Introduction
A major problem in pediatric intensive care units (PICUs) is candidal sepsis, which causes a lot of harm, people to stay in the hospital longer, and costs more money [
1,
2]. The most common kind of bloodstream infection in pediatric hospital settings is Candida spp., which accounts for 10%-15% of these cases in PICUs in the US and Europe [
3]. Additional research conducted by Mellinghoff et al. suggests that bloodstream infections caused by Candida greatly increase mortality rates to 20-49% and are responsible for about 15% of all PICU deaths [
4]. An alarming 207% increase in cases of fungal sepsis from 1979 to 2000 underscores the growing threat that this infection poses [
5]. Noting a notable rise in such infections over the last decade, Pfaller, and Ostrosky-Zeichner et al. have also added to the existing body of research [
6,
7]. Candida bloodstream infections continue to be a significant concern in pediatric settings, as shown by Piqueras et al. who recorded an incidence rate of 1.97 per 1,000 patient-days [
8]. In contrast, Hsu et al. (2018) found that candidemia typically occurs in pediatric patients at a rate of 0.21 to 10.5 occurrences per thousand admissions [
9].
In pediatric intensive care units (PICUs), candidiasis is associated with numerous risk factors and becomes more severe after seven to ten days in the ICU.Immunosuppression, steroid use, dialysis, total parenteral nutrition (TPN), central venous catheterization, recent major surgery, and antibiotic consumption in the past are primary factors [
4,
10]. Factors unique to children include extended periods of neutropenia, exposure to large doses of steroids, and, more recently, advancing age [
11]. Prematurity, urinary catheter use, antacids, H2 blockers, malnutrition, immunosuppressive and cytotoxic treatments, immunodeficiency, burns, diabetes mellitus, and immunodeficiency are all known to enhance the risk of colonization and candidiasis [
12]. Furthermore, Candida bloodstream infections (CBSIs) are more common in specific PICU populations, including newborns, children younger than one year old, and children with hematologic malignancies or severely sick situations (8). Aplastic anemia, graft-versus-host disease, steroid usage, and unrelated bone marrow or cord blood transplants are additional risk factors in hematological patients. Candidemia can develop after three days of taking vancomycin or another antibiotic that kills anaerobic bacteria, having a malignant disease, or using a central venous catheter [
13]. An uptick in patients who are immunosuppressed or in critical care, as well as the widespread use of broad-spectrum antibiotics, may be causing an increase in Candida infections, as pointed out by Hsu et al. [
9].
Candida bloodstream infections (CBSIs) are still a major issue, causing a lot of illness and death, even though infection management and antimicrobial stewardship have gotten better. On top of that, low sensitivity of blood cultures and symptoms that aren't always specific can cause diagnostic delays. Moreover, variable incidence rates, ranging from 0.21 to 10.5 cases per 1,000 admissions, hinder the development of standardized treatment protocols [
8,
9]. A notable gap in data from PICUs in developing countries exacerbates these challenges, indicating a critical need for global insights into this issue [
2]. Although the potential benefits of machine learning in healthcare are significant [
14,
15], there is currently a lack of predictive models specifically designed for the early prediction of pediatric fungemia-associated mortality. Developing such models is crucial, as they can significantly enhance early detection, enable personalized medicine [
16,
17], optimize resource allocation, and provide insights into disease progression. These models could potentially transform patient outcomes by allowing for tailored interventions based on real-time risk assessments and improving the efficiency of clinical trials. However, the success of these models hinges on the availability of high-quality, diverse datasets and the integration of machine learning predictions into clinical practice. Addressing these challenges and ethical considerations, such as ensuring patient privacy and data security, is essential for the successful implementation and acceptance of predictive models in medical settings.
Predicting mortality due to fungemia in pediatric intensive care units (PICUs) may be possible with the help of the Candida Score. The Candida Score is useful for predicting candidiasis in adult intensive care units [
18], but it doesn't work when applied to children's needs since their immune systems and pathophysiology are different. Current models, such as the EQUAL Candida score, are designed for adults and do not consider aspects that are unique to children, like the length of time they are on immunosuppressive medication or mechanical breathing [
19,
20]. Considering these caveats, the purpose of our research is to adapt the Candida Score into a pediatric-specific predictive model by adding relevant pediatric-specific characteristics. Two essential components of fungemia management—improving mortality risk prediction and meeting a significant demand in pediatric intensive care—are expected to be addressed by this adjustment.
Methods
Study Design and Setting
This retrospective cohort study was conducted in pediatric intensive care units (PICUs) at King Abdulaziz University Hospital, Jeddah, Saudi Arabia. The study included pediatric patients admitted between 2016 and 2020.
Study Population
The study cohort comprised 85 pediatric patients diagnosed with fungemia. Inclusion criteria were patients aged 1months to 18 years with a positive blood culture for Candida species. Exclusion criteria included patients with incomplete medical records and those who were not treated in the PICU. For the PICU fungemia cohort study, inclusion criteria encompassed pediatric patients aged 0 to 18 years who were admitted to the PICU within the study timeframe and had a confirmed diagnosis of fungemia via blood culture. Only those with comprehensive medical records providing the necessary clinical data for the study's duration were considered. Excluded were individuals older than 18, those who developed fungemia after transferring out of the PICU or had antifungal treatment prior to PICU admission. Also excluded were patients with incomplete data, those with co-existing conditions influencing mortality risk independent of fungemia, such as terminal illnesses, and patients with PICU stays shorter than 24 hours to ensure adequate data collection for the study’s predictive modeling objectives. This delineation of criteria aimed to ensure a homogenous study population for accurate analysis of fungemia risk and outcomes in a pediatric intensive care context.
Data Collection
In the retrospective cohort study within a pediatric intensive care unit setting, data were meticulously gathered to inform a multivariate predictive model for fungemia. Data collection encompassed patient demographics, clinical parameters, and microbiological findings. The Candida Score was calculated for each patient to assess the risk of invasive candidiasis, incorporating elements such as total parenteral nutrition, surgical history, multifocal Candida colonization, and the presence of severe sepsis. Ages and weights at PICU admission were noted, along with the Pediatric Risk of Mortality scores, to gauge illness severity. The duration of mechanical ventilation and length of PICU stay were documented, as well as the duration of antibiotic treatment preceding fungal infection. The presence fungus was identified from blood cultures. The overall length of hospitalization prior to fungal infection onset was recorded. Gender was categorized as male or female. Additionally, outcomes were classified into survival or mortality to correlate clinical data with patient prognosis, and the isolation of the specific type of fungus was noted to discern patterns in fungal prevalence.
Diagnosis of Fungemia
We used the BacT/Alert automated system (Organon, Teknika, USA) to conduct blood cultures. Five milliliters of blood were added to a pediatric culture container for every sample. When the BacT/Alert system identified microbial growth or after five days of incubation without growth, the culture bottles were placed inside and left to continue. The contents of the bottles were Gram stained for preliminary identification once growth was detected. One step further was to subculture the bottles that showed yeast cells onto Sabouraud dextrose agar (SDA) from Saudi Prepared Media Laboratories in Riyadh, KSA. Yeast cells were quickly recognized using VITEK MS the same day when there was sufficient growth on the SDA. Using the VITEK®2 system (bioMérieux, Inc., France), the antifungal susceptibility testing, and species identification of Candida were carried out. This all-encompassing method guaranteed that fungemia cases were properly identified and treated with targeted antifungal medication.
Candida Score
The Candida Score is a clinical tool designed to assess the risk of invasive candidiasis in critically ill patients, particularly those in intensive care units. It helps clinicians decide when to start antifungal therapy in high-risk patients who do not have confirmed candidiasis. The score includes four factors: the administration of total parenteral nutrition (TPN), a history of recent surgery (especially abdominal surgery), multifocal Candida colonization (such as colonization in the urine, respiratory tract, or wounds), and the presence of severe sepsis. Each factor contributes to the score as follows: 0.908 for TPN, 0.997 for surgery, 1.112 for multifocal colonization, and 1.112 for severe sepsis. The scores for each patient are summed, and a total score of 3 or higher indicates a high risk of invasive candidiasis. This score was used in the model as risk factors for mortality.
Outcome Measures
The primary outcome was mortality within 30 days of the diagnosis of fungemia.
Statistical Analysis
Descriptives Statistics
The study used descriptive statistics to summarize the demographic and clinical features of the pediatric patients that were part of the research. Depending on the data distribution, continuous variables were either shown as medians with interquartile ranges or as means with standard deviations. The distribution of categorical variables within the study population was made clear by expressing them as percentages and frequencies. The data was displayed in a tabular format using tableone R tools alone, with outcomes (survival versus fatality) stratified.
Creating the Model
Logistic regression analysis was the first step in developing the model that could predict the likelihood of death. The purpose of this research was to determine which of the several clinical and laboratory factors were significant predictors of death. After being included in the univariate analysis, variables that showed a significant connection with mortality (p-value <0.05) were eventually added to the multivariate logistic regression model. This methodical procedure improved the model's prediction power by ensuring that only the most important predictors were included in the final version. To enhance our predictive capabilities, we compared three advanced statistical models: Random Forest, Gradient Boosting, and Logistic Regression.
Assessment of the Model
Area under the receiver operating characteristic curve (AUC) was the primary metric used to assess the prediction models' discriminative capacity. To find out which model had the greatest predictive performance, we computed and compared the AUC values for Logistic Regression, Random Forest, and Gradient Boosting. A model with good discrimination has an AUC value closer to 1, but a value closer to 0.5 implies about the same performance as chance.
Statistical Programs
Appropriate statistical software applications, including R, were used to conduct the studies. This study's extensive data analyses were made possible by the computational power and flexibility given by these technologies. A statistically significant link between the factors and the result of interest was shown by a p-value less than 0.05 with CI 95%. ChatGPT 4 statistical tool was used to create image by Python 3.8 version with appropriate libraries.
Verifying the Model
The prediction models were subjected to internal validation using bootstrapping approaches to guarantee their robustness and dependability. To accomplish this, we created several bootstrap samples by resampling the initial dataset with replacement. Next, the samples were subjected to the models, and the AUC was determined for every one of them. After adjusting for optimism, the average of these AUC values gave a prediction of how well the models would do on previously unknown data, proving their validity.
Model Validation
Model validation is crucial to ensure that the logistic regression model performs well on unseen data and is not overfitting the training data. Cross-validation is a robust method for assessing model performance. Variance Inflation Factor (VIF) and Hosmer-Lemeshow goodness-of-fit test were done for model validation.
Ethical Considerations
This study was conducted in strict accordance with the ethical standards outlined in the Declaration of Helsinki (1964 and its subsequent amendments). Ethical approval was obtained from the Institutional Review Board (IRB) of King Abdulaziz University Hospital, Jeddah, Saudi Arabia, under Reference No. 566-23. Due to the retrospective nature of our analysis, the IRB waived the requirement for informed consent. To ensure rigorous protection of patient confidentiality and anonymity, several data anonymization and security measures were implemented. Firstly, all patient identifiers were removed from the dataset prior to analysis and replaced with unique anonymization codes. This process ensured that the data could not be traced back to individual patients. Additionally, all electronic data were encrypted and stored on secure, password-protected servers accessible only to authorized research personnel. Physical copies of data, if any, were kept in locked cabinets in secure facilities. These precautions were taken to adhere to the highest standards of privacy and ethical integrity, minimizing any risk of data breaches while maintaining the utility of the data for research purposes. This approach not only protects individual patient privacy but also ensures the integrity of the research process.
Results
Varying Results for Survival and Non-Survival
We found statistically significant differences between fungemia survivors and non-survivors in our pediatric patient sample. Particularly, the median number of days that non-survivor spent on mechanical breathing (DOMV) was 20.00 compared to 11.00 for survivors (p = 0.011), indicating a statistically significant difference. Likewise, DOAbBFI, the length of time antibiotics was taken before fungal infection, was 18.00 days for non-survivors and 13.00 days for survivors (p = 0.024). Survivors had a median Candida Score (Cscore) of 2.00, while non-survivors had a higher median Candida Score (Cscore) of 4.00 (p < 0.001). Although there were no significant variations observed in other characteristics such age, weight, PRISM scores, length of PICU stay, and the presence of Candida Albicans, the results highlight the importance of DOMV, DOAbBFI, and Cscore as important factors that differentiate patient outcomes. (Table-1)
Isolation of Fungus
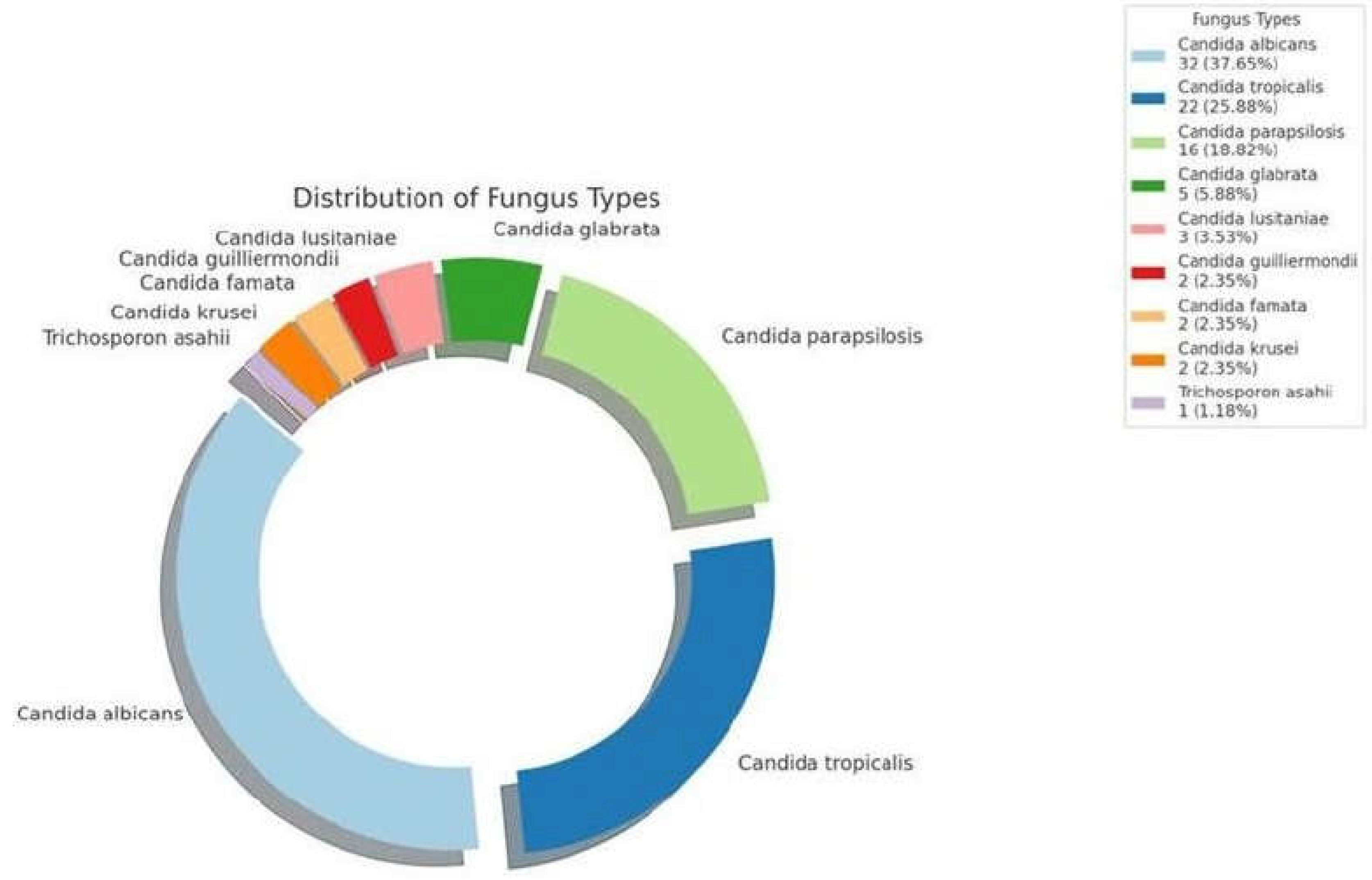
Candida albicans was the most isolated species, accounting for 32 cases (37.65%). This was followed by Candida tropicalis with 19 cases (22.35%). Other notable species included Candida parapsilosis with 14 cases (16.18%) and Candida glabrata with 5 cases (5.88%). Additionally, isolations included Candida lusitaniae (3 cases, 3.53%), Candida guilliermondii (2 cases, 2.35%), Candida famata (2 cases, 2.35%), Candida krusei (2 cases, 2.35%), and Trichosporon asahii (1 case, 1.18%). This diversity highlights the importance of careful pathogen monitoring and customized antifungal strategies in the management of pediatric fungemia. (
Figure 1)
Logistic Regression Analysis
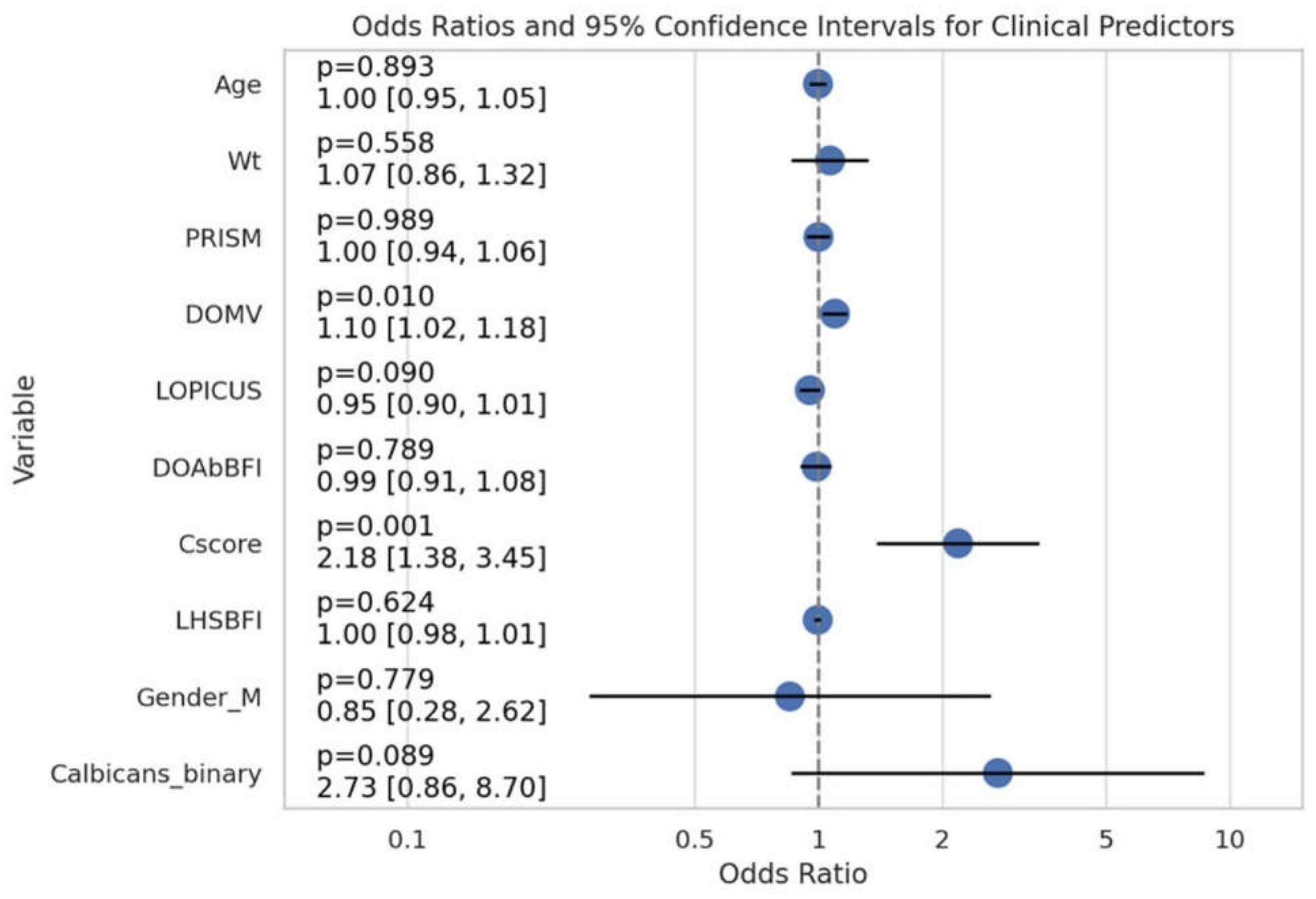
(Figure-2) The logistic regression model compared the likelihood of death (Outcome=1) and survival (Outcome=0) by analyzing the relationships between the predictor variables. Mortality was not significantly associated with age (OR: 0.997, 95% CI: 0.949-1.046, p = 0.894) or weight (OR: 1.066, 95% CI: 0.860-1.322, p = 0.558) in the study. Just like PRISM, it failed to demonstrate significance (OR: 1.000, 95% CI: 0.938-1.065, p = 0.989). Nevertheless, DOMV was determined to be a strong predictor of mortality; a 10% increase in the odds of mortality was related with each unit increase (OR: 1.096, 95% CI: 1.022-1.175, p = 0.010). Patients with a Cscore 2.18 times higher were also significantly more likely to die (95% CI: 1.383-3.452, p = 0.001). Males had an odds ratio (OR) of 0.851 (95% CI: 0.276-2.621, p = 0.779) higher than females, indicating that gender did not play a significant role in mortality. Additional variables that failed to demonstrate a significant correlation with mortality were LOPICUS, DOAbBFI, and LHSBFI. Patients with positive Candida albicans, had a 2.73-fold increased risk of death (95% CI: 0.857-8.703, p = 0.089), suggesting a trend towards statistical significance. (
Figure 3) Research into the predictive roles of DOMV, Cscore, and maybe Calbicans for mortality in this cohort is warranted considering these results.
Comparative Evaluation of Predictive Models
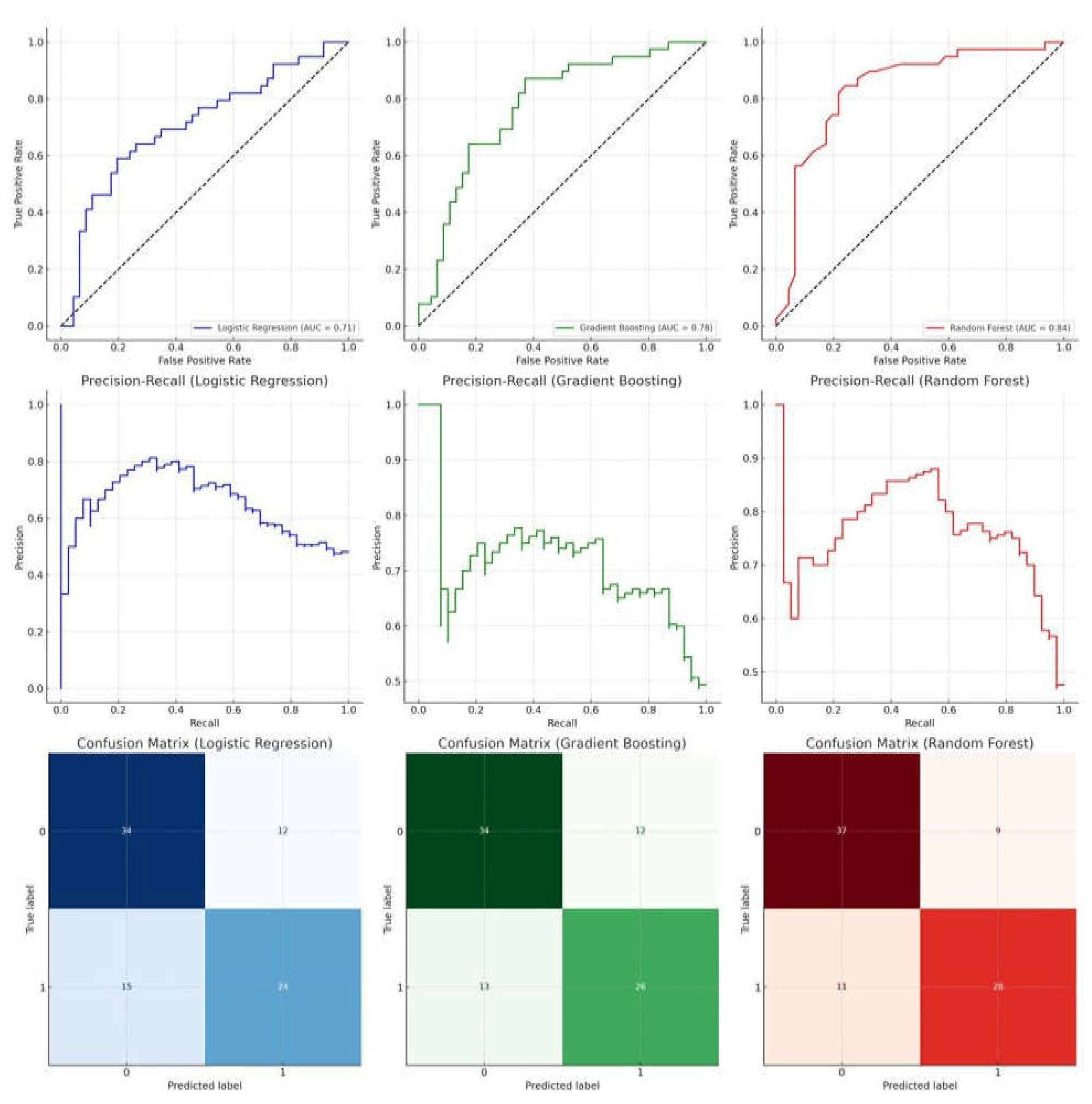
The Random Forest model outperformed the others when it came to predicting the likelihood of mortality in children hospitalized with fungemia. Its AUC of 0.8386 (with a 95% confidence interval of 0.7389 to 0.9175) was higher than the other examined models, demonstrating excellent sensitivity and specificity. Alternatively, the AUC for the Logistic Regression model was 0.7085 (95% CI: 0.5894-0.8129) and for the Gradient Boosting model it was 0.7759 (95% CI: 0.6678-0.8777). Outperforming Logistic Regression and Gradient Boosting models in predicting mortality, the Random Forest model shows promise as a strong decision support tool in pediatric intensive care settings, as evidenced by its higher AUC values, Precision-Recall curves, and Confusion Matrices. (
Figure 3)
Important Features for Predictive Models
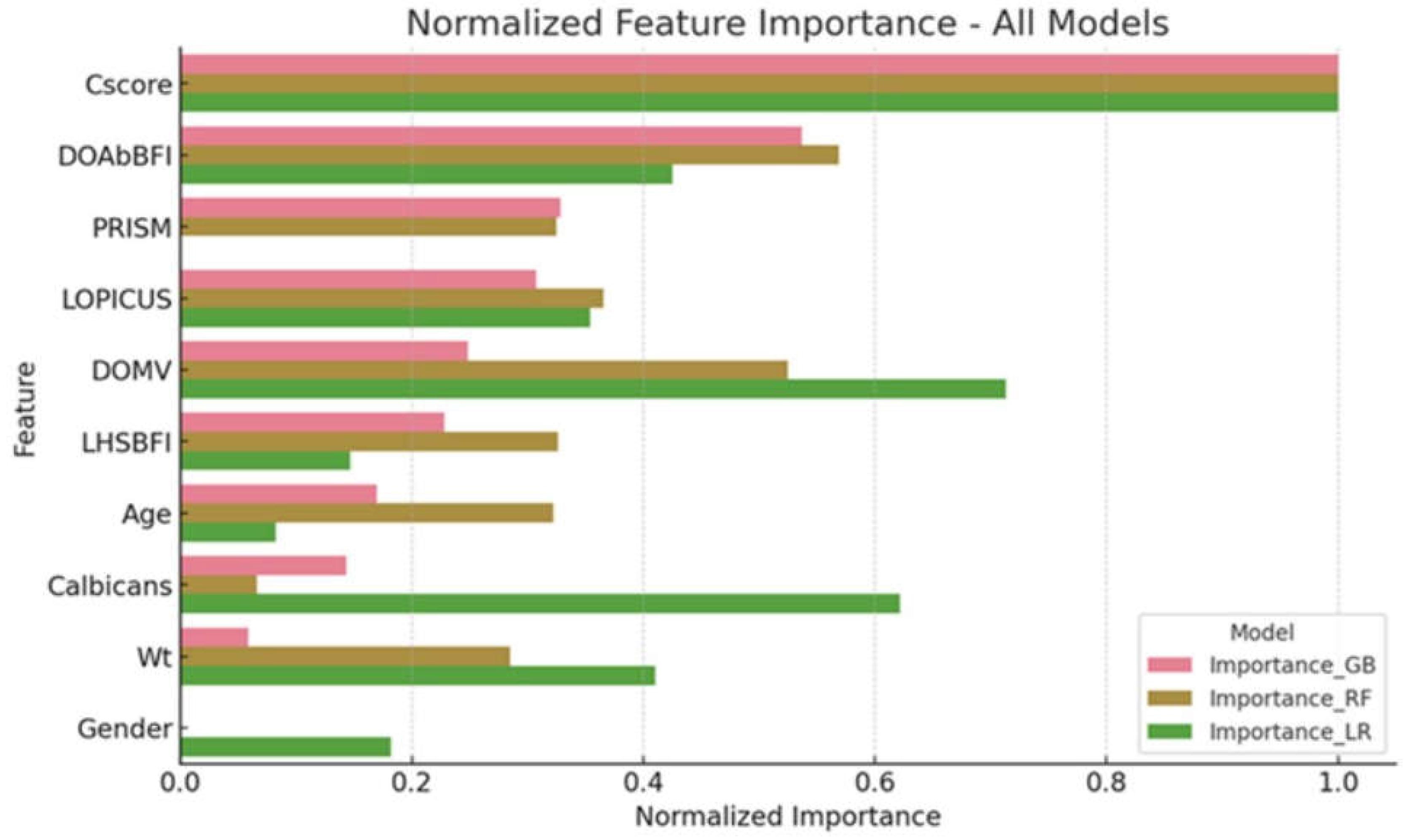
The Candida Score played a crucial impact in the Random Forest model (importance > 0.8), according to a thorough analysis of feature importance across the models. Models also differed with respect to the importance of other clinical characteristics, such as PRISM and LOPICUS scores. To improve tools for monitoring the risk of mortality from fungemia in pediatric intensive care settings, it is crucial to examine the value of features. This will shed light on the aspects that have the most impact on these models' predictive capacities. The significance of features as assessed by the Random Forest and Gradient Boosting models are summarized in the
Figure 4.
Verifying the Model
To determine whether the regression model has multicollinearity among the predictor variables, the study used the Variance Inflation Factor (VIF). As larger values may suggest multicollinearity issues, VIF values below 5 are usually considered acceptable. The VIF values for Age and Wt were 5.86 and 6.02, respectively, which is slightly higher than 5, indicating a little amount of multicollinearity. However, none of the other predictors had VIF values below 5, therefore there were no major problems with multicollinearity. The results show that the model is correct. Additionally, the Hosmer-Lemeshow goodness-of-fit test yielded a chi-squared statistic of 11.818 with 8 degrees of freedom and a p-value of 0.1595. The non-significant p-value suggests that the logistic regression model fits the data adequately. Moreover, to ensure our logistic regression model was strong and trustworthy, we used a five-fold K-Fold Cross-Validation procedure. In this method, the dataset is partitioned into five equal parts. The model is trained on four parts, which make up 80% of the data, and tested on the fifth part, which makes up 20% of the data. This way, each part is used as a testing set once during the process. Because of this, we can use every single piece of data for our training and testing. Important metrics including recall, accuracy, precision, and the Area Under the Receiver Operating Characteristic Curve (ROC-AUC) were used to assess the model's efficacy. By correctly identifying true positives and true negatives in about 72% of the instances, the logistic regression model attained an average accuracy of 71.76%, suggesting a good degree of prediction correctness. It is critical to identify patients at high risk of death in clinical settings, and the model's ability to predict right outcomes nearly 67% of the time was reflected in the stated accuracy of 67.49%. In addition, the model had a recall of 74.64%, which means it correctly identified 75% of all positive instances. This is important for clinical applications since it allows us to catch the majority of patients who are at danger. Finally, the model's ability to distinguish between survival and non-survival groups across multiple threshold settings is excellent, as indicated by the ROC-AUC of approximately 79.90%.
Discussion
High Mortality Rates in Pediatric Fungemia
Our study reports a concerning 45.9% mortality rate (39 out of 85 patients) in pediatric intensive care units due to fungemia, which significantly exceeds previous findings. This rate contrasts starkly with earlier studies: Zeng et al. (2020) reported the lowest mortality rate of 8.1% [
21] and Rajendran et al. (2016) at 20.9% [
22], and again Zeng et al. (2020) at 20.4% [
23]. Other studies such as Karacaer et al. (2014) noted a 27% mortality rate [
24], Chakrabarti et al. (2020) at 29.4% [
25], Keighley et al. (2019) at 31% [
26], Zaoutis et al. (2010) at 44% [
27], and Ghrenassia et al. (2019) reported 60% among immunocompromised patients [
28], with Dimopoulos et al. (2008) observing the highest rate of 66% [
29]. These varied figures underline the severe challenges in treating children with fungemia and the substantial differences in mortality rates across studies.
Understanding Pediatric Fungemia Risk Factors
Our research delves into the complex dynamics of pediatric fungemia in intensive care settings, uncovering a detailed landscape of risk factors and outcomes. While extensive studies have identified several contributing factors, pinpointing the exact drivers behind the severity and mortality of candidiasis remains complex. Established risk factors such as immunosuppression, steroid use, dialysis, and total parenteral nutrition (TPN) are confirmed in our findings, consistent with both adult and pediatric studies [
4,
10]. Specific to pediatrics, extended neutropenia and high-dose steroid exposure were highlighted [
11]. Our logistic regression analysis explored the impact of various demographic factors on mortality, revealing that age and weight did not significantly influence outcomes—a finding corroborated by several studies [
3,
25,
27,
30,
31], although Hegazi et al. (2014) suggest age may still be relevant in certain candida infections [
32].
Limitations of the PRISM Score in Predicting Fungemia Mortality
The PRISM (Pediatric Risk of Mortality) score is a widely recognized tool used to assess mortality risk in pediatric intensive care units (PICUs), typically calculated upon admission [
33]. While its utility is well established for initial clinical assessments, our study raises concerns about its effectiveness in predicting mortality from fungemia, an infection that usually develops during the ICU stay and beyond the acute phase initially assessed by the PRISM score. The findings from our analysis showed no significant relationship between the PRISM score and mortality related to fungemia, with an odds ratio (OR) of 1.000 and a confidence interval (95% CI) from 0.938 to 1.065 (p = 0.989), suggesting that the PRISM score may not effectively capture the risk of nosocomial infections that occur post-admission [
34].This limitation highlights the necessity for additional predictive tools that are tailored to the evolving conditions within the ICU. The Candida score, for instance, has shown significant predictive value for mortality in our cohort, underscoring its potential utility in this specific context [
35]. Integrating the Candida score or similar ICU-specific tools could improve the accuracy of mortality predictions and enable more precise clinical interventions for pediatric patients with ICU-acquired infections, potentially enhancing overall prognosis and outcomes (Vincent et al. [
36]. The adaptation of existing models or development of new models to better suit the dynamic ICU environment could bridge the gap in current predictive capabilities, ensuring that assessments remain relevant throughout the patient's ICU stay [
37]. As such, there is a pressing need to reevaluate the applicability of the PRISM score in the context of late-onset complications such as fungemia and to consider supplementary models that address these limitations [
38].
Impact of Mechanical Ventilation on Mortality Risk
The study highlights two pivotal determinants of mortality in pediatric intensive care patients with fungemia: the duration of mechanical ventilation and the Candida Score. Our analysis indicates that each additional day of mechanical ventilation correlates with an 11.5% increase in the odds of mortality (odds ratio [OR] 1.115; 95% confidence interval [CI] 1.025-1.212; p=0.011), suggesting that prolonged mechanical ventilation is a marker of severe underlying conditions or complications, thereby elevating the risk of adverse outcomes. Xiao et al. (2019) noted a significant disparity in the duration of mechanical ventilation between survivors and non-survivors, with survivors requiring shorter periods (5 days) compared to non-survivors (11.5 days) [
46]. Similarly, Paiva et al. (2016) identified mechanical ventilation as an independent predictor of mortality, with a notably high OR of 8.86 [
47]. Conversely, Singh et al. (2016) contended that the duration and complexity of mechanical ventilation do not directly impact fungal disease outcomes [
48], a view that contrasts with findings from Peres-Bota et al. (2004), who confirmed mechanical ventilation as an independent predictor of mortality in Candida infections [
49]. Supporting this, Morrell et al. (2005) suggested that longer durations of mechanical ventilation correlate with more severe Candida bloodstream infections, thus increasing mortality rates [
50].
Candida Score as a Predictor of Mortality
Our study demonstrates that a higher Candida Score significantly increases the odds of mortality, with an odds ratio of 2.205 and a confidence interval ranging from 1.359 to 3.578. This underscores the predictive utility of the Candida Score in assessing patient outcomes in pediatric ICU settings, suggesting that a higher score—often reflective of severe clinical scenarios like multifocal colonization and severe sepsis—substantially elevates mortality risk. Unlike previous research, our study introduces a predictive model that incorporates the Candida Score along with other relevant predictors specifically within a pediatric ICU context. In comparison, Juneja et al. (2022) documented a more pronounced association in an adult medical ICU, where a Candida Score greater than 3 was linked to an approximately 13.2-fold increase in mortality risk, with a wide but statistically significant confidence interval ranging from 1.3 to 125 [
51]. This points to the importance of the Candida Score in predicting outcomes, though prior models, including those by Juneja et al. (2022), have not been adapted for pediatric ICU populations, marking a significant gap that our study aims to fill.
Mortality Risks: Candida Albicans vs. Non-Albicans Candida
In the context of candidemia in pediatric intensive care units, our study identified a 2.73-fold increased risk of mortality associated with Candida albicans infections, though this did not reach statistical significance (95% CI: 0.857-8.703, p = 0.089). This finding contrasts with several other studies, which report higher mortality and morbidity rates associated with non-albicans Candida (NAC) infections. Behera et al. (2020) observed that all recorded deaths in their study were linked to NAC infections, suggesting a potentially higher virulence or resistance profile for these species [
52]. Similarly, Caggiano et al. (2017) found that infants with NAC infections experienced significantly longer stays in neonatal intensive care units compared to those infected by C. albicans, indicating more severe outcomes or complicated clinical courses with NAC infections [
53]. Furthermore, Dutta et al. (2012) reported that NAC species are often linked with higher morbidity due to their resistance to common antifungal treatments like fluconazole [
54]. However, Rajeshwari et al. (2022) found no significant difference in survival rates between infections caused by Candida albicans and non-albicans species [
55], suggesting that the impact of Candida species on mortality may vary across different study settings and populations. Tsai et al. (2017) and Filioti et al. (2007) both highlight the significant challenges posed by NAC species in managing candidemia, due to their distinct resistance profiles and associations with higher morbidity compared to C. albicans [
56,
57]. The variation in findings across these studies, including ours, underscores the complexity of candidemia in critically ill pediatric populations. It highlights the need for a nuanced understanding of the impact of different Candida species on clinical outcomes. This suggests that while Candida albicans remains a significant pathogen, the increasing prevalence and complexity of NAC species infections require tailored therapeutic strategies and highlight the importance of integrating Candida species identification into risk models for better management and prognosis of pediatric patients in intensive care settings. Our study contributes to this body of knowledge by exploring the differential impacts of Candida species on mortality, underscoring the critical need for ongoing surveillance and adaptive treatment approaches in the face of evolving fungal epidemiology.
Comparative Evaluation of Predictive Models
This study demonstrated the superior performance of the Random Forest model over Logistic Regression and Gradient Boosting models in predicting mortality among children hospitalized with fungemia. Our Random Forest model achieved an AUC of 0.8386, indicating not only high accuracy but also robustness in handling complex interactions and heterogeneity inherent in clinical data [
58,
59].The Logistic Regression model, with an AUC of 0.7085, though useful for its interpretability and simplicity, may have underperformed due to its linear nature and inability to capture more complex patterns in the data without extensive feature engineering [
60]. On the other hand, Gradient Boosting showed a moderate performance with an AUC of 0.7759, which aligns with literature suggesting its efficacy in handling varied data types and distributions, yet possibly falling short due to overfitting concerns in smaller or more imbalanced datasets [
61].The findings align with recent studies emphasizing the effectiveness of ensemble methods like Random Forest in clinical predictions, which benefit from their capacity to model non-linear interactions and their robustness against overfitting, especially in datasets with many variables and complex underlying structures [
62,
63]. Additionally, the precision-recall curves and confusion matrices suggest that Random Forest not only predicts more accurately but also maintains a balance between sensitivity and specificity, crucial for clinical decision-making in pediatric settings [
64,
65]. These results support the integration of Random Forest models into decision-support tools in pediatric intensive care, offering potentially lifesaving insights with high reliability. Future research should focus on validating these models in multicentric settings and across broader demographic groups to enhance generalizability and applicability [
66,
67].
Predictive Model Validation
Our study's logistic regression model appears robust and adequately fitted to predict clinical outcomes, as demonstrated through various statistical tests and validation techniques. The use of the Variance Inflation Factor (VIF) to assess multicollinearity among predictors revealed slightly elevated VIF values for Age and Wt (5.86 and 6.02, respectively). Although these values exceed the commonly accepted threshold of 5, suggesting mild multicollinearity, they do not indicate severe problems that could undermine the model's reliability significantly. Research suggests that VIF values up to 10 may not distort the regression estimates significantly, though they should be interpreted with caution [
68]. Furthermore, the Hosmer-Lemeshow goodness-of-fit test yielded a chi-squared statistic of 11.818 with 8 degrees of freedom and a non-significant p-value (p=0.1595), supporting the model's fit [
60]. This indicates that the model's predictions are not significantly deviant from the observed outcomes, suggesting that the model is appropriate for the data. The employment of a five-fold K-Fold Cross-Validation method enhances the model's credibility by utilizing every data segment both for training and testing, ensuring generalizability and robustness of the predictive accuracy [
69]. This method of validation is crucial in clinical predictive modeling, as it helps avoid overfitting and ensures that the model performs well across different subsets of data. Performance metrics such as recall, precision, accuracy, and ROC-AUC are crucial for evaluating the clinical utility of a predictive model. Our model demonstrated a high level of predictive accuracy with an average accuracy of 71.76%, and a precision of approximately 67.49%, indicating a good predictive balance [
70]. The recall rate of 74.64% is particularly noteworthy for clinical applications, as it implies that the model can identify many positive cases, which is essential for early intervention in clinical settings [
71]. Lastly, the ROC-AUC score of approximately 79.90% is indicative of the model’s excellent capacity to distinguish between the patient survival and non-survival across various threshold settings [
6]. Such a high AUC value confirms the model’s effectiveness in handling binary classification problems typical in medical outcome predictions. The model shows slight multicollinearity for some predictors, its overall statistical validation and cross-validation through k-fold testing confirm its robustness and reliability for clinical use. Future studies might explore the implications of multicollinearity in more depth and refine the model to enhance its predictive accuracy and precision.
Implications for Clinical Practice, Challenges and Future Directions
The study introduces a model that combines the Candida Score with key predictors to assess mortality risk in pediatric patients with fungemia, marking a significant improvement in pediatric intensive care. This approach aids in making precise predictions that can guide more effective treatments. However, integrating such predictive models into daily medical practice is challenging. The accuracy of predictions depends heavily on the quality of data, which varies significantly across different regions, especially in developing countries. Ensuring the model's seamless integration into clinical workflows without disrupting existing practices is another hurdle. Moreover, the ethical implications concerning patient privacy and data security must be addressed to prevent misuse of sensitive health information.
Clinical Implication and Future Challenges
This study develops an innovative model to enhance mortality risk assessments in pediatric patients with fungemia in intensive care units by integrating the Candida Score with crucial clinical predictors. This model promises significant clinical utility by potentially reducing fungemia-related mortality through more precise risk stratification. Early identification of high-risk patients could lead to earlier and more tailored interventions, possibly improving patient outcomes. Moreover, this approach demonstrates the feasibility of applying machine learning tools in pediatric healthcare, potentially leading to broader applications for similar predictive modeling in other complex health conditions. The future application of this model faces several hurdles that need addressing. Firstly, external validation of the model across varied healthcare settings is crucial to confirm its effectiveness and reliability beyond the study's initial environment. Secondly, integrating such predictive models into routine clinical workflows could be challenging, requiring adjustments to current practices and training for healthcare providers. Additionally, ethical concerns regarding the management of patient data, including privacy and security, are paramount as predictive modeling typically involves handling sensitive information. Lastly, ongoing adaptations of the model may be necessary to keep pace with evolving pathogens and resistance patterns, ensuring sustained relevance and accuracy.
Strengths
The study's primary strength lies in its innovative integration of the Candida Score with specific clinical predictors to refine mortality risk assessments for pediatric fungemia patients. This approach leverages the predictive power of machine learning, demonstrated by the superior performance of the Random Forest model, which notably outperforms traditional statistical methods. The rigorous methodological framework and comprehensive data analysis strengthen the study's findings, providing a robust foundation for potential clinical application. The research also fills a significant gap by addressing the need for specialized predictive tools in pediatric intensive care settings.
Weaknesses
The study's limited sample size of 85 patients may restrict the generalizability of its findings, as it represents a relatively small cohort from a single institution. This limitation could impact the model's applicability to broader pediatric populations or other clinical environments. The study's retrospective design might also introduce selection biases or influence the accuracy of data collection. Furthermore, the study has only been internally validated, necessitating further research to assess its effectiveness in external settings. The complexity of the machine learning techniques used could also pose a barrier to widespread adoption, requiring specific expertise and resources that may not be readily available in all clinical settings.
Mitigating the Weaknesses
To address these weaknesses, the study could be improved by expanding the number of participants to include a more diverse group of pediatric patients. This expansion would help in assessing the model’s effectiveness more broadly. Conducting external validation studies in various healthcare settings can also confirm the model's applicability and effectiveness across different environments. Incorporating real-time data collection could refine the model’s accuracy, making it more responsive to immediate changes in patient conditions. Lastly, simplifying the title and possibly the terminology within the article could make the findings more accessible to a wider audience, including non-specialists, thereby increasing the impact and understanding of the study’s contributions to pediatric care.
Conclusion
This study presents a novel predictive model that incorporates the Candida Score with essential clinical predictors to refine mortality risk assessments in pediatric intensive care patients with fungemia. The study revealed a concerning mortality rate of 45.9%, highlighting the critical nature of fungemia management in pediatric ICUs. The Random Forest model, with an AUC of 0.838, demonstrated superior accuracy in predicting mortality, outperforming other models due to its robust handling of complex clinical data. By adapting the Candida Score for pediatric use, the model offers an effective tool for early risk assessment. The need for external validation and clinical integration of this model is crucial, as it has the potential to improve clinical outcomes through precise and timely interventions in pediatric fungemia cases.
Acknowledgments
Khouloud Abdulrahman Al-Sofyani, MD was involved in the conceptualization and design of the study, data collection, and initial drafting of the manuscript. She also contributed significantly to the analysis and interpretation of data. Mohammed Shahab Uddin, MRCPCH(UK), FRCP (Edin, Glasgow) played a key role in the study design, was heavily involved in data interpretation, revised the manuscript critically for important intellectual content, and gave final approval of the version to be published. He also serves as the corresponding author for this paper. We would like to extend our heartfelt gratitude to the staff of the Pediatric Intensive Care Unit (PICU) for their invaluable support and assistance throughout the study. Their dedication and commitment were crucial to the successful completion of this research.
Financial Disclosures
None received.
Conflicts of Interest
None declared.
References
- Brissaud, O.; Guichoux, J.; Harambat, J.; Tandonnet, O.; Zaoutis, T. Invasive fungal disease in PICU: epidemiology and risk factors. Ann. Intensiv. Care 2012, 2, 6–6. [Google Scholar] [CrossRef] [PubMed]
- Arslankoylu, A.E.; Kuyucu, N.; Yilmaz, B.S.; Erdogan, S. Symptomatic and asymptomatic candidiasis in a pediatric intensive care unit. Ital. J. Pediatr. 2011, 37, 56–6. [Google Scholar] [CrossRef] [PubMed]
- Matic T, Novak M, Braovac D, Vinkovic M, Cicak AM, Milosevic M, et al. Characteristics, Risk Factors and Predictors for Candidemia in the Pediatric Intensive Care Unit at the University Hospital Centre Zagreb in Croatia: A 9-Year Retrospective Study. Pediatric Infectious Disease Journal. 2021 Nov 1;40(11):981–6.
- Mellinghoff, S.C.; Cornely, O.A.; Jung, N. Essentials in Candida bloodstream infection. Infection 2018, 46, 897–899. [Google Scholar] [CrossRef] [PubMed]
- Zaoutis, T.E.; Prasad, P.A.; Localio, A.R.; Coffin, S.E.; Bell, L.M.; Walsh, T.J.; Gross, R. Risk Factors and Predictors for Candidemia in Pediatric Intensive Care Unit Patients: Implications for Prevention. Clin. Infect. Dis. 2010, 51, e38–e45. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A. Nosocomial Candidiasis: Emerging Species, Reservoirs, and Modes of Transmission. Clin. Infect. Dis. 1996, 22, S89–S94. [Google Scholar] [CrossRef] [PubMed]
- Ostrosky-Zeichner, L.; Pappas, P.G. Invasive candidiasis in the intensive care unit. Crit. Care Med. 2006, 34, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Piqueras, A.; Ganapathi, L.; Carpenter, J.F.; Rubio, T.; Sandora, T.J.; Flett, K.B.; Köhler, J.R. Trends in Pediatric Candidemia: Epidemiology, Anti-Fungal Susceptibility, and Patient Characteristics in a Children’s Hospital. J. Fungi 2021, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.-F.; Lai, M.-Y.; Lee, C.-W.; Chu, S.-M.; Wu, I.-H.; Huang, H.-R.; Lee, I.-T.; Chiang, M.-C.; Fu, R.-H.; Tsai, M.-H. Comparison of the incidence, clinical features and outcomes of invasive candidiasis in children and neonates. BMC Infect. Dis. 2018, 18, 194. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues LS, Motta FA, Picharski GL, Vasconcelos TM, Riccieri MC, Dalla-Costa LM. Invasive candidiasis: risk factor for mortality in a pediatric tertiary care hospital in south of Brazil. Medicine. e: 2019;98(23), 2019.
- Fisher, B.T.; Robinson, P.D.; Lehrnbecher, T.; Steinbach, W.J.; Zaoutis, T.E.; Phillips, B.; Sung, L. Risk Factors for Invasive Fungal Disease in Pediatric Cancer and Hematopoietic Stem Cell Transplantation: A Systematic Review. J. Pediatr. Infect. Dis. Soc. 2017, 7, 191–198. [Google Scholar] [CrossRef]
- Singhi, S.M.; Rao, D.S.V.R.; Chakrabarti, A.M. Candida colonization and candidemia in a pediatric intensive care unit. Pediatr. Crit. Care Med. 2008, 9, 91–95. [Google Scholar] [CrossRef]
- Cugno, C.; Cesaro, S. Epidemiology, Risk Factors, and Therapy of Candidemia in Pediatric Hematological Patients. Pediatr. Rep. 2012, 4, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Alanazi, A. Using machine learning for healthcare challenges and opportunities. Informatics Med. Unlocked 2022, 30, 100924. [Google Scholar] [CrossRef]
- Javaid, M.; Haleem, A.; Singh, R.P.; Suman, R.; Rab, S. Significance of machine learning in healthcare: Features, pillars and applications. Int. J. Intell. Networks 2022, 3, 58–73. [Google Scholar] [CrossRef]
- Vicente, A.M.; Ballensiefen, W.; Jönsson, J.-I. How personalised medicine will transform healthcare by 2030: the ICPerMed vision. J. Transl. Med. 2020, 18, 1–4. [Google Scholar] [CrossRef]
- Savard, J. Personalised Medicine: A Critique on the Future of Health Care. J. Bioethical Inq. 2013, 10, 197–203. [Google Scholar] [CrossRef]
- Risandy, D.A.; Rusmawatiningtyas, D.; Makrufardi, F.; Herini, E.S. ; Nurnaningsih Predicting Candida Infection in Pediatric Intensive Care Unit using Candida Score in a Low-Resource Setting. Glob. Pediatr. Heal. 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-Y.; Lu, P.-L.; Wang, Y.-L.; Chen, T.-C.; Chang, K.; Lin, S.-Y. Usefulness of EQUAL Candida Score for predicting outcomes in patients with candidaemia: a retrospective cohort study. Clin. Microbiol. Infect. 2020, 26, 1501–1506. [Google Scholar] [CrossRef]
- El Zakhem, A.; El Eid, R.; Istambouli, R.; Tamim, H.; Kanj, S.S. The Utility of EQUAL Candida Score in Predicting Mortality in Patients with Candidemia. J. Fungi 2022, 8, 238. [Google Scholar] [CrossRef]
- Zeng, Z.; Ding, Y.; Tian, G.; Yang, K.; Deng, J.; Li, G.; Liu, J. A seven-year surveillance study of the epidemiology, antifungal susceptibility, risk factors and mortality of candidaemia among paediatric and adult inpatients in a tertiary teaching hospital in China. Antimicrob. Resist. Infect. Control. 2020, 9, 1–12. [Google Scholar] [CrossRef]
- Rajendran, R.; Sherry, L.; Deshpande, A.; Johnson, E.M.; Hanson, M.F.; Williams, C.; Munro, C.A.; Jones, B.L.; Ramage, G. A Prospective Surveillance Study of Candidaemia: Epidemiology, Risk Factors, Antifungal Treatment and Outcome in Hospitalized Patients. Front. Microbiol. 2016, 7, 915. [Google Scholar] [CrossRef]
- Zeng, Z.; Tian, G.; Ding, Y.; Yang, K.; Deng, J.; Liu, J. Epidemiology, antifungal susceptibility, risk factors and mortality of invasive candidiasis in neonates and children in a tertiary teaching hospital in Southwest China. Mycoses 2020, 63, 1164–1174. [Google Scholar] [CrossRef] [PubMed]
- Karacaer Z, Oncul O, Turhan V, Gorenek L, Ozyurt M. A surveillance of nosocomial candida infections: epidemiology and influences on mortality in intensive care units. Pan Afr Med J. 2014;19(1).
- Chakrabarti, A.; Sood, P.; Rudramurthy, S.M.; Chen, S.; Jillwin, J.; Iyer, R.; Sharma, A.; Harish, B.N.; Roy, I.; Kindo, A.J.; et al. Characteristics, outcome and risk factors for mortality of paediatric patients with ICU-acquired candidemia in India: A multicentre prospective study. Mycoses 2020, 63, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Keighley, C.; Chen, S.C.-A.; Marriott, D.; Pope, A.; Chapman, B.; Kennedy, K.; Bak, N.; Underwood, N.; Wilson, H.L.; McDonald, K.; et al. Candidaemia and a risk predictive model for overall mortality: a prospective multicentre study. BMC Infect. Dis. 2019, 19, 445. [Google Scholar] [CrossRef] [PubMed]
- Zaoutis, T.E.; Prasad, P.A.; Localio, A.R.; Coffin, S.E.; Bell, L.M.; Walsh, T.J.; Gross, R. Risk Factors and Predictors for Candidemia in Pediatric Intensive Care Unit Patients: Implications for Prevention. Clin. Infect. Dis. 2010, 51, e38–e45. [Google Scholar] [CrossRef] [PubMed]
- Ghrenassia, E.; Mokart, D.; Mayaux, J.; Demoule, A.; Rezine, I.; Kerhuel, L.; Calvet, L.; De Jong, A.; Azoulay, E.; Darmon, M. Candidemia in critically ill immunocompromised patients: report of a retrospective multicenter cohort study. Ann. Intensiv. Care 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dimopoulos, G.; Ntziora, F.; Rachiotis, G.; Armaganidis, A.; Falagas, M.E. Candida Albicans Versus Non-Albicans Intensive Care Unit-Acquired Bloodstream Infections: Differences in Risk Factors and Outcome. Obstet. Anesthesia Dig. 2008, 106, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Viudes A, Pemán J, Cantón E, Úbeda P, López-Ribot JL, Gobernado M. Candidemia at a tertiary-care hospital: Epidemiology, treatment, clinical outcome and risk factors for death. European Journal of Clinical Microbiology and Infectious Diseases. 2002;21(11):767–74.
- Lona-Reyes, J.C.; Gómez-Ruiz, L.M.; Cordero-Zamora, A.; Cortés-González, S.I.; Quiles-Corona, M.; Pérez-Ramírez, R.O.; Pinto-Macedo, H. Incidence and factors associated with invasive candidiasis in a neonatal intensive care unit in Mexico. 97, 86. [CrossRef]
- Hegazi, M.; Abdelkader, A.; Zaki, M.; El-Deek, B. Characteristics and risk factors of candidemia in pediatric intensive care unit of a tertiary care children's hospital in Egypt. J. Infect. Dev. Ctries. 2014, 8, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Pollack MM, Patel KM, Ruttimann UE. PRISM III: an updated Pediatric Risk of Mortality score. Crit Care Med. 1996;24(5):743-752.
- Knaus WA, Wagner DP, Draper EA, Zimmerman JE, Bergner M, Bastos PG, et al. The APACHE III prognostic system: Risk prediction of hospital mortality for critically III hospitalized adults. Chest. 1991;100(6):1619–36.
- León, C.; Ruiz-Santana, S.; Saavedra, P.; Galván, B.; Blanco, A.; Castro, C.; Balasini, C.; Utande-Vázquez, A.; de Molina, F.J.G.; Blasco-Navalproto, M.A.; et al. Usefulness of the “Candida score” for discriminating between Candida colonization and invasive candidiasis in non-neutropenic critically ill patients: A prospective multicenter study. Crit. Care Med. 2009, 37, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Vincent J-L, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. 1996;707-710.
- Moreno, R.; Vincent, J.-L.; Matos, R.; Mendonça, A.; Cantraine, F.; Thijs, L.; Takala, J.; Sprung, C.; Antonelli, M.; Bruining, H.; et al. The use of maximum SOFA score to quantify organ dysfunction/failure in intensive care. Results of a prospective, multicentre study. Intensiv. Care Med. 1999, 25, 686–696. [Google Scholar] [CrossRef]
- Suresh, S.; Varadarajan, P.; Sangareddi, S.; Gandhi, J. Pediatric index of mortality 2 score as an outcome predictor in pediatric Intensive Care Unit in India. Indian J. Crit. Care Med. 2013, 17, 288–291. [Google Scholar] [CrossRef]
- Bruyère, R.; Quenot, J.-P.; Prin, S.; Dalle, F.; Vigneron, C.; Aho, S.; Leon, C.; Charles, P.-E. Empirical antifungal therapy with an echinocandin in critically-ill patients: prospective evaluation of a pragmatic Candida score-based strategy in one medical ICU. BMC Infect. Dis. 2014, 14, 385–385. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Kumar, S.; Rajpal, K.; Sinha, R.; Kumar, R.; Muni, S.; Kumari, N. Candidemia Among ICU Patients: Species Characterisation, Resistance Pattern and Association With Candida Score: A Prospective Study. Cureus 2022, 14, e24612. [Google Scholar] [CrossRef]
- Leroy, G.; Lambiotte, F.; Thévenin, D.; Lemaire, C.; Parmentier, E.; Devos, P.; Leroy, O. Evaluation of "Candida score" in critically ill patients: a prospective, multicenter, observational, cohort study. Ann. Intensiv. Care 2011, 1, 50–50. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Kumar, S.; Rajpal, K.; Sinha, R.; Kumar, R.; Muni, S.; Kumari, N. Candidemia Among ICU Patients: Species Characterisation, Resistance Pattern and Association With Candida Score: A Prospective Study. Cureus 2022, 14, e24612. [Google Scholar] [CrossRef] [PubMed]
- Juneja, D.; Jain, R.; Singh, O.; Borah, A.K. Candida Score: a Predictor of Mortality in Patients with Candidemia. . 2022, 70, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, G.G.; Menegueti, M.G.; Auxiliadora-Martins, M.; Basile-Filho, A.; Martinez, R. Evaluation of the predictive indices for candidemia in an adult intensive care unit. Rev. da Soc. Bras. de Med. Trop. 2015, 48, 77–82. [Google Scholar] [CrossRef]
- León, C.; Ruiz-Santana, S.; Saavedra, P.; Almirante, B.; Nolla-Salas, J.; Álvarez-Lerma, F.; Garnacho-Montero, J.; León, M.A.; EPCAN Study Group. A bedside scoring system (“Candida score”) for early antifungal treatment in nonneutropenic critically ill patients with Candida colonization. Crit. Care Med. 2006, 34, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Wang, Q.; Zhu, F.; An, Y. Epidemiology, species distribution, antifungal susceptibility and mortality risk factors of candidemia among critically ill patients: a retrospective study from 2011 to 2017 in a teaching hospital in China. Antimicrob. Resist. Infect. Control. 2019, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Paiva, J.-A.; Pereira, J.M.; Tabah, A.; Mikstacki, A.; de Carvalho, F.B.; Koulenti, D.; Ruckly, S.; Çakar, N.; Misset, B.; Dimopoulos, G.; et al. Characteristics and risk factors for 28-day mortality of hospital acquired fungemias in ICUs: data from the EUROBACT study. Crit. Care 2016, 20, 1–13. [Google Scholar] [CrossRef]
- Pitoyo, C.W.; Aditianingsih, D.; Rumende, C.M. Risk factors for early invasive fungal disease in critically ill patients. Indian J. Crit. Care Med. 2016, 20, 633–639. [Google Scholar] [CrossRef]
- Peres-Bota, D.; Rodriguez-Villalobos, H.; Dimopoulos, G.; Melot, C.; Vincent, J.-L. Potential risk factors for infection with Candida spp. in critically ill patients. Clin. Microbiol. Infect. 2004, 10, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Morrell, M.; Fraser, V.J.; Kollef, M.H. Delaying the Empiric Treatment of Candida Bloodstream Infection until Positive Blood Culture Results Are Obtained: a Potential Risk Factor for Hospital Mortality. Antimicrob. Agents Chemother. 2005, 49, 3640–5. [Google Scholar] [CrossRef] [PubMed]
- Juneja, D.; Jain, R.; Singh, O.; Borah, A.K. Candida Score: a Predictor of Mortality in Patients with Candidemia. . 2022, 70, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Behera, C.; Jena, P.K.; Mishra, S.; Sahoo, B.; Patnaik, S.B.; Jain, M. Candidemia in the pediatric intensive care unit in Eastern India. J. Pediatr. Crit. Care 2020, 7, 237. [Google Scholar] [CrossRef]
- Caggiano, G.; Lovero, G.; De Giglio, O.; Barbuti, G.; Montagna, O.; Laforgia, N.; Montagna, M.T. Candidemia in the Neonatal Intensive Care Unit: A Retrospective, Observational Survey and Analysis of Literature Data. BioMed Res. Int. 2017, 2017, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Zaoutis, T.E.; Palazzi, D.L. An Update on the Epidemiology of Candidemia in Children. Curr. Fungal Infect. Rep. 2012, 6, 296–302. [Google Scholar] [CrossRef]
- Karuppusami, R.; Rajeshwari, R.; Vyasam, S.; Chandran, J.; Porwal, S.; Ebenezer, K.; Thokchom, M.; James, E.J. Risk Factors for Candida Infection among Children Admitted to a Pediatric Intensive Care Unit in a Tertiary Care Centre in Southern India. Indian J. Crit. Care Med. 2022, 26, 717–722. [Google Scholar] [CrossRef]
- Tsai, M.-H.; Hsu, J.-F.; Chu, S.-M.; Chang, P.-J.; Lai, M.-Y.; Wu, I.-H.; Huang, H.-R.; Chiang, M.-C.; Fu, R.-H.; Lu, J.-J. Clinical and microbiological characteristics, and impact of therapeutic strategies on the outcomes of children with candidemia. Sci. Rep. 2017, 7, 1083. [Google Scholar] [CrossRef] [PubMed]
- Filioti, J.; Spiroglou, K.; Roilides, E. Invasive candidiasis in pediatric intensive care patients: epidemiology, risk factors, management, and outcome. Intensiv. Care Med. 2007, 33, 1272–1283. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach Learn. 2001;45:5-32.
- Caruana R, Niculescu-Mizil A. An empirical comparison of supervised learning algorithms. In: Proceedings of the 23rd International Conference on Machine Learning. 2006:161-168.
- Hosmer, D.W.; Hosmer, T.; LE Cessie, S.; Lemeshow, S. A COMPARISON OF GOODNESS-OF-FIT TESTS FOR THE LOGISTIC REGRESSION MODEL. Stat. Med. 1997, 16, 965–980. [Google Scholar] [CrossRef]
- Natekin A, Knoll A. Gradient boosting machines, a tutorial. Front Neurorobot. 2013;7:21.
- Biau, G. Analysis of a random forests model. J Mach Learn Res. 2012;13(1):1063-1095.
- Chen, X.; Ishwaran, H. Random forests for genomic data analysis. Genomics 2012, 99, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, T. An Introduction to ROC analysis. Pattern Recogn. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Saito, T.; Rehmsmeier, M. The Precision-Recall Plot Is More Informative than the ROC Plot When Evaluating Binary Classifiers on Imbalanced Datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar] [CrossRef] [PubMed]
- Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: A framework for traditional and novel measures. Epidemiology. 2010; 21:128-138.
- Collins, G.S.; Moons, K.G.M. Reporting of artificial intelligence prediction models. Lancet 2019, 393, 1577–1579. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, R.M. A Caution Regarding Rules of Thumb for Variance Inflation Factors. Qual. Quant. 2007, 41, 673–690. [Google Scholar] [CrossRef]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In: Proceedings of the 14th International Joint Conference on Artificial Intelligence (IJCAI). 1995;14(2):1137-1145.
- Fawcett, T. An Introduction to ROC analysis. Pattern Recogn. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Altman DG, Bland JM. Diagnostic tests. 1: Sensitivity and specificity. BMJ. 1994;308(6943):1552. [CrossRef]
Figure 1.
Fungal Pathogen Distribution in Pediatric Intensive Care Fungemia Cases. The Donut chart illustrates the proportion of different fungal pathogens isolated from a cohort of 85 pediatric patients diagnosed with fungemia in the intensive care unit between 2016 and 2020, presented with frequency and percentage of each type of Fungus.
Figure 1.
Fungal Pathogen Distribution in Pediatric Intensive Care Fungemia Cases. The Donut chart illustrates the proportion of different fungal pathogens isolated from a cohort of 85 pediatric patients diagnosed with fungemia in the intensive care unit between 2016 and 2020, presented with frequency and percentage of each type of Fungus.
Figure 2.
Odds Ratios and Confidence Intervals for Clinical Predictors of Outcomes. This figure illustrates the odds ratios and their corresponding 95% confidence intervals for various clinical and demographic predictors influencing the outcome, as determined by logistic regression analysis. Each horizontal line represents the confidence interval for a predictor, with the point denoting the odds ratio. Labels on the extreme left display the p-value, the odds ratio, and the confidence interval range for each predictor. The vertical dashed line at odds ratio 1 indicates no effect.
Figure 2.
Odds Ratios and Confidence Intervals for Clinical Predictors of Outcomes. This figure illustrates the odds ratios and their corresponding 95% confidence intervals for various clinical and demographic predictors influencing the outcome, as determined by logistic regression analysis. Each horizontal line represents the confidence interval for a predictor, with the point denoting the odds ratio. Labels on the extreme left display the p-value, the odds ratio, and the confidence interval range for each predictor. The vertical dashed line at odds ratio 1 indicates no effect.
Figure 3.
Diagnostic and Predictive Performance of Machine Learning Models in PICU Fungemia Risk Assessment. ROC curves, Precision-Recall curves, and Confusion Matrices for Logistic Regression, Gradient Boosting, and Random Forest are displayed. Random Forest shows superior performance with an AUC of 0.86 compared to Logistic Regression (AUC = 0.72) and Gradient Boosting (AUC = 0.79). The model demonstrates high sensitivity and specificity, with favorable precision-recall balance, making it a more effective tool for mortality risk prediction in pediatric intensive care. Confusion Matrices highlight Random Forest's accuracy and lower misclassification rate, supporting its use as a robust decision support system in improving patient management and outcomes.
Figure 3.
Diagnostic and Predictive Performance of Machine Learning Models in PICU Fungemia Risk Assessment. ROC curves, Precision-Recall curves, and Confusion Matrices for Logistic Regression, Gradient Boosting, and Random Forest are displayed. Random Forest shows superior performance with an AUC of 0.86 compared to Logistic Regression (AUC = 0.72) and Gradient Boosting (AUC = 0.79). The model demonstrates high sensitivity and specificity, with favorable precision-recall balance, making it a more effective tool for mortality risk prediction in pediatric intensive care. Confusion Matrices highlight Random Forest's accuracy and lower misclassification rate, supporting its use as a robust decision support system in improving patient management and outcomes.
Figure 4.
Normalized Feature Importance Across Mortality Prediction Models in PICU. This figure compares feature importance in Logistic Regression (LR), Gradient Boosting (GB), and Random Forest (RF) models. Key predictors include Candida Score (CScore), most prominent in RF (importance > 0.8), PRISM, and LOPICUS scores, showing varied significance. Lesser predictors such as age and gender in LR, and moderate predictors like weight (Wt) and C. albicans in GB and RF, are also assessed. This visualization highlights the relative impact of each predictor on mortality risk assessment in pediatric intensive care, with RF notably valuing comprehensive clinical parameters, essential for enhancing prognostic accuracy in critical care.
Figure 4.
Normalized Feature Importance Across Mortality Prediction Models in PICU. This figure compares feature importance in Logistic Regression (LR), Gradient Boosting (GB), and Random Forest (RF) models. Key predictors include Candida Score (CScore), most prominent in RF (importance > 0.8), PRISM, and LOPICUS scores, showing varied significance. Lesser predictors such as age and gender in LR, and moderate predictors like weight (Wt) and C. albicans in GB and RF, are also assessed. This visualization highlights the relative impact of each predictor on mortality risk assessment in pediatric intensive care, with RF notably valuing comprehensive clinical parameters, essential for enhancing prognostic accuracy in critical care.
Table 1.
Clinical and Demographic Characteristics by Outcome in Pediatric ICU Patients. This table compares median and interquartile ranges for age, weight (Wt), Pediatric Risk of Mortality (PRISM) scores, duration of mechanical ventilation (DOMV), PICU stay length (LOPICUS), pre-fungal infection antibiotic duration (DOAbBFI), fungal score (Cscore), Candida Albicans presence (Calbicans), and hospital stay length before fungal infection (LHSBFI) between survivors (Outcome=0) and non-survivors. Non-parametric tests assess statistical significance, with p-values highlighting key outcome determinants in pediatric critical care.
Table 1.
Clinical and Demographic Characteristics by Outcome in Pediatric ICU Patients. This table compares median and interquartile ranges for age, weight (Wt), Pediatric Risk of Mortality (PRISM) scores, duration of mechanical ventilation (DOMV), PICU stay length (LOPICUS), pre-fungal infection antibiotic duration (DOAbBFI), fungal score (Cscore), Candida Albicans presence (Calbicans), and hospital stay length before fungal infection (LHSBFI) between survivors (Outcome=0) and non-survivors. Non-parametric tests assess statistical significance, with p-values highlighting key outcome determinants in pediatric critical care.
| Variable |
Survive |
Mortality |
P value |
| N |
46 |
39 |
|
| Age (median [IQR]) |
7.00 [2.00, 20.25] |
4.00 [2.00, 11.00] |
0.362 |
| Gender = Male (%) |
29 (63.0) |
24 (61.5) |
1.000 |
| Wt (median [IQR]) |
5.00 [3.41, 8.00] |
4.80 [3.60, 6.80] |
0.958 |
| PRISM (median [IQR]) |
9.50 [5.00, 13.00] |
10.00 [6.50, 15.50] |
0.422 |
| DOMV (median [IQR]) |
11.00 [1.50, 24.00] |
20.00 [11.50, 30.00] |
0.011** |
| LOPICUS (median [IQR]) |
25.50 [14.00, 35.75] |
29.00 [18.50, 38.00] |
0.514 |
| DOAbBFI (median [IQR]) |
13.00 [10.00, 16.00] |
18.00 [10.00, 21.50] |
0.024* |
| Cscore (median [IQR]) |
2.00 [1.00, 2.00] |
4.00 [2.00, 4.00] |
<0.001*** |
| Candida albicans (%) |
14 (30.4) |
20 (51.3) |
0.083 |
| Non-Albicans (%) |
32(69.6) |
19(48.7) |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).