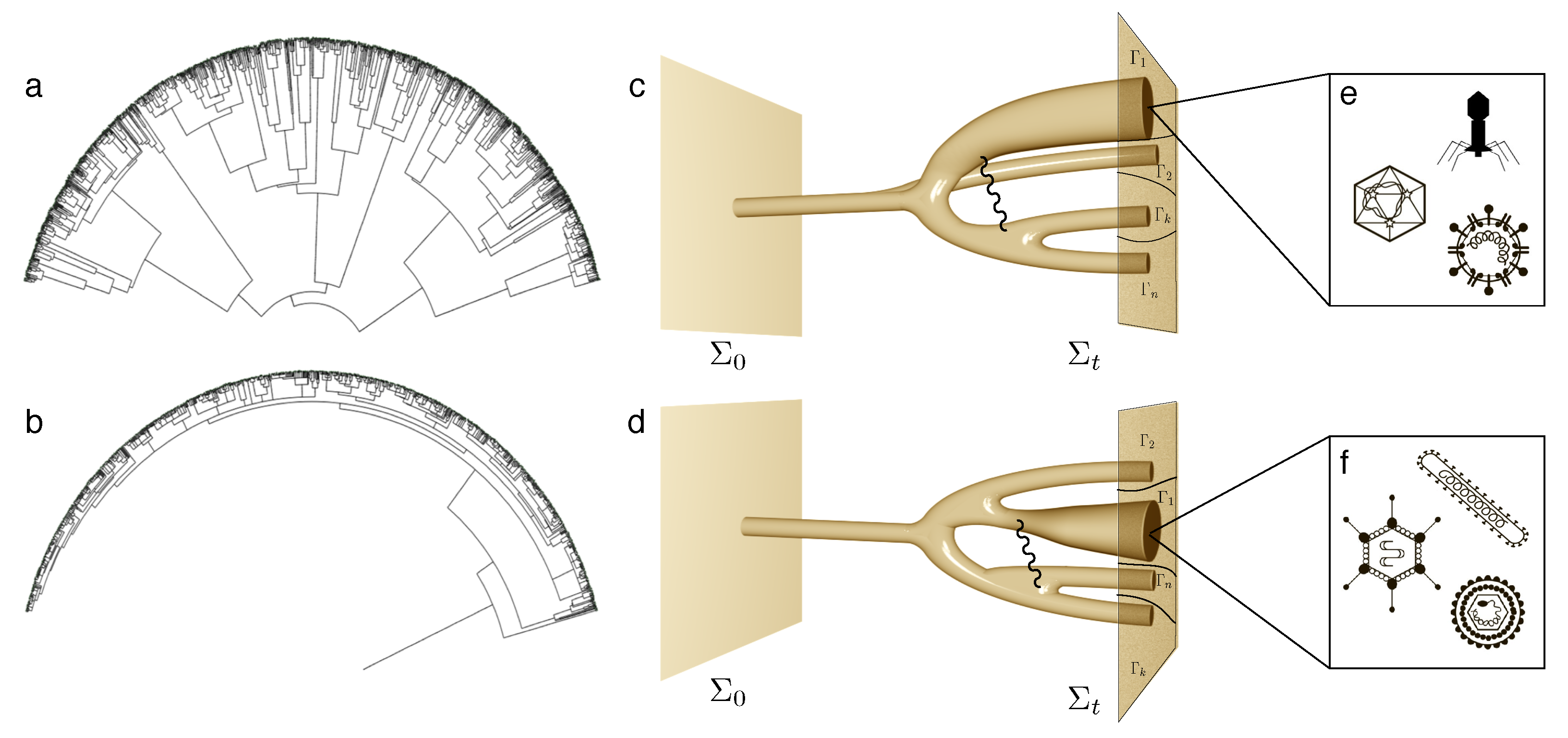
5. Multicellularity and Development: on Growth, Form and Life Cycles
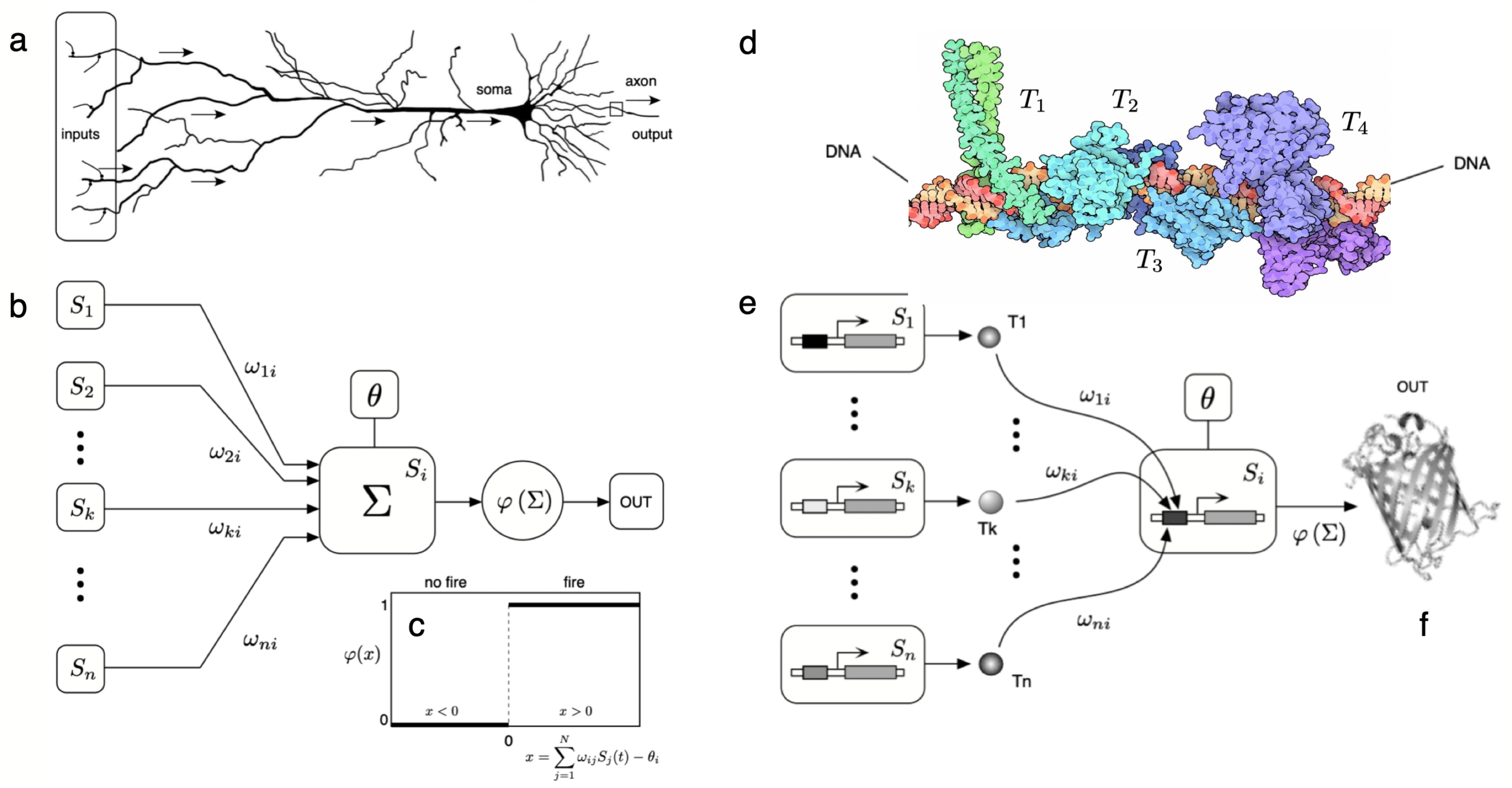
6. Cognitive Networks, Thresholds and Brains
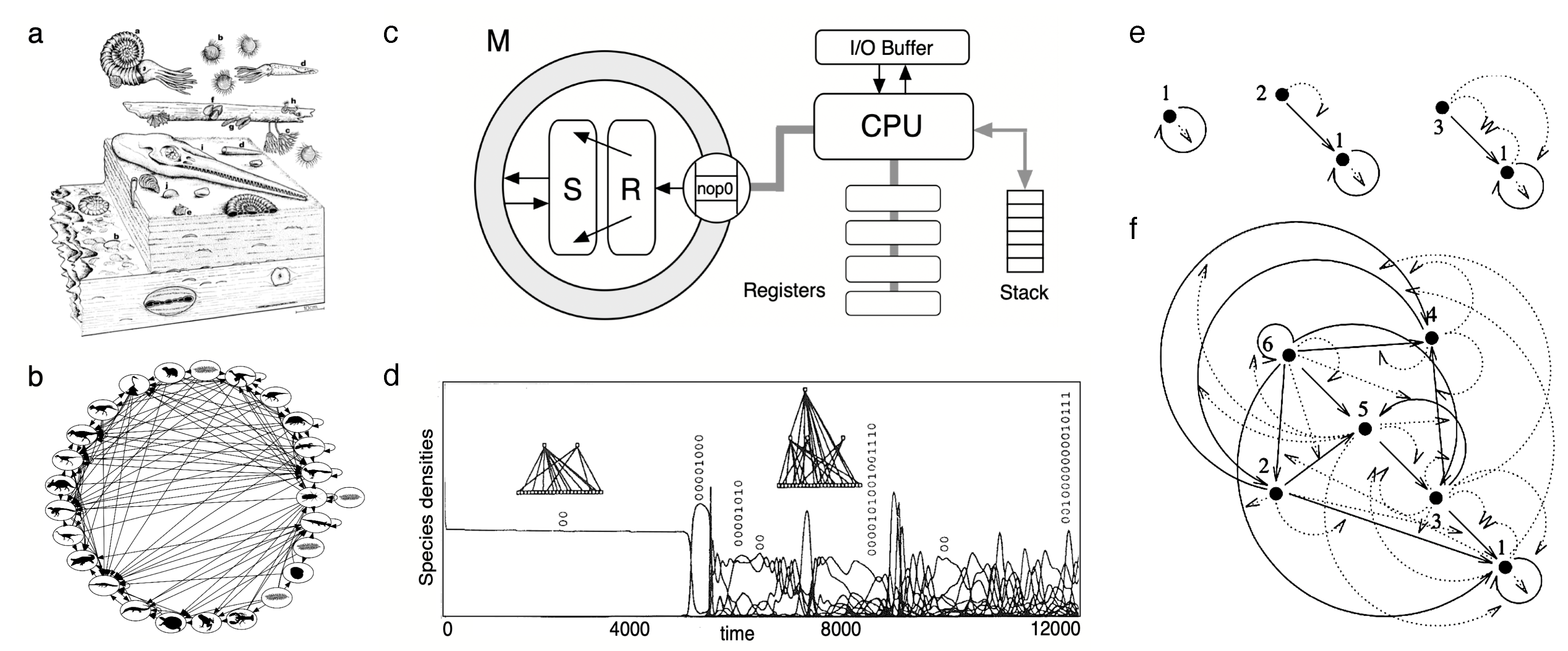
7. Ecology: Inevitable Parasites and Functional Trees
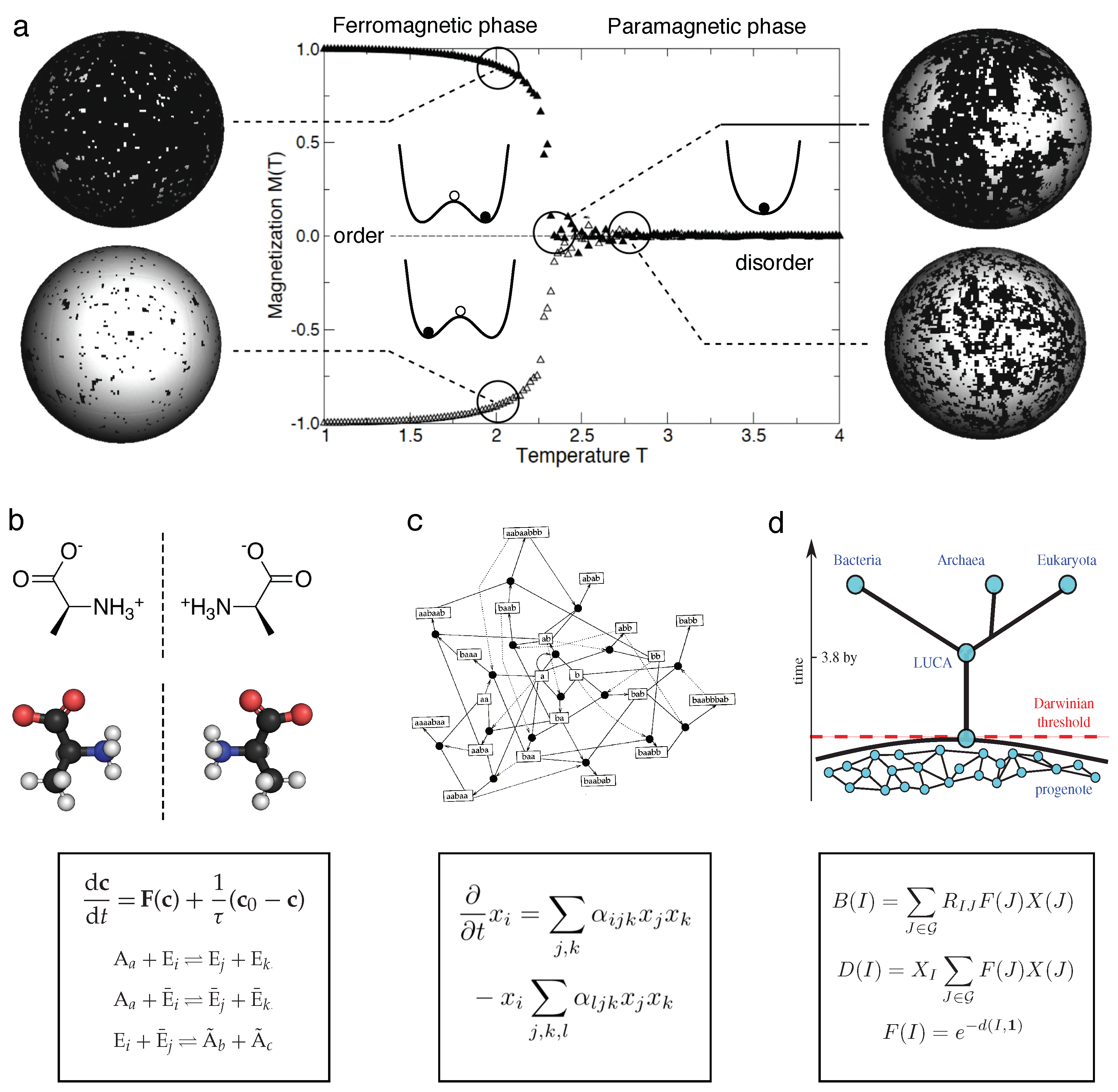
8. Phase Transitions and Critical States
9. Discussion
- Internal entropy-reducing processes characterise the thermodynamic logic of living systems. Such processes are enabled by coupling processes that produce greater entropy in the environment, likely in the form of generated heat. Life is also expected to store and employ energy intermediates to drive internal processes and to decouple from environmental conditions, thereby attaining a degree of thermodynamic autonomy. Finally, the internal metabolic process will be organized around cyclic transformations.
- Linear heteropolymers formed by sets of units (symbols) having near-equivalent energies are the expected substrate for carrying molecular information. They allow the exploration of vast combinatorial spaces, and the physical constraints associated with linearity might pose severe limitations to the repertoire of possible monomer candidates.
- Closed cell compartments equipped with a von Neumann replication logic are needed for self-reproducing living forms capable of evolution. The compartment allows the concentration of required molecules and defines a boundary between internal and external environments connected through a membrane that can play a part in the constructor roles by exploiting physical instabilities. Such a closed container can be achieved using a specific class of molecules (the amphiphiles) and is thus constrained to a subset of chemical candidates.
- Multicellularity allows the emergence of new kinds of organisation out of simpler units. One universal precondition for this innovation is the presence of some physically embodied process that guarantees the closeness of cells. While the group provides mechanisms of efficient collective reproduction, these new units of selection (from cell clusters to organisms) need to deal with cheaters through ratcheting. The potential diversity of basic morphological designs might be strongly constrained by a finite number of physico-genetic motifs, whose combinations might generate the whole repertoire of basic developmental programs sharing deep common morphological motifs.
- Beyond information coding on coded strings, cognitive systems require threshold-like units that allow reliable integration and decision-making. Complex cognition has been unfolding by evolving different (but formally equivalent) circuits based on threshold functions that integrate surrounding signals. In multicellular systems, this means evolving cells that display polarisation and provide the means for rapid sensing and propagation of information. Because of these features, complex cognition might have been constrained to evolve towards multilayer systems.
- Ecosystem architectures are deeply constrained within a finite set of possible classes of ecological interactions. Current and past ecosystems reveal such a discrete repertoire of possibilities, and in silico models of evolving ecologies support this constrained repertoire. Among other regularities, the widespread presence of parasites suggests that they are an inevitable outcome of complex adaptive systems.
Acknowledgments
| a | Those estimates are based on evidence from the fossil record that reveals a sharp transition, whereas molecular clock studies (where divergence times are obtained from sequence comparison) locate the origin of animals at about 780 Ma. See [22] and references therein. |
| b | The mapping between genes and phenotypes is poorly captured by the linear connection. The nature of the genotype-phenotype mapping [29] is highly compositional and algorithmic, reflecting the inevitable and essential interdependence of the molecular components that constitute organisms and enact their physiology and development. |
| c |
In statistical physics, an ergodic system is one which, over sufficiently long timescales, explores all possible microstates that are consistent with its macroscopic properties. Mathematically, for any physical observable , where is the microstate that specifies all of the system’s coordinates, the long-time average of converges to the ensemble average, so
In intuitive terms, an ergodic system explores (over time) all possible ways it can exist. It suggests that, given enough time, the system will visit all the available configurations or states.
|
| d | In nonequilibrium thermodynamics [73], the rate of entropy production per time can be expressed as an integral
|
| e | Once helicases unwind the double-stranded DNA helix. |
| f | To a large extent, the complementary nature of DNA is what allows the enormous flexibility in building structures and machines. A given strand will match only its complement and weakly interact with other sequences. In this way, many components can be assembled together in two or three dimensions while keeping the control on interaction strengths between every pair of elements. |
| g | A distinction needs to be made between replicators and reproducers. As suggested by E. Szathmáry, reproduction of a cell implies making a copy of the whole that is not limited to the genome, in contrast with viral particles or other putative “naked” molecular replicators in early life. |
| h | Some crucial insights into this problem were advanced within the context of Turing machines. If Universal Turing Machines can have access to its own code (a form of self-description), it is possible to show that one can re-write any program so that it will print out a copy of itself before it starts running. In formal terms, for any Turing machine , there exists a such that prints out a description of on its tape and then behaves in the same way as . Conceptually, this is reminiscent of the UC, which makes a copy of the Instructions and then carries out the Instructions. The mathematician Stephen Kleene first developed these ideas in his second recursion theorem [132,133]. |
| i | Simple MC (SMC) refers to a form of multicellular organisation where cells form clusters or colonies with limited differentiation and coordination. Unlike complex multicellularity, where cells exhibit specialised functions and are organised into distinct tissues and organs, SMC involves groups of similar or identical cells working together, often with minimal communication or structural integration. |
| j | In some cases, as it occurs with fractal branching patterns in plants [219] or growing corals [220], there is a set of generative rules (that sometimes can be expressed in terms of grammar) that successfully reproduce the whole repertoire of forms, which can often be classified within a parameter space in terms of seperated phases. For intriguing arguments that fractal growth with environmentally regulated parameters governed the Ediacaran fauna that may have been a transition stage to non-self-similar development, see [221] |
| k | Alternative implementations may employ smooth step functions, such as
|
| l | Three of these combinations, and are subdivided when applied to real cases. Predation and parasitism, for example, share the pair, but the kind of interaction and associated life cycles are different. |
| m | It is worth mentioning that there is a branch of theoretical ecology that deals with the qualitative stability of communities as derived from the sign matrix of pairwise interactions [279,280]. This indicates that much can be inferred solely based on the qualitative effects of member species on each other. |
| n | Importantly, thermodynamic arguments based on the Maximum Entropy formalism also reveal the power of constraints when dealing with the overal statistical patterns of ecosystem organization (instead of the internal logic) such as species– area relationships or abundance distributions in macroecology [285]. |
| o | In this case, by making the summation over k we obtain as the associated dilution term. |
| p | Ignoring spatial effects, a mean-field approximation can be made for the average magnetisation, which follows the differential equation . It is easy to show that three fixed points exist, associated with a symmetry-breaking (pitchfork) bifurcation [350]. A potential function (as defined in one-dimensional dynamical systems, see [351]) can be derived from , i.e., from , which in this case, gives a fourth-order expression that is a symmetric function, i. e. . For , the only stable state is , whereas for , the zero state is unstable, whereas two possible, completely symmetric solutions exist, namely . These potential functions are displayed as insets in Figure 10a. |
| q |
References
- Summons, R.E.; Albrecht, P.; McDonald, G.; Moldowan, J.M. Molecular biosignatures. <italic>Strategies of Life Detection</italic> <bold>2008</bold>, pp. 133–159.
- Marshall, S.M.; Mathis, C.; Carrick, E.; Keenan, G.; Cooper, G.J.; Graham, H.; Craven, M.; Gromski, P.S.; Moore, D.G.; Walker, S.I.; others. Identifying molecules as biosignatures with assembly theory and mass spectrometry. Nature communications 2021, 12, 3033. [Google Scholar] [CrossRef]
- Chan, M.A.; Hinman, N.W.; Potter-McIntyre, S.L.; Schubert, K.E.; Gillams, R.J.; Awramik, S.M.; Boston, P.J.; Bower, D.M.; Des Marais, D.J.; Farmer, J.D.; others. Deciphering biosignatures in planetary contexts. Astrobiology 2019, 19, 1075–1102. [Google Scholar] [CrossRef]
- Seager, S.; Bains, W.; Petkowski, J. Toward a list of molecules as potential biosignature gases for the search for life on exoplanets and applications to terrestrial biochemistry. Astrobiology 2016, 16, 465–485. [Google Scholar] [CrossRef]
- Johnson, S.S.; Anslyn, E.V.; Graham, H.V.; Mahaffy, P.R.; Ellington, A.D. Fingerprinting non-terran biosignatures. Astrobiology 2018, 18, 915–922. [Google Scholar] [CrossRef]
- Cleaves, H.J.; Hystad, G.; Prabhu, A.; Wong, M.L.; Cody, G.D.; Economon, S.; Hazen, R.M. A robust, agnostic molecular biosignature based on machine learning. Proceedings of the National Academy of Sciences 2023, 120, e2307149120. [Google Scholar] [CrossRef]
- Solé, R.V.; Munteanu, A. The large-scale organization of chemical reaction networks in astrophysics. Europhysics Letters 2004, 68, 170. [Google Scholar] [CrossRef]
- Estrada, E. Returnability as a criterion of disequilibrium in atmospheric reactions networks. Journal of Mathematical Chemistry 2012, 50, 1363–1372. [Google Scholar] [CrossRef]
- Bartlett, S.; Li, J.; Gu, L.; Sinapayen, L.; Fan, S.; Natraj, V.; Jiang, J.H.; Crisp, D.; Yung, Y.L. Assessing planetary complexity and potential agnostic biosignatures using epsilon machines. Nature Astronomy 2022, 6, 387–392. [Google Scholar] [CrossRef]
- McGhee, G.R. <italic>Convergent evolution: limited forms most beautiful</italic>; MIT press, 2011.
- Grefenstette, N.; Chou, L.; Colón-Santos, S.; Fisher, T.M.; Mierzejewski, V.; Nural, C.; Sinhadc, P.; Vidaurri, M.; Vincent, L.; Weng, M.M. Chapter 9: Life as We Don’t Know It. Astrobiology 2024, 24, S–186. [Google Scholar] [CrossRef]
- Church, G.M.; Regis, E. <italic>Regenesis: how synthetic biology will reinvent nature and ourselves</italic>; Basic Books, 2014.
- Solé, R. Synthetic transitions: towards a new synthesis. Philosophical Transactions of the Royal Society B: Biological Sciences 2016, 371, 20150438. [Google Scholar] [CrossRef]
- Ebrahimkhani, M.R.; Levin, M. Synthetic living machines: A new window on life. Iscience 2021, 24. [Google Scholar] [CrossRef]
- Davies, J.; Levin, M. Synthetic morphology with agential materials. Nature Reviews Bioengineering 2023, 1, 46–59. [Google Scholar] [CrossRef]
- Monod, J. <italic>On chance and necessity</italic>; Springer, 1974.
- Gould, S.J. <italic>Wonderful life: the Burgess Shale and the nature of history</italic>; WW Norton & Company, 1989.
- Carroll, S.B. <italic>A series of fortunate events: chance and the making of the planet, life, and you</italic>; Princeton University Press, 2020.
- Jacob, F. Evolution and tinkering. Science 1977, 196, 1161–1166. [Google Scholar] [CrossRef]
- Anderson, P.W. More Is Different: Broken symmetry and the nature of the hierarchical structure of science. Science 1972, 177, 393–396. [Google Scholar] [CrossRef]
- Artime, O.; De Domenico, M. From the origin of life to pandemics: Emergent phenomena in complex systems, 2022.
- Erwin, D.H.; Valentine, J.W. The cambrian explosion. <italic>Genwodd Village, Colorado: Roberts and Company</italic> <bold>2013</bold>.
- Carroll, S.B. Chance and necessity: the evolution of morphological complexity and diversity. Nature 2001, 409, 1102–1109. [Google Scholar] [CrossRef]
- Erwin, D.H. Was the Ediacaran–Cambrian radiation a unique evolutionary event? Paleobiology 2015, 41, 1–15. [Google Scholar] [CrossRef]
- Blount, Z.D.; Lenski, R.E.; Losos, J.B. Contingency and determinism in evolution: Replaying life’s tape. Science 2018, 362, eaam5979. [Google Scholar] [CrossRef]
- Morris, S.C. <italic>Life’s solution: inevitable humans in a lonely universe</italic>; Cambridge university press, 2003.
- Solé, R.V.; Valverde, S.; Rodriguez-Caso, C. Convergent evolutionary paths in biological and technological networks. Evolution: Education and Outreach 2011, 4, 415–426. [Google Scholar] [CrossRef]
- Alberch, P. The logic of monsters: Evidence for internal constraint in development and evolution. Geobios 1989, 22, 21–57. [Google Scholar] [CrossRef]
- Manrubia, S.; Cuesta, J.A.; Aguirre, J.; Ahnert, S.E.; Altenberg, L.; Cano, A.V.; Catalán, P.; Diaz-Uriarte, R.; Elena, S.F.; García-Martín, J.A.; others. From genotypes to organisms: State-of-the-art and perspectives of a cornerstone in evolutionary dynamics. Physics of Life Reviews 2021, 38, 55–106. [Google Scholar] [CrossRef]
- Raup, D.M. Geometric analysis of shell coiling: general problems. Journal of paleontology 1966, 1178–1190. [Google Scholar]
- Corominas-Murtra, B.; Goñi, J.; Solé, R.V.; Rodríguez-Caso, C. On the origins of hierarchy in complex networks. Proceedings of the National Academy of Sciences 2013, 110, 13316–13321. [Google Scholar] [CrossRef]
- Avena-Koenigsberger, A.; Goni, J.; Solé, R.; Sporns, O. Network morphospace. Journal of the Royal Society Interface 2015, 12, 20140881. [Google Scholar] [CrossRef]
- Saavedra, S.; Rohr, R.P.; Bascompte, J.; Godoy, O.; Kraft, N.J.; Levine, J.M. A structural approach for understanding multispecies coexistence. Ecological Monographs 2017, 87, 470–486. [Google Scholar] [CrossRef]
- Long, C.; Deng, J.; Nguyen, J.; Liu, Y.Y.; Alm, E.J.; Solé, R.; Saavedra, S. Structured community transitions explain the switching capacity of microbial systems. Proc. Natl. Acad. Sci. USA 2024, 121. [Google Scholar] [CrossRef]
- Deng, J.; Taylor, W.; Saavedra, S. Understanding the impact of third-party species on pairwise coexistence. PLoS Comput. Biol. 2022, 18, e1010630. [Google Scholar] [CrossRef]
- Waddington, C.H. Canalization of development and the inheritance of acquired characters. Nature 1942, 150, 563–565. [Google Scholar] [CrossRef]
- Wagner, A. Robustness and Evolvability in Living Systems; Princeton Univ. Press: NJ, 2007. [Google Scholar]
- Medeiros, L.P.; Boege, K.; del Val, E.; Zaldivar-Riverón, A.; Saavedra, S. Observed ecological communities are formed by species combinations that are among the most likely to persist under changing environments. The American Naturalist 2021, 197, E17–E29. [Google Scholar] [CrossRef]
- Kauffman. <italic>Investigations</italic>; Oxford University Press, 2000.
- Kauffman, S.A.; Roli, A. A third transition in science? Interface Focus 2023, 13, 20220063. [Google Scholar] [CrossRef]
- Kauffman, S.A. Prolegomenon to patterns in evolution. Biosystems 2014, 123, 3–8. [Google Scholar] [CrossRef]
- Dryden, D.T.; Thomson, A.R.; White, J.H. How much of protein sequence space has been explored by life on Earth? Journal of The Royal Society Interface 2008, 5, 953–956. [Google Scholar] [CrossRef]
- Kempes, C.P.; Koehl, M.; West, G.B. The scales that limit: the physical boundaries of evolution. Frontiers in Ecology and Evolution 2019, 7, 242. [Google Scholar] [CrossRef]
- Kempes, C.P.; Krakauer, D.C. The multiple paths to multiple life. Journal of molecular evolution 2021, 89, 415–426. [Google Scholar] [CrossRef]
- Aguirre, J.; Catalán, P.; Cuesta, J.A.; Manrubia, S. On the networked architecture of genotype spaces and its critical effects on molecular evolution. Open biology 2018, 8, 180069. [Google Scholar] [CrossRef]
- Manrubia, S. The simple emergence of complex molecular function. Philosophical Transactions of the Royal Society A 2022, 380, 20200422. [Google Scholar] [CrossRef]
- Alberch, P. From genes to phenotype: dynamical systems and evolvability. Genetica 1991, 84, 5–11. [Google Scholar] [CrossRef]
- Gavrilets, S.; Gravner, J. Percolation on the fitness hypercube and the evolution of reproductive isolation. Journal of theoretical biology 1997, 184, 51–64. [Google Scholar] [CrossRef]
- Fontana, W. Modelling ‘evo-devo’with RNA. BioEssays 2002, 24, 1164–1177. [Google Scholar] [CrossRef]
- Schultes, E.A.; Bartel, D.P. One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science 2000, 289, 448–452. [Google Scholar] [CrossRef]
- Li, H.; Helling, R.; Tang, C.; Wingreen, N. Emergence of preferred structures in a simple model of protein folding. Science 1996, 273, 666–669. [Google Scholar] [CrossRef]
- Li, H.; Tang, C.; Wingreen, N. Are protein folds atypical? Proc. Nat. Acad. Sci. USA 1998, 95, 4987–4990. [Google Scholar] [CrossRef]
- Denton, M. The Protein Folds as Platonic Forms: New Support for the Pre-Darwinian Conception of Evolution by Natural Law. J. Theor. Biol. 2002, 219, 325–342. [Google Scholar] [CrossRef]
- Banavar, J.R.; Maritan, A. Colloquium: Geometrical approach to protein folding: a tube picture. Reviews of Modern Physics 2003, 75, 23. [Google Scholar] [CrossRef]
- Ahnert, S.E.; Johnston, I.G.; Fink, T.M.; Doye, J.P.; Louis, A.A. Self-assembly, modularity, and physical complexity. Physical Review E 2010, 82, 026117. [Google Scholar] [CrossRef]
- Sharma, A.; Czégel, D.; Lachmann, M.; Kempes, C.P.; Walker, S.I.; Cronin, L. Assembly theory explains and quantifies selection and evolution. Nature 2023, 622, 321–328. [Google Scholar] [CrossRef]
- Cleland, C.E. <italic>The Quest for a Universal Theory of Life: Searching for Life as we don’t know it</italic>; Vol. 11, Cambridge University Press, 2019.
- Goldenfeld, N.; Woese, C. Life is physics: evolution as a collective phenomenon far from equilibrium. Annu. Rev. Condens. Matter Phys. 2011, 2, 375–399. [Google Scholar] [CrossRef]
- Goldenfeld, N.; Biancalani, T.; Jafarpour, F. Universal biology and the statistical mechanics of early life. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences 2017, 375, 20160341. [Google Scholar] [CrossRef]
- Walker, S.I.; Packard, N.; Cody, G. Re-conceptualizing the origins of life. Philosophical transactions. Series A, Mathematical, physical, and engineering sciences 2017, 375. [Google Scholar] [CrossRef]
- Walker, S.I. Origins of life: a problem for physics, a key issues review. Reports on Progress in Physics 2017, 80, 092601. [Google Scholar] [CrossRef]
- Davies, P.C.; Walker, S.I. The hidden simplicity of biology. Reports on Progress in Physics 2016, 79, 102601. [Google Scholar] [CrossRef]
- Walker, S.I.; Bains, W.; Cronin, L.; DasSarma, S.; Danielache, S.; Domagal-Goldman, S.; Kacar, B.; Kiang, N.Y.; Lenardic, A.; Reinhard, C.T.; others. Exoplanet biosignatures: future directions. Astrobiology 2018, 18, 779–824. [Google Scholar] [CrossRef]
- Langton, C.G. Self-reproduction in cellular automata. Physica D: Nonlinear Phenomena 1984, 10, 135–144. [Google Scholar] [CrossRef]
- von Neumann, John; Ed. Burks, A. <italic>Theory of self-reproducing automata</italic>; University of IIlinois Press, Illinois, 1966.
- Langton, C.G.; Taylor, C.; Farmer, J.D.; Rasmussen, S. <italic>Artificial Life II: Santa Fe Institute Studies in the Sciences of Complexity, Proceedings Vol. 10</italic>; Addison-Wesley Redwood City, CA, 1992.
- Langton, C.G.; Taylor, C.; Farmer, J.D.; Rasmussen, S. <italic>Artificial Life III, Santa Fe Institute Studies in the Science of Complexity</italic>; Addison-Wesley Redwood City, CA, 1994.
- Küppers, B.O. <italic>Information and the Origin of Life</italic>; MIT Press, 1990.
- Yockey, H.P. Information theory, evolution, and the origin of life; Cambridge University Press, 2005.
- Walker, S.I.; Davies, P.C. The algorithmic origins of life. Journal of the Royal Society Interface 2013, 10, 20120869. [Google Scholar] [CrossRef]
- Flack, Jessica; Eds. Walker, S.I.; Davies, P.C.; Ellis, G.F., From Matter to Life: Information and Causality; Cambridge University Press, 2017; chapter Life’s Information Hierarchy, p. 283.
- Kempes, C.P.; Wolpert, D.; Cohen, Z.; Pérez-Mercader, J. The thermodynamic efficiency of computations made in cells across the range of life. Phil. Trans. Roy. Soc. A 2017, arXiv:1706.05043v1 [q-bio.OT]375, 20160343–arXiv:1706. [Google Scholar] [CrossRef]
- Glansdorff, P.; Prigogine, I. On a general evolution criterion in macroscopic physics. Physica 1964, 30, 351–374. [Google Scholar] [CrossRef]
- Schrödinger, E. <italic>What is life? The physical aspect of the living cell and mind</italic>; Cambridge university press, 1944.
- Boltzmann, L. The second law of thermodynamics, Populare Schriften, Essay no. 3, address to the Imperial Academy of Sciences 1886. In <italic>Theoretical Physics and Philosophical Problems, Selected Writings of L. Boltzmann</italic>; D. Reidel, 1974; pp. 13–32.
- Lengeler, J.W.; Drews, G.; Schlegel, H.G. Biology of the Prokaryotes; Blackwell Science: New York, 1999. [Google Scholar]
- Liu, J.; Marison, I.W.; Von Stockar, U. Microbial growth by a net heat up-take: A calorimetric and thermodynamic study on acetotrophic methanogenesis by Methanosarcina barkeri. Biotechnology and Bioengineering 2001, 75, 170–180. [Google Scholar] [CrossRef]
- Heijnen, J.J.; Van Dijken, J.P. In search of a thermodynamic description of biomass yields for the chemotrophic growth of microorganisms. Biotechnology and Bioengineering 1992, 39, 833–858. [Google Scholar] [CrossRef]
- Barlow, C.; Volk, T. Open systems living in a closed biosphere: a new paradox for the Gaia debate. BioSystems 1990, 23, 371–384. [Google Scholar] [CrossRef]
- Budyko, M.I. <italic>The evolution of the biosphere</italic>; Vol. 9, Springer Science & Business Media, 2012.
- Smith, E.; Morowitz, H.J. The origin and nature of life on Earth: the emergence of the fourth geosphere; Cambridge U. Press: London, 2016; ISBN 9781316348772. [Google Scholar]
- Benner, S.A.; Ellington, A.D.; Tauer, A. Modern metabolism as a palimpsest of the RNA world <bold>1989</bold>. <italic>18</italic>, 7054–7058.
- Branscomb, E.; Russell, M.J. Turnstiles and bifurcators: the disequilibrium converting engines that put metabolism on the road. Biochim. Biophys. Acta 2013, 1827, 62–78. [Google Scholar] [CrossRef]
- mechanisms, E.; the conversion of disequilibria; the engines of creation. Branscomb, E. and Biancalani, T. and Goldenfeld, N. and Russell, M. Phys. Rep. 2017, 677, 1–60. [Google Scholar]
- Pasek, M.A.; Kee, T.P. On the Origin of Phosphorylated Biomolecules. In Origins of Life: The Primal Self-Organization; Egel, R., Lankenau, D.H., Mulkidjanian, A.Y., Eds.; Springer-Verlag: Berlin, 2011; pp. 57–84. [Google Scholar]
- Braakman, R.; Smith, E. The compositional and evolutionary logic of metabolism. Physical Biology 2013, 10, 011001. [Google Scholar] [CrossRef] [PubMed]
- Moreno Bergareche, A.; Ruiz-Mirazo, K. Metabolism and the problem of its universalization. BioSystems 1999, 49, 45–61. [Google Scholar] [CrossRef]
- Braakman, R.; Smith, E. The emergence and early evolution of biological carbon fixation. PLoS Comp. Biol. 2012, 8, e1002455. [Google Scholar] [CrossRef] [PubMed]
- Nunoura, T.; Chikaraishi, Y.; Izaki, R.; Suwa, T.; Sato, T.; Harada, T.; Mori, K.; Kato, Y.; Miyazaki, M.; Shimamura, S.; Yanagawa, K.; Shuto, A.; Ohkouchi, N.; Fujita, N.; Takaki, Y.; Atomi, H.; Takai, K. A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science 2018, 359, 559–563. [Google Scholar] [CrossRef]
- Koschmieder, E.; Pallas, S. Heat transfer through a shallow, horizontal convecting fluid layer. International Journal of Heat and Mass Transfer 1974, 17, 991–1002. [Google Scholar] [CrossRef]
- Onsager, L. Reciprocal Relations in Irreversible Processes. I. Physical Review 1931, 37, 405–426. [Google Scholar] [CrossRef]
- Morowitz, H.J. Physical background of cycles in biological systems. Journal of Theoretical Biology 1966, 13, 60–62. [Google Scholar] [CrossRef]
- Smith, E.; Morowitz, H.J. Universality in intermediary metabolism. Proc. Nat. Acad. Sci. USA 2004, 101, 13168–13173. [Google Scholar] [CrossRef] [PubMed]
- Morowitz, H.; Smith, E. Energy flow and the organization of life. Complexity 2007, 13, 51–59. [Google Scholar] [CrossRef]
- Schnakenberg, J. Network theory of microscopic and macroscopic behavior of master equation systems. Reviews of Modern physics 1976, 48, 571. [Google Scholar] [CrossRef]
- England, J.L. Statistical physics of self-replication. J. Chem. Phys. 2013, 139, 121923. [Google Scholar] [CrossRef]
- Calow, P. Conversion efficiencies in heterotrophic organisms. Biol. Rev. 1977, 52, 385–409. [Google Scholar] [CrossRef]
- Morowitz, H.J. Some Order-Disorder Considerations in Living Systems. Bull. Math. Biophys. 1955, 17, 81–86. [Google Scholar] [CrossRef]
- West, G.B.; Brown, J.H.; Enquist, B.J. A general model for the origin of allometric scaling laws in biology. Science 1997, 276, 122–126. [Google Scholar] [CrossRef]
- Kempes, C.P.; Dutkiewicz, S.; Follows, M.J. Growth, metabolic partitioning, and the size of microorganisms. Proceedings of the National Academy of Sciences 2012, 109, 495–500. [Google Scholar] [CrossRef]
- Prigogine, I.; Nicolis, G. Biological order, structure and instabilities. Quarterly Reviews of Biophysics 1971, 4, 107–148. [Google Scholar] [CrossRef]
- Cronin, L.; Krasnogor, N.; Davis, B.G.; Alexander, C.; Robertson, N.; Steinke, J.H.; Schroeder, S.L.; Khlobystov, A.N.; Cooper, G.; Gardner, P.M.; others. The imitation game - a computational chemical approach to recognizing life. Nature biotechnology 2006, 24, 1203–1206. [Google Scholar] [CrossRef]
- Koltsov, N. Physical-chemical fundamentals of morphology. Prog Exp Biol 1927, 3–31. [Google Scholar]
- Koltsov, N. Hereditary molecules. Sci. Life 1935, 5, 4–314. [Google Scholar]
- Soyfer, V.N. The consequences of political dictatorship for Russian science. Nature Reviews Genetics 2001, 2, 723–729. [Google Scholar] [CrossRef]
- Hopcroft, J.E. Turing machines. Scientific American 1984, 250, 86–E9. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Franklin, R.E.; Gosling, R.G. Evidence for 2-chain helix in crystalline structure of sodium deoxyribonucleate. Nature 1953, 172, 156–157. [Google Scholar] [CrossRef]
- Amos, M. <italic>Cellular computing</italic>; Systems Biology, 2004.
- Al-Hashimi, H.M. Turing, von Neumann, and the computational architecture of biological machines. Proceedings of the National Academy of Sciences 2023, 120, e2220022120. [Google Scholar] [CrossRef]
- Runnels, C.; Lanier, K.A.; Williams, J.K.; Bowman, J.C.; Petrov, A.S.; Hud, N.V.; Williams, L.D. Folding, Assembly, and Persistence: The Essential Nature and Origins of Biopolymers. J. Mol. Evol. 2018, 86, 598–610. [Google Scholar] [CrossRef]
- Pattee, H. How does a molecule become a message? Communication in Development, Anton L (ed) Developmental Biology Supplement, 1969.
- Kull, K. A sign is not alive—a text is. Σημειωτκη´-Sign Systems Studies 2002, 30, 327–336. [Google Scholar] [CrossRef]
- Barbieri, M. <italic>The organic codes: an introduction to semantic biology</italic>; Cambridge University Press, 2003.
- Pattee, H.H. Physical and functional conditions for symbols, codes, and languages. Biosemiotics 2008, 1, 147–168. [Google Scholar] [CrossRef]
- Rocha, L.M.; Hordijk, W. Material representations: From the genetic code to the evolution of cellular automata. Artificial life 2005, 11, 189–214. [Google Scholar] [CrossRef]
- Goodwin, B. Biology and meaning. Towards a Theoretical Biology (Edinburgh: Edinburgh University Press, 1972) 2017, 4, 259–275. [Google Scholar]
- Seeman, N.C. DNA in a material world. Nature 2003, 421, 427–431. [Google Scholar] [CrossRef]
- Ramezani, H.; Dietz, H. Building machines with DNA molecules. Nature Reviews Genetics 2020, 21, 5–26. [Google Scholar] [CrossRef]
- Sowerby, S.J.; Stockwell, P.A.; Heckl, W.M.; Petersen, G.B. Self-programmable, self-assembling two-dimensional genetic matter. Origins of Life and Evolution of the Biosphere 2000, 30, 81–99. [Google Scholar] [CrossRef]
- Sowerby, S.J.; Holm, N.G.; Petersen, G.B. Origins of life: a route to nanotechnology. Biosystems 2001, 61, 69–78. [Google Scholar] [CrossRef]
- Cairns-Smith, A.G. The chemistry of materials for artificial Darwinian systems. International Reviews in Physical Chemistry 1988, 7, 209–250. [Google Scholar] [CrossRef]
- Milo, R.; Phillips, R. <italic>Cell Biology by the Numbers</italic>; 2015.
- Landauer, R. Irreversibility and heat generation in the computing process. IBM journal of research and development 1961, 5, 183–191. [Google Scholar] [CrossRef]
- Kolchinsky, A.; Corominas-Murtra, B. Decomposing information into copying versus transformation. Journal of the Royal Society Interface 2020, 17, 20190623. [Google Scholar] [CrossRef]
- Kempes, C.P.; Wolpert, D.; Cohen, Z.; Pérez-Mercader, J. The thermodynamic efficiency of computations made in cells across the range of life. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences 2017, 375, 20160343. [Google Scholar] [CrossRef]
- Munteanu, A.; Attolini, C.S.O.; Rasmussen, S.; Ziock, H.; Solé, R.V. Generic Darwinian selection in catalytic protocell assemblies. Philosophical Transactions of the Royal Society B: Biological Sciences 2007, 362, 1847–1855. [Google Scholar] [CrossRef]
- Koonin, E.V. The origins of cellular life. Antonie Van Leeuwenhoek 2014, 106, 27–41. [Google Scholar] [CrossRef]
- Flack, J.C. Multiple time-scales and the developmental dynamics of social systems. Phil. Trans. R. Soc. B 2012, 367, 1802–1810. [Google Scholar] [CrossRef]
- Flack, J.C.; Erwin, D.; Elliot, T.; Krakauer, D.C. Timescales, symmetry, and uncertainty reduction in the origins of hierarchy in biological systems. In Evolution Cooperation and Complexity; Sterelny, K., Joyce, R., Calcott, B., Fraser, B., Eds.; MIT Press: Cambridge, MA, 2013; pp. 45–74. [Google Scholar]
- Odling-Smee, F.J.; Laland, K.N.; Feldman, M.W. Niche Construction: The Neglected Process in Evolution; Princeton U. Press: Princeton, N. J, 2003. [Google Scholar]
- Kleene, S.C. On notation for ordinal numbers. The Journal of Symbolic Logic 1938, 3, 150–155. [Google Scholar] [CrossRef]
- Marion, J.Y. From Turing machines to computer viruses. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences 2012, 370, 3319–3339. [Google Scholar] [CrossRef]
- Pattee, H.H. Dynamic and linguistic modes of complex systems. International Journal Of General System 1977, 3, 259–266. [Google Scholar] [CrossRef]
- Rocha, L.M. Evolution with material symbol systems. Biosystems 2001, 60, 95–121. [Google Scholar] [CrossRef]
- McMullin, B. John von Neumann and the evolutionary growth of complexity: Looking backward, looking forward. Artificial life 2000, 6, 347–361. [Google Scholar] [CrossRef]
- Ruiz-Mirazo, K.; Umerez, J.; Moreno, A. Enabling conditions for ‘open-ended evolution’. Biology & Philosophy 2008, 23, 67–85. [Google Scholar]
- Ruiz-Mirazo, K.; Peretó, J.; Moreno, A. A universal definition of life: autonomy and open-ended evolution. Origins of Life and Evolution of the Biosphere 2004, 34, 323–346. [Google Scholar] [CrossRef]
- Packard, N.; Bedau, M.A.; Channon, A.; Ikegami, T.; Rasmussen, S.; Stanley, K.O.; Taylor, T. An overview of open-ended evolution: Editorial introduction to the open-ended evolution ii special issue. Artificial life 2019, 25, 93–103. [Google Scholar] [CrossRef]
- Sipper, M. Fifty years of research on self-replication: An overview. Artificial life 1998, 4, 237–257. [Google Scholar] [CrossRef]
- Mouritsen, O.G. <italic>Life-as a matter of fat</italic>; Vol. 538, Springer, 2005.
- Hamley, I.W. <italic>Introduction to soft matter: synthetic and biological self-assembling materials</italic>; John Wiley & Sons, 2007.
- Rasmussen, S.; Chen, L.; Deamer, D.; Krakauer, D.C.; Packard, N.H.; Stadler, P.F.; Bedau, M.A. Transitions from nonliving to living matter. Science 2004, 303, 963–965. [Google Scholar] [CrossRef]
- Stano, P.; Luisi, P.L. Achievements and open questions in the self-reproduction of vesicles and synthetic minimal cells. Chemical Communications 2010, 46, 3639–3653. [Google Scholar] [CrossRef]
- Ruiz-Mirazo, K.; Briones, C.; de la Escosura, A. Prebiotic systems chemistry: new perspectives for the origins of life. Chemical reviews 2014, 114, 285–366. [Google Scholar] [CrossRef]
- Luisi, P.L. <italic>The emergence of life: from chemical origins to synthetic biology</italic>; Cambridge University Press, 2016.
- Serra, R.; Villani, M.; others. <italic>Modelling protocells</italic>; Springer, 2017.
- Rasmussen, S.; Bedau, M.A.; McCaskill, J.S.; Packard, N.H. A roadmap to protocells. Protocells: bridging nonliving and living matter. MIT Press, Cambridge.
- Sole, R.V. Evolution and self-assembly of protocells. The international journal of biochemistry & cell biology 2009, 41, 274–284. [Google Scholar]
- Kahana, A.; Lancet, D. Self-reproducing catalytic micelles as nanoscopic protocell precursors. Nature Reviews Chemistry 2021, 5, 870–878. [Google Scholar] [CrossRef]
- Morowitz, H.J.; Heinz, B.; Deamer, D.W. The chemical logic of a minimum protocell. Origins of Life and Evolution of the Biosphere 1988, 18, 281–287. [Google Scholar] [CrossRef]
- Deamer, D. <italic>First life: Discovering the connections between stars, cells, and how life began</italic>; Univ of California Press, 2011.
- Solé, R.V.; Munteanu, A.; Rodriguez-Caso, C.; Macía, J. Synthetic protocell biology: from reproduction to computation. Philosophical Transactions of the Royal Society B: Biological Sciences 2007, 362, 1727–1739. [Google Scholar] [CrossRef]
- Ruiz-Herrero, T.; Fai, T.G.; Mahadevan, L. Dynamics of growth and form in prebiotic vesicles. Physical review letters 2019, 123, 038102. [Google Scholar] [CrossRef]
- Fellermann, H.; Rasmussen, S.; Ziock, H.J.; Solé, R.V. Life cycle of a minimal protocell—a dissipative particle dynamics study. Artificial Life 2007, 13, 319–345. [Google Scholar] [CrossRef]
- Macía, J.; Solé, R.V. Protocell self-reproduction in a spatially extended metabolism–vesicle system. Journal of theoretical biology 2007, 245, 400–410. [Google Scholar] [CrossRef]
- Macia, J.; Solé, R.V. Synthetic Turing protocells: vesicle self-reproduction through symmetry-breaking instabilities. Philosophical Transactions of the Royal Society B: Biological Sciences 2007, 362, 1821–1829. [Google Scholar] [CrossRef]
- Corominas-Murtra, B. Thermodynamics of Duplication Thresholds in Synthetic Protocell Systems. Life 2019, 9(1), 9. [Google Scholar] [CrossRef]
- Fellermann, H.; Solé, R.V. Minimal model of self-replicating nanocells: a physically embodied information-free scenario. Philosophical Transactions of the Royal Society B: Biological Sciences 2007, 362, 1803–1811. [Google Scholar] [CrossRef]
- Vasas, V.; Szathmáry, E.; Santos, M. Lack of evolvability in self-sustaining autocatalytic networks constraints metabolism-first scenarios for the origin of life. Proceedings of the National Academy of Sciences 2010, 107, 1470–1475. [Google Scholar] [CrossRef]
- Darwin, C. On the Origin of Species; John Murray: London, 1859. [Google Scholar]
- Wilson, E.O. <italic>The diversity of life</italic>; WW Norton & Company, 1999.
- Mombach, J.C.; Glazier, J.A.; Raphael, R.C.; Zajac, M. Quantitative comparison between differential adhesion models and cell sorting in the presence and absence of fluctuations. Physical Review Letters 1995, 75, 2244. [Google Scholar] [CrossRef]
- Ollé-Vila, A.; Duran-Nebreda, S.; Conde-Pueyo, N.; Montañez, R.; Solé, R. A morphospace for synthetic organs and organoids: the possible and the actual. Integrative Biology 2016, 8, 485–503. [Google Scholar] [CrossRef]
- Márquez-Zacarías, P.; Pineau, R.M.; Gomez, M.; Veliz-Cuba, A.; Murrugarra, D.; Ratcliff, W.C.; Niklas, K.J. Evolution of cellular differentiation: from hypotheses to models. Trends in Ecology & Evolution 2021, 36, 49–60. [Google Scholar]
- Grosberg, R.K.; Strathmann, R.R. The evolution of multicellularity: a minor major transition? Annu. Rev. Ecol. Evol. Syst. 2007, 38, 621–654. [Google Scholar] [CrossRef]
- Bonner, J.T. <italic>First signals: the evolution of multicellular development</italic>; Princeton University Press, 2009.
- Knoll, A.H. The multiple origins of complex multicellularity. Annual Review of Earth and Planetary Sciences 2011, 39, 217–239. [Google Scholar] [CrossRef]
- Ratcliff, W.C.; Denison, R.F.; Borrello, M.; Travisano, M. Experimental evolution of multicellularity. Proceedings of the National Academy of Sciences 2012, 109, 1595–1600. [Google Scholar] [CrossRef]
- Ratcliff, W.C.; Fankhauser, J.D.; Rogers, D.W.; Greig, D.; Travisano, M. Origins of multicellular evolvability in snowflake yeast. Nature communications 2015, 6, 6102. [Google Scholar] [CrossRef]
- Solé, R.; Ollé-Vila, A.; Vidiella, B.; Duran-Nebreda, S.; Conde-Pueyo, N. The road to synthetic multicellularity. Current Opinion in Systems Biology 2018, 7, 60–67. [Google Scholar] [CrossRef]
- Toda, S.; Blauch, L.R.; Tang, S.K.; Morsut, L.; Lim, W.A. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science 2018, 361, 156–162. [Google Scholar] [CrossRef]
- Michod, R.E. Evolution of individuality during the transition from unicellular to multicellular life. Proceedings of the National Academy of Sciences 2007, 104, 8613–8618. [Google Scholar] [CrossRef]
- West, S.A.; Fisher, R.M.; Gardner, A.; Kiers, E.T. Major evolutionary transitions in individuality. Proceedings of the National Academy of Sciences 2015, 112, 10112–10119. [Google Scholar] [CrossRef]
- Libby, E.; Rainey, P.B. A conceptual framework for the evolutionary origins of multicellularity. Physical biology 2013, 10, 035001. [Google Scholar] [CrossRef]
- Carmel, Y. Human societal development: is it an evolutionary transition in individuality? Philosophical Transactions of the Royal Society B 2023, 378, 20210409. [Google Scholar] [CrossRef]
- Rainey, P.B. Major evolutionary transitions in individuality between humans and AI. Philosophical Transactions of the Royal Society B 2023, 378, 20210408. [Google Scholar] [CrossRef]
- Krakauer, D.; Bertschinger, N.; Olbrich, E.; Flack, J.C.; Ay, N. The information theory of individuality. Theory in Biosciences 2020, 139, 209–223. [Google Scholar] [CrossRef]
- Queller, D.C.; Strassmann, J.E. Beyond society: the evolution of organismality. Philosophical Transactions of the Royal Society B: Biological Sciences 2009, 364, 3143–3155. [Google Scholar] [CrossRef]
- Van Gestel, J.; Tarnita, C.E. On the origin of biological construction, with a focus on multicellularity. Proceedings of the National Academy of Sciences 2017, 114, 11018–11026. [Google Scholar] [CrossRef]
- Queller, D.C. Relatedness and the fraternal major transitions. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 2000, 355, 1647–1655. [Google Scholar] [CrossRef]
- Libby, E.; Ratcliff, W.C. Lichens and microbial syntrophies offer models for an interdependent route to multicellularity. The Lichenologist 2021, 53, 283–290. [Google Scholar] [CrossRef]
- Andersson, R.; Isaksson, H.; Libby, E. Multi-species multicellular life cycles. In <italic>The Evolution of Multicellularity</italic>; CRC Press, 2022; pp. 343–356.
- Aktipis, C.A.; Boddy, A.M.; Jansen, G.; Hibner, U.; Hochberg, M.E.; Maley, C.C.; Wilkinson, G.S. Cancer across the tree of life: cooperation and cheating in multicellularity. Philosophical Transactions of the Royal Society B: Biological Sciences 2015, 370, 20140219. [Google Scholar] [CrossRef]
- Márquez-Zacarías, P.; Conlin, P.L.; Tong, K.; Pentz, J.T.; Ratcliff, W.C. Why have aggregative multicellular organisms stayed simple? Current Genetics 2021, 67, 871–876. [Google Scholar] [CrossRef]
- Tarnita, C.E.; Taubes, C.H.; Nowak, M.A. Evolutionary construction by staying together and coming together. Journal of theoretical biology 2013, 320, 10–22. [Google Scholar] [CrossRef]
- Sandoz, K.M.; Mitzimberg, S.M.; Schuster, M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proceedings of the National Academy of Sciences 2007, 104, 15876–15881. [Google Scholar] [CrossRef]
- Aktipis, A.; Maley, C.C. Cooperation and cheating as innovation: insights from cellular societies. Philosophical Transactions of the Royal Society B: Biological Sciences 2017, 372, 20160421. [Google Scholar] [CrossRef]
- Libby, E.; Conlin, P.L.; Kerr, B.; Ratcliff, W.C. Stabilizing multicellularity through ratcheting. Philosophical Transactions of the Royal Society B: Biological Sciences 2016, 371, 20150444. [Google Scholar] [CrossRef]
- Libby, E.; Ratcliff, W.C. Ratcheting the evolution of multicellularity. Science 2014, 346, 426–427. [Google Scholar] [CrossRef]
- Sebé-Pedrós, A.; Degnan, B.M.; Ruiz-Trillo, I. The origin of Metazoa: a unicellular perspective. Nature Reviews Genetics 2017, 18, 498–512. [Google Scholar] [CrossRef]
- Ruiz-Trillo, I.; Kin, K.; Casacuberta, E. The origin of metazoan multicellularity: a potential microbial black swan event. Annual Review of Microbiology 2023, 77, 499–516. [Google Scholar] [CrossRef]
- Murray, J.D.; Murray, J.D. <italic>Mathematical biology: II: spatial models and biomedical applications</italic>; Vol. 18, Springer, 2003.
- Turing, A.M. The chemical basis of morphogenesis. Bulletin of mathematical biology 1990, 52, 153–197. [Google Scholar] [CrossRef]
- Marcon, L.; Sharpe, J. Turing patterns in development: what about the horse part? Current opinion in genetics & development 2012, 22, 578–584. [Google Scholar]
- Prigogine, I.; Nicolis, G. On symmetry-breaking instabilities in dissipative systems. The Journal of Chemical Physics 1967, 46, 3542–3550. [Google Scholar] [CrossRef]
- Odell, G.; Oster, G.; Burnside, B.; Alberch, P. A mechanical model for epithelial morphogenesis. Journal of mathematical biology 1980, 9, 291–295. [Google Scholar] [CrossRef]
- Collinet, C.; Lecuit, T. Programmed and self-organized flow of information during morphogenesis. Nature Reviews Molecular Cell Biology 2021, 22, 245–265. [Google Scholar] [CrossRef]
- Waddington, C.H. <italic>The strategy of the genes</italic>; Routledge, 2014.
- Fontana, W.; Schuster, P. Shaping space: the possible and the attainable in RNA genotype–phenotype mapping. Journal of Theoretical Biology 1998, 194, 491–515. [Google Scholar] [CrossRef]
- Solé, R.V.; Fernández, P.; Kauffman, S.A. Adaptive walks in a gene network model of morphogenesis: insights into the Cambrian explosion. <italic>arXiv preprint q-bio/0311013</italic> <bold>2003</bold>.
- Borenstein, E.; Krakauer, D.C. An end to endless forms: epistasis, phenotype distribution bias, and nonuniform evolution. PLoS computational biology 2008, 4, e1000202. [Google Scholar] [CrossRef]
- Munteanu, A.; Sole, R.V. Neutrality and robustness in evo-devo: emergence of lateral inhibition. PLoS computational biology 2008, 4, e1000226. [Google Scholar] [CrossRef]
- Cotterell, J.; Sharpe, J. An atlas of gene regulatory networks reveals multiple three-gene mechanisms for interpreting morphogen gradients. Molecular systems biology 2010, 6, 425. [Google Scholar] [CrossRef]
- Fernández, P.; Solé, R.V. Neutral fitness landscapes in signalling networks. Journal of The Royal Society Interface 2007, 4, 41–47. [Google Scholar] [CrossRef]
- Catalán, P.; Arias, C.F.; Cuesta, J.A.; Manrubia, S. Adaptive multiscapes: an up-to-date metaphor to visualize molecular adaptation. Biology Direct 2017, 12, 1–15. [Google Scholar] [CrossRef]
- Deutsch, A.; Dormann, S. <italic>Mathematical modeling of biological pattern formation</italic>; Springer, 2005.
- Thomson, J.A. On growth and form, 1917.
- Ball, P. The self-made tapestry: pattern formation in nature <bold>1999</bold>.
- Graner, F.; Riveline, D. ‘The Forms of Tissues, or Cell-aggregates’: D’Arcy Thompson’s influence and its limits. Development 2017, 144, 4226–4237. [Google Scholar] [CrossRef]
- Forgacs, G.; Newman, S.A. <italic>Biological physics of the developing embryo</italic>; Cambridge University Press, 2005.
- Stolarska, M.A.; Kim, Y.; Othmer, H.G. Multi-scale models of cell and tissue dynamics. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences 2009, 367, 3525–3553. [Google Scholar] [CrossRef]
- Lenne, P.F.; Trivedi, V. Sculpting tissues by phase transitions. Nature Communications 2022, 13, 664. [Google Scholar] [CrossRef]
- Newman, S.A.; Bhat, R. Dynamical patterning modules: a" pattern language" for development and evolution of multicellular form. International Journal of Developmental Biology 2009, 53, 693. [Google Scholar] [CrossRef]
- Newman, S.A. Physico-genetic determinants in the evolution of development. Science 2012, 338, 217–219. [Google Scholar] [CrossRef]
- Graner, F.; Glazier, J.A. Simulation of biological cell sorting using a two-dimensional extended Potts model. Physical review letters 1992, 69, 2013. [Google Scholar] [CrossRef]
- Brodland, G.W. Computational modeling of cell sorting, tissue engulfment, and related phenomena: A review. Appl. Mech. Rev. 2004, 57, 47–76. [Google Scholar] [CrossRef]
- Steinberg, M.S. Adhesion-guided multicellular assembly. Journal of theoretical biology 1975, 55, 431–443. [Google Scholar] [CrossRef]
- Prusinkiewicz, P.; Lindenmayer, A. <italic>The algorithmic beauty of plants</italic>; Springer Science & Business Media, 2012.
- Kaandorp, J.A. <italic>Fractal Modelling: Growth and Form in Biology</italic>; Springer Science & Business Media, 1994.
- Hoyal Cuthill, J.F.; Conway Morris, S. Fractal branching organizations of Ediacaranrangeomorph fronds reveal a lost Proterozoicbody plan <bold>2014</bold>. <italic>111</italic>, 13122–13126.
- Wolpert, L. <italic>Principles of Development</italic>; Oxford University Press, 2015.
- Staple, D.; Farhadifar, R.; Röper, J.C.; Aigouy, B.; Eaton, S.; Jülicher, F. Mechanics and remodelling of cell packings in epithelia. The European physical journal. E, Soft matter 2010, 33, 117–27. [Google Scholar] [CrossRef]
- Bi, D.; Lopez, J.; Schwarz, J.; Manning, M.L. A density-independent glass transition in biological tissues. Nature Physics 2014, 11. [Google Scholar] [CrossRef]
- Jablonka, E.; Lamb, M.J. The evolution of information in the major transitions. Journal of theoretical biology 2006, 239, 236–246. [Google Scholar] [CrossRef]
- Lyon, P.; Keijzer, F.; Arendt, D.; Levin, M. Reframing cognition: getting down to biological basics, 2021.
- Wagensberg, J. Complexity versus uncertainty: The question of staying alive. Biology and philosophy 2000, 15, 493–508. [Google Scholar] [CrossRef]
- Llinás, R.R. <italic>I of the vortex: From neurons to self</italic>; MIT press, 2002.
- Seoane, L.F.; Solé, R.V. Information theory, predictability and the emergence of complex life. Royal Society open science 2018, 5, 172221. [Google Scholar] [CrossRef]
- Paulin, M.G.; Cahill-Lane, J. Events in early nervous system evolution. Topics in Cognitive Science 2021, 13, 25–44. [Google Scholar] [CrossRef]
- Sarpeshkar, R. Analog versus digital: extrapolating from electronics to neurobiology. Neural computation 1998, 10, 1601–1638. [Google Scholar] [CrossRef]
- Sjöström, P.J.; Turrigiano, G.G.; Nelson, S.B. Rate, timing, and cooperativity jointly determine cortical synaptic plasticity. Neuron 2001, 32, 1149–1164. [Google Scholar] [CrossRef]
- Shagrir, O. <italic>The nature of physical computation</italic>; Oxford University Press, 2022.
- Maley, C.J. Analogue computation and representation. The British Journal for the Philosophy of Science 2023, 74, 739–769. [Google Scholar] [CrossRef]
- Kristan, W.B. Early evolution of neurons. Current Biology 2016, 26, R949–R954. [Google Scholar] [CrossRef]
- Jékely, G. Origin and early evolution of neural circuits for the control of ciliary locomotion. Proceedings of the Royal Society B: Biological Sciences 2011, 278, 914–922. [Google Scholar] [CrossRef]
- Jékely, G. The chemical brain hypothesis for the origin of nervous systems. Philosophical Transactions of the Royal Society B 2021, 376, 20190761. [Google Scholar] [CrossRef]
- McCulloch, W.S.; Pitts, W. A logical calculus of the ideas immanent in nervous activity. The bulletin of mathematical biophysics 1943, 5, 115–133. [Google Scholar] [CrossRef]
- Cowan, J.D. Discussion: McCulloch-Pitts and related neural nets from 1943 to 1989. Bulletin of mathematical biology 1990, 52, 73–97. [Google Scholar] [CrossRef]
- Anderson, J.A.; Rosenfeld, E. <italic>Talking nets: An oral history of neural networks</italic>; MiT Press, 2000.
- Churchland, P.S.; Sejnowski, T.J. <italic>The computational brain</italic>; MIT press, 1992.
- Rojas, R. <italic>Neural networks: a systematic introduction</italic>; Springer Science & Business Media, 2013.
- Fukushima, K. Neocognitron: A self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biological cybernetics 1980, 36, 193–202. [Google Scholar] [CrossRef]
- Glass, L.; Kauffman, S.A. The logical analysis of continuous, non-linear biochemical control networks. Journal of theoretical Biology 1973, 39, 103–129. [Google Scholar] [CrossRef]
- Kurten, K. Correspondence between neural threshold networks and Kauffman Boolean cellular automata. Journal of Physics A: Mathematical and General 1988, 21, L615. [Google Scholar] [CrossRef]
- Luque, B.; Solé, R.V. Phase transitions in random networks: Simple analytic determination of critical points. Physical Review E 1997, 55, 257. [Google Scholar] [CrossRef]
- Bornholdt, S. Boolean network models of cellular regulation: prospects and limitations. Journal of the Royal Society Interface 2008, 5, S85–S94. [Google Scholar] [CrossRef]
- Parisi, G. A simple model for the immune network. Proceedings of the National Academy of Sciences 1990, 87, 429–433. [Google Scholar] [CrossRef]
- Perelson, A.S.; Weisbuch, G. Immunology for physicists. Reviews of modern physics 1997, 69, 1219. [Google Scholar] [CrossRef]
- Agliari, E.; Barra, A.; Del Ferraro, G.; Guerra, F.; Tantari, D. Anergy in self-directed B lymphocytes: a statistical mechanics perspective. Journal of theoretical biology 2015, 375, 21–31. [Google Scholar] [CrossRef]
- Deneubourg, J.L.; Goss, S.; Franks, N.; Sendova-Franks, A.; Detrain, C.; Chrétien, L. The dynamics of collective sorting robot-like ants and ant-like robots. From animals to animats: proceedings of the first international conference on simulation of adaptive behavior, 1991, pp. 356–365.
- Solé, R.V.; Miramontes, O.; Goodwin, B.C. Oscillations and chaos in ant societies. Journal of theoretical Biology 1993, 161, 343–357. [Google Scholar] [CrossRef]
- Theraulaz, G.; Bonabeau, E.; Nicolis, S.C.; Solé, R.V.; Fourcassié, V.; Blanco, S.; Fournier, R.; Joly, J.L.; Fernández, P.; Grimal, A.; others. Spatial patterns in ant colonies. Proceedings of the National Academy of Sciences 2002, 99, 9645–9649. [Google Scholar] [CrossRef]
- Couzin, I.D. Collective cognition in animal groups. Trends in cognitive sciences 2009, 13, 36–43. [Google Scholar] [CrossRef]
- McMillen, P.; Levin, M. Collective intelligence: A unifying concept for integrating biology across scales and substrates. Communications Biology 2024, 7, 378. [Google Scholar] [CrossRef]
- Yusufaly, T.I.; Boedicker, J.Q. Mapping quorum sensing onto neural networks to understand collective decision making in heterogeneous microbial communities. Physical biology 2017, 14, 046002. [Google Scholar] [CrossRef]
- Solé, R.; Moses, M.; Forrest, S. Liquid brains, solid brains, 2019.
- Piñero, J.; Solé, R. Statistical physics of liquid brains. Philosophical Transactions of the Royal Society B 2019, 374, 20180376. [Google Scholar] [CrossRef]
- Mjolsness, E.; Sharp, D.H.; Reinitz, J. A connectionist model of development. Journal of theoretical Biology 1991, 152, 429–453. [Google Scholar] [CrossRef]
- Alon, U. <italic>An introduction to systems biology: design principles of biological circuits</italic>; CRC press, 2019.
- Sneppen, K. <italic>Models of life</italic>; Cambridge University Press, 2014.
- Ginsburg, S.; Jablonka, E. The evolution of associative learning: A factor in the Cambrian explosion. Journal of theoretical biology 2010, 266, 11–20. [Google Scholar] [CrossRef]
- Piccinini, G. The First computational theory of mind and brain: a close look at mcculloch and pitts’s “logical calculus of ideas immanent in nervous activity”. Synthese 2004, 141, 175–215. [Google Scholar] [CrossRef]
- Shigeno, S. Brain evolution as an information flow designer: The ground architecture for biological and artificial general intelligence. <italic>Brain Evolution by Design: From Neural Origin to Cognitive Architecture</italic> <bold>2017</bold>, pp. 415–438.
- Shigeno, S.; Andrews, P.L.; Ponte, G.; Fiorito, G. Cephalopod brains: an overview of current knowledge to facilitate comparison with vertebrates. Frontiers in Physiology 2018, 952. [Google Scholar] [CrossRef]
- Garcia-Lopez, P.; Garcia-Marin, V.; Freire, M. The histological slides and drawings of Cajal. Frontiers in neuroanatomy 2010, 4, 1156. [Google Scholar] [CrossRef]
- North, G.; Greenspan, R.J. Invertebrate neurobiology. <italic>(No Title)</italic> <bold>2007</bold>.
- Seth, A.K.; Bayne, T. Theories of consciousness. Nature Reviews Neuroscience 2022, 23, 439–452. [Google Scholar] [CrossRef]
- Dennett, D.C. <italic>Kinds of minds: Toward an understanding of consciousness</italic>; Basic Books, 2008.
- Powell, R.; Mikhalevich, I.; Logan, C.; Clayton, N.S. Convergent minds: the evolution of cognitive complexity in nature, 2017.
- Dennett, D.C. <italic>From bacteria to Bach and back: The evolution of minds</italic>; WW Norton & Company, 2017.
- Solé, R.; Seoane, L.F. Evolution of brains and computers: the roads not taken. Entropy 2022, 24, 665. [Google Scholar] [CrossRef]
- Adami, C. <italic>Introduction to artificial life</italic>; Springer Science & Business Media, 1998.
- Kristian, L.; Nordahl, M.G. Artificial Food Webs. <italic>ARTIFICIAL LIFE III, Santa Fe Institute, Addison-Wesley</italic> <bold>1994</bold>.
- Banzhaf, W. Self-organisation in a system of binary strings <bold>1994</bold>.
- Margalef, R. Our biosphere. <italic>(No Title)</italic> <bold>1997</bold>.
- Ghilarov, A.M. Ecosystem functioning and intrinsic value of biodiversity. Oikos 2000, 90, 408–412. [Google Scholar] [CrossRef]
- Odum, E.P.; Barrett, G.W.; others. <italic>Fundamentals of ecology</italic>; Vol. 3, Saunders Philadelphia, 1971.
- May, R.M. Qualitative stability in model ecosystems. Ecology 1973, 54, 638–641. [Google Scholar] [CrossRef]
- Marzloff, M.P.; Dambacher, J.M.; Johnson, C.R.; Little, L.R.; Frusher, S.D. Exploring alternative states in ecological systems with a qualitative analysis of community feedback. Ecological Modelling 2011, 222, 2651–2662. [Google Scholar] [CrossRef]
- Poulin, R. <italic>Evolutionary ecology of parasites</italic>; Princeton university press, 2011.
- Sasselov, D.D.; Grotzinger, J.P.; Sutherland, J.D. The origin of life as a planetary phenomenon. Sci. Adv. 2020, 6, eaax3419. [Google Scholar] [CrossRef]
- Batalha, N.M. Exploring exoplanet populations with NASA’s Kepler Mission <bold>2014</bold>. <italic>111</italic>, 12647–12654.
- McKay, C.P. Requirements and limits for life in the context of exoplanets. Proc. Nat. Acad. Sci. USA 2014, 111, 12628–12633. [Google Scholar] [CrossRef]
- Harte, J.; Newman, E.A. Maximum information entropy: a foundation for ecological theory. Trends in ecology & evolution 2014, 29, 384–389. [Google Scholar]
- Gause, G.F. Experimental studies on the struggle for existence. Journal of Experimental Biology 1932, 9, 389–402. [Google Scholar] [CrossRef]
- May, R.M. Will a large complex system be stable? Nature 1972, 238, 413–414. [Google Scholar] [CrossRef]
- Pimm, S.L. <italic>The balance of nature?: ecological issues in the conservation of species and communities</italic>; University of Chicago Press, 1991.
- Roopnarine, P.D. Extinction cascades and catastrophe in ancient food webs. Paleobiology 2006, 32, 1–19. [Google Scholar] [CrossRef]
- Mitchell, J.S.; Roopnarine, P.D.; Angielczyk, K.D. Late Cretaceous restructuring of terrestrial communities facilitated the end-Cretaceous mass extinction in North America. Proceedings of the National Academy of Sciences 2012, 109, 18857–18861. [Google Scholar] [CrossRef]
- Dunne, J.A.; Williams, R.J.; Martinez, N.D.; Wood, R.A.; Erwin, D.H. Compilation and network analyses of Cambrian food webs. PLoS biology 2008, 6, e102. [Google Scholar] [CrossRef]
- Barricelli, N.A. Numerical testing of evolution theories: part I theoretical introduction and basic tests. Acta Biotheoretica 1962, 16, 69–98. [Google Scholar] [CrossRef]
- Barricelli, N.A. Numerical testing of evolution theories: Part II preliminary tests of performance. symbiogenesis and terrestrial life. Acta Biotheoretica 1963, 16, 99–126. [Google Scholar] [CrossRef]
- Dyson, G. <italic>Turing’s cathedral: the origins of the digital universe</italic>; Vintage, 2012.
- Langton, C.G. Studying artificial life with cellular automata. Physica D: Nonlinear Phenomena 1986, 22, 120–149. [Google Scholar] [CrossRef]
- Rocha, L. Evolutionary systems and artificial life. <italic>Lecture Notes, Los Alamos National Laboratory</italic> <bold>1997</bold>.
- Rasmussen, S.; Knudsen, C.; Feldberg, R.; Hindsholm, M. The coreworld: Emergence and evolution of cooperative structures in a computational chemistry. Physica D: Nonlinear Phenomena 1990, 42, 111–134. [Google Scholar] [CrossRef]
- Solé, R.; Elena, S.F. <italic>Viruses as complex adaptive systems</italic>; Vol. 15, Princeton University Press, 2018.
- Lloyd, A.L.; May, R.M. How viruses spread among computers and people. Science 2001, 292, 1316–1317. [Google Scholar] [CrossRef]
- Ray, T.S. An Approach to the Synthesis of Life, Artificial Life II. Santa Fe Institute Studies in the Sciences of Complexity, Proc. Addison-Wesley, 1991, pp. 371–408.
- Ray, T.S. Evolution, complexity, entropy and artificial reality. Physica D: Nonlinear Phenomena 1994, 75, 239–263. [Google Scholar] [CrossRef]
- Lachman, M. Origins of Evolution <bold>1991</bold>.
- Hillis, W.D. Co-evolving parasites improve simulated evolution as an optimization procedure. Physica D: Nonlinear Phenomena 1990, 42, 228–234. [Google Scholar] [CrossRef]
- Dennett, D.C. <italic>Darwin’s dangerous idea</italic>; Allen Press, 1995.
- Wilke, C.O.; Adami, C. The biology of digital organisms. TRENDS in ecology & evolution 2002, 17, 528–532. [Google Scholar]
- Solé, R.V.; Valverde, S. Macroevolution in silico: scales, constraints and universals. Palaeontology 2013, 56, 1327–1340. [Google Scholar] [CrossRef]
- Eldredge, N.; Gould, S.J. Punctuated equilibria: an alternative to phyletic gradualism. Models in paleobiology 1972, 1972, 82–115. [Google Scholar]
- Lindgren, K. Evolutionary phenomena in simple dynamics. Artificial life II 1991, 10, 295–312. [Google Scholar]
- Zaman, L.; Meyer, J.R.; Devangam, S.; Bryson, D.M.; Lenski, R.E.; Ofria, C. Coevolution drives the emergence of complex traits and promotes evolvability. PLoS Biology 2014, 12, e1002023. [Google Scholar] [CrossRef]
- Dittrich, P.; Ziegler, J.; Banzhaf, W. Artificial chemistries—a review. Artificial life 2001, 7, 225–275. [Google Scholar] [CrossRef]
- Banzhaf, W.; Yamamoto, L. <italic>Artificial chemistries</italic>; MIT Press, 2015.
- Stadler, P.F.; Fontana, W.; Miller, J.H. Random catalytic reaction networks. Physica D: Nonlinear Phenomena 1993, 63, 378–392. [Google Scholar] [CrossRef]
- Eigen, M. <italic>From strange simplicity to complex familiarity: a treatise on matter, information, life and thought</italic>; OUP Oxford, 2013.
- Smith, E.; Krishnamurthy, S. Symmetry and Collective Fluctuations in Evolutionary Games; IOP Press: Bristol, 2015. [Google Scholar]
- Fontana, W.; Wagner, G.; Buss, L.W. Beyond Digital Naturalism. Artificial Life 1994, 1, 211–227. [Google Scholar] [CrossRef]
- Fontana, W.; Buss, L.W. The barrier of objects: From dynamical systems to bounded organizations. In Boundaries and Barriers; Casti, J., Karlqvist, A., Eds.; Addison-Wesley: New York, 1996; pp. 56–116. [Google Scholar]
- Eigen, M. Selforganization of Matter and the Evolution of Biological Macromolecules. Naturwissenschaften 1971, 33, 465–522. [Google Scholar] [CrossRef]
- Eigen, M.; Schuster, P. The hypercycle, Part A: The emergence of the hypercycle. Naturwissenschaften 1977, 64, 541–565. [Google Scholar] [CrossRef]
- Eigen, M.; Schuster, P. The hypercycle, Part C: The realistic hypercycle. Naturwissenschaften 1978, 65, 341–369. [Google Scholar] [CrossRef]
- Hickinbotham, S.J.; Stepney, S.; Hogeweg, P. Nothing in evolution makes sense except in the light of parasitism: evolution of complex replication strategies. Royal Society Open Science 2021, 8, 210441. [Google Scholar] [CrossRef]
- Seoane, L.F.; Solé, R. How Turing parasites expand the computational landscape of digital life. Physical Review E 2023, 108, 044407. [Google Scholar] [CrossRef]
- Hudson, P.J.; Dobson, A.P.; Lafferty, K.D. Is a healthy ecosystem one that is rich in parasites? Trends in ecology & evolution 2006, 21, 381–385. [Google Scholar]
- Kempes, C.P.; Follows, M.J.; Smith, H.; Graham, H.; House, C.H.; Levin, S.A. Generalized stoichiometry and biogeochemistry for astrobiological applications. Bulletin of Mathematical Biology 2021, 83, 73. [Google Scholar] [CrossRef]
- Gagler, D.C.; Karas, B.; Kempes, C.P.; Malloy, J.; Mierzejewski, V.; Goldman, A.D.; Kim, H.; Walker, S.I. Scaling laws in enzyme function reveal a new kind of biochemical universality. Proceedings of the National Academy of Sciences 2022, 119, e2106655119. [Google Scholar] [CrossRef]
- Brown, J.H.; Gillooly, J.F.; Allen, A.P.; Savage, V.M.; West, G.B. Toward a metabolic theory of ecology. Ecology 2004, 85, 1771–1789. [Google Scholar] [CrossRef]
- Szathmáry, E.; Smith, J.M. The major evolutionary transitions. Nature 1995, 374, 227–232. [Google Scholar] [CrossRef]
- Szathmáry, E. Toward major evolutionary transitions theory 2.0. Proceedings of the National Academy of Sciences 2015, 112, 10104–10111. [Google Scholar] [CrossRef]
- Walker, S.I. Origins of life: a problem for physics,a key issues review. Rep. Prof. Phys. 2017, 80, 092601. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Katsnelson, M.I.; Koonin, E.V. Physical foundations of biological complexity. Proceedings of the National Academy of Sciences 2018, 115, E8678–E8687. [Google Scholar] [CrossRef]
- Eldredge, N. <italic>Unfinished synthesis: biological hierarchies and modern evolutionary thought</italic>; Oxford University Press, USA, 1985.
- Tëmkin, I.; Eldredge, N. Networks and hierarchies: Approaching complexity in evolutionary theory. <italic>Macroevolution: Explanation, interpretation and evidence</italic> <bold>2015</bold>, pp. 183–226.
- DeLong, J.P.; Okie, J.G.; Moses, M.E.; Sibly, R.M.; Brown, J.H. Shifts in metabolic scaling, production, and efficiency across major evolutionary transitions of life. Proceedings of the National Academy of Sciences 2010, 107, 12941–12945. [Google Scholar] [CrossRef]
- Kempes, C.P.; Wang, L.; Amend, J.P.; Doyle, J.; Hoehler, T. Evolutionary tradeoffs in cellular composition across diverse bacteria. The ISME journal 2016, 10, 2145–2157. [Google Scholar] [CrossRef]
- Foëx, G.; Weiss, P. Le magnétisme; Armand Colin: Paris, 1926. [Google Scholar]
- Kochmański, M.; Paszkiewicz, T.; Wolski, S. Curie-Weiss magnet – a simple model of phase transition. Eur. J. Phys. 2013, 34, 1555–1573. [Google Scholar] [CrossRef]
- Landau, L.D. Theory of phase transformations. Zh. Eksp. Teor. Fiz. 1937, 7, 19–32. [Google Scholar]
- Gell-Mann, M.; Low, F.E. Quantum electrodynamics at small distances. Phys. Rev. 1954, 95, 1300–1312. [Google Scholar] [CrossRef]
- Wilson, K.G.; Kogut, J. The renormalization group and the ε expansion. Phys. Rep., Phys. Lett. 1974, 12C, 75–200. [Google Scholar] [CrossRef]
- Weinberg, S. Phenomenological Lagrangians. Physica A 1979, 96, 327–340. [Google Scholar] [CrossRef]
- Polchinski, J.G. Renormalization group and effective lagrangians. Nuclear Physics B 1984, 231, 269–295. [Google Scholar] [CrossRef]
- Wiener, N. Cybernetics: Or the Control and Communication in the Animal and the Machine, second ed.; MIT Press: Cambridge, MA, 1965. [Google Scholar]
- Haken, H. Synergetics: Introduction and Advanced Topics, first ed.; Springer Verlag: New York, 2010. [Google Scholar]
- Li, R.; Bowerman, B. Symmetry breaking in biology. Cold Spring Harbor perspectives in biology 2010, 2, a003475. [Google Scholar] [CrossRef]
- Krakauer, D.C. Symmetry–simplicity, broken symmetry–complexity. Interface Focus 2023, 13, 20220075. [Google Scholar] [CrossRef]
- Wilson, K.G. Problems in physics with many scales of length. Scientific American 1979, 241, 158–179. [Google Scholar] [CrossRef]
- Yeomans, J.M. <italic>Statistical mechanics of phase transitions</italic>; Clarendon Press, 1992.
- Hughes, R.I. The Ising model, computer simulation, and universal physics. Ideas In Context 1999, 52, 97–145. [Google Scholar]
- Wikipedia. Chirality (chemistry) — Wikipedia, The Free Encyclopedia, 2024.
- Farmer, J.D.; Kauffman, S.A.; Packard, N.H. Autocatalytic replication of polymers. Physica D: Nonlinear Phenomena 1986, 22, 50–67. [Google Scholar] [CrossRef]
- Solé, R. <italic>Phase transitions</italic>; Princeton University Press, 2011.
- Leuthäusser, I. An exact correspondence between Eigen’s evolution model and a two-dimensional Ising system. The Journal of chemical physics 1986, 84, 1884–1885. [Google Scholar] [CrossRef]
- Leuthäusser, I. Statistical mechanics of Eigen’s evolution model. Journal of statistical physics 1987, 48, 343–360. [Google Scholar] [CrossRef]
- Tarazona, P. Error thresholds for molecular quasispecies as phase transitions: From simple landscapes to spin-glass models. Physical Review A 1992, 45, 6038. [Google Scholar] [CrossRef]
- Duke, T.; Bray, D. Heightened sensitivity of a lattice of membrane receptors. Proceedings of the National Academy of Sciences 1999, 96, 10104–10108. [Google Scholar] [CrossRef]
- Weber, M.; Buceta, J. The cellular Ising model: a framework for phase transitions in multicellular environments. Journal of The Royal Society Interface 2016, 13, 20151092. [Google Scholar] [CrossRef]
- Simpson, K.; L’Homme, A.; Keymer, J.; Federici, F. Spatial biology of Ising-like synthetic genetic networks. BMC biology 2023, 21, 185. [Google Scholar] [CrossRef]
- Schlicht, R.; Iwasa, Y. Forest gap dynamics and the Ising model. Journal of theoretical Biology 2004, 230, 65–75. [Google Scholar] [CrossRef]
- De las Cuevas, G.; Cubitt, T.S. Simple universal models capture all classical spin physics. Science 2016, 351, 1180–1183. [Google Scholar] [CrossRef]
- Fraiman, D.; Balenzuela, P.; Foss, J.; Chialvo, D.R. Ising-like dynamics in large-scale functional brain networks. Physical Review E 2009, 79, 061922. [Google Scholar] [CrossRef]
- Nowak, M.A.; Ohtsuki, H. Prevolutionary dynamics and the origin of evolution. Proc. Nat. Acad. Sci. USA 2008, 105, 14924–14927. [Google Scholar] [CrossRef]
- Eigen, M. Natural selection: a phase transition? Biophysical chemistry 2000, 85, 101–123. [Google Scholar] [CrossRef]
- Saito, Y.; Hyuga, H. Colloquium: Homochirality: Symmetry breaking in systems driven far from equilibrium. Reviews of Modern Physics 2013, 85, 603. [Google Scholar] [CrossRef]
- Jafarpour, F.; Biancalani, T.; Goldenfeld, N. Noise-induced mechanism for biological homochirality of early life self-replicators. Physical review letters 2015, 115, 158101. [Google Scholar] [CrossRef]
- Ribó, J.M.; Hochberg, D.; Crusats, J.; El-Hachemi, Z.; Moyano, A. Spontaneous mirror symmetry breaking and origin of biological homochirality. Journal of The Royal Society Interface 2017, 14, 20170699. [Google Scholar] [CrossRef]
- Kauffman, S.A. Autocatalytic sets of proteins. Journal of theoretical biology 1986, 119, 1–24. [Google Scholar] [CrossRef]
- Kauffman. The Origins of Order: Self-Organization and Selection in Evolution <bold>1993</bold>.
- Hanel, R.; Kauffman, S.A.; Thurner, S. Phase transition in random catalytic networks. Physical Review E 2005, 72, 036117. [Google Scholar] [CrossRef]
- Xavier, J.C.; Hordijk, W.; Kauffman, S.; Steel, M.; Martin, W.F. Autocatalytic chemical networks at the origin of metabolism. Proceedings of the Royal Society B 2020, 287, 20192377. [Google Scholar] [CrossRef]
- Vetsigian, K.; Woese, C.; Goldenfeld, N. Collective evolution and the genetic code. Proc. Nat. Acad. Sci. USA 2006, 103, 10696–10701. [Google Scholar] [CrossRef]
- Woese, C.R. The Genetic Code: The Molecular Basis for Genetic Expression; Harper and Row: New York, 1967. [Google Scholar]
- Crick, F.H.C. The origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Crutchfield, J.P. Between order and chaos. Nature Physics 2012, 8, 17–24. [Google Scholar] [CrossRef]
- Langton, C.G. Computation at the edge of chaos: Phase transitions and emergent computation. Physica D: nonlinear phenomena 1990, 42, 12–37. [Google Scholar] [CrossRef]
- Tlusty, T. A model for the emergence of the genetic code as a transition in a noisy information channel. Journal of theoretical biology 2007, 249, 331–342. [Google Scholar] [CrossRef]
- Schuster, P. Genotypes with phenotypes: Adventures in an RNA toy world. Biophysical chemistry 1997, 66, 75–110. [Google Scholar] [CrossRef]
- Kamp, C.; Bornholdt, S. Coevolution of quasispecies: B-cell mutation rates maximize viral error catastrophes. Physical Review Letters 2002, 88, 068104. [Google Scholar] [CrossRef]
- Chialvo, D.R. Emergent complex neural dynamics. Nature physics 2010, 6, 744–750. [Google Scholar] [CrossRef]
- Schuster, H.G. <italic>Criticality in neural systems</italic>; John Wiley & Sons, 2014.
- Solé, R.V.; Alonso, D.; McKane, A. Self–organized instability in complex ecosystems. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 2002, 357, 667–681. [Google Scholar] [CrossRef]
- Biroli, G.; Bunin, G.; Cammarota, C. Marginally stable equilibria in critical ecosystems. New Journal of Physics 2018, 20, 083051. [Google Scholar] [CrossRef]
- Beggs, J.M. The criticality hypothesis: how local cortical networks might optimize information processing. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences 2008, 366, 329–343. [Google Scholar] [CrossRef]
- Shew, W.L.; Plenz, D. The functional benefits of criticality in the cortex. The neuroscientist 2013, 19, 88–100. [Google Scholar] [CrossRef]
- Mora, T.; Bialek, W. Are biological systems poised at criticality? Journal of Statistical Physics 2011, 144, 268–302. [Google Scholar] [CrossRef]
- Touchette, H. The large deviation approach to statistical mechanics. Phys. Rep. 2009, 478, 1–69. [Google Scholar] [CrossRef]
- Smith, E. Emergent order in processes: the interplay of complexity, robustness, correlation, and hierarchy in the biosphere. Complexity and the Arrow of Time; Lineweaver, C., Davies, P., Ruse, M., Eds.; Cambridge U. Press: Cambridge, MA, 2013. [Google Scholar]
- Batterman, R.W. The Devil in the Details: Asymptotic Reasoning in Explanation, Reduction, and Emergence; Oxford University Press: New York, 2002. [Google Scholar]
- Lawson-Keister, E.; Manning, M.L. Jamming and arrest of cell motion in biological tissues. Current Opinion in Cell Biology 2021, 72, 146–155. [Google Scholar] [CrossRef]
- Hannezo, E.; Heisenberg, C.P. Rigidity transitions in development and disease. Trends in Cell Biology 2022, 32, 433–444. [Google Scholar] [CrossRef]
- Corominas-Murtra, B.; Petridou, N. Viscoelastic Networks: Forming Cells and Tissues. Frontiers in Physics 2021, 9. [Google Scholar] [CrossRef]
- Petridou, N.I.; Corominas-Murtra, B.; Heisenberg, C.P.; Hannezo, E. Rigidity percolation uncovers a structural basis for embryonic tissue phase transitions. Cell 2021, 184, 1914–1928. [Google Scholar] [CrossRef]
- Kim, J.H.; Pegoraro, A.F.; Das, A.; Koehler, S.A.; Ujwary, S.A.; Lan, B.; Mitchel, J.A.; Atia, L.; He, S.; Wang, K.; Bi, D.; Zaman, M.H.; Park, J.A.; Butler, J.P.; Lee, K.H.; Starr, J.R.; Fredberg, J.J. Unjamming and collective migration in MCF10A breast cancer cell lines. Biochemical and Biophysical Research Communications 2020, 521, 706–715. [Google Scholar] [CrossRef]
- Levin, S.R.; Scott, T.W.; Cooper, H.S.; West, S.A. Darwin’s aliens. International Journal of Astrobiology 2019, 18, 1–9. [Google Scholar] [CrossRef]
- Langton, C.G. Artificial life. In <italic>Artificial life</italic>; Routledge, 1989; pp. 1–47.
- Bedau, M.A.; McCaskill, J.S.; Packard, N.H.; Rasmussen, S.; Adami, C.; Green, D.G.; Ikegami, T.; Kaneko, K.; Ray, T.S. Open problems in artificial life. Artificial life 2000, 6, 363–376. [Google Scholar] [CrossRef]
- Bedau, M.A. Artificial life. In <italic>Philosophy of biology</italic>; Elsevier, 2007; pp. 585–603.
- Lehman, J.; Clune, J.; Misevic, D. The surprising creativity of digital evolution. Artificial Life Conference Proceedings. MIT Press One Rogers Street, Cambridge, MA 02142-1209, USA journals-info …, 2018, pp. 55–56.


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




