1. Introduction
The regulatory role of nutrients on the secondary metabolism in
Pseudomonas spp. has been researched mainly to provide data on enhancing the production of the most valuable metabolites – antimicrobials [
1]. The broader research showed, however, that the availability of the carbon sources influences the secondary metabolism of pseudomonads in a complex compound- and strain-dependent manner. In
Pseudomonas sp. F113 (now classified as a novel species of
Pseudomonas ogarae sp. nov. nom. rev. [
2]), the production of 2,4-diacetylphloroglucinol (2,4-DAPG) was elevated in the presence of sucrose and fructose while reduced in the presence of glucose [
3]. However, a study by Duffy and Défago (1999) [
4] on various strains of
P. fluorescens (currently some of them classified as
P. protegens) revealed that this relationship is strain-dependent, as some of the strains included in their work exhibited a glucose-stimulated increase in 2,4-DAPG yield. Thus, more studies investigated the inconsistent regulatory nature of carbon sources in producing various secondary metabolites by
Pseudomonas spp. [
5,
6,
7,
8].
Among the most studied carbon sources in the context of
Pseudomonas spp. secondary metabolism are glucose and glycerol [
5,
6,
7,
8,
9,
10]. Glucose is one of the primary energy sources for most living organisms and is the most ubiquitous carbohydrate on Earth.
Pseudomonas spp. metabolize glucose efficiently; however, when the environment is abundant in organic acids or amino acids, they will be metabolized primarily by pseudomonads due to carbon catabolic repression (CCR) [
11].
The second mentioned carbon source, glycerol, is a common organic compound, a simple polyol primarily found in nature as a component of glyceride lipids. It can be assimilated as a carbon source by most Gram-negative bacteria, including
Pseudomonas spp. The primary glycerol metabolism in pseudomonads has been researched, mainly utilizing
P. putida KT2440 as a model [
12,
13]. Recently, our study revealed a significant relationship between these two carbon sources and
P. donghuensis P482 antibacterial activity governed by 7-hydroxytropolone (7-HT) [
14].
7-HT is the most often reported metabolite of
P. donghuensis; it has been described as a broad-range antimicrobial, nematicide, and iron scavenger [
15,
16,
17]. Its biosynthesis has been linked to a defined gene cluster, the inactivation of which attenuated all the
P. donghuensis traits attributed to 7-HT; as such, mutants did not produce this compound under rich medium conditions [
17,
18]. Likewise, the importance of 7-HT was also reported for the
P. donghuensis HYS and SVBP6 strains [
16,
19].
In our earlier studies, we demonstrated that inactivation of the genes involved in the synthesis of 7-HT in
P. donghuensis strain P482 entirely abolished the antibacterial activity of this strain when tested against the pathogens
Dickeya solani and
Pectobacterium brasiliense on a non-defined-rich-medium (LB) [
17]. However, when nutrient availability was limited, and glucose was the only carbon source in the minimal medium, the effect of mutations on the antibacterial activity, surprisingly, was not as distinct as in the rich medium, and the mutants retained nearly half of the P482 wild type (wt) activity [
14]. Moreover, the presence of glycerol in the minimal medium allowed the mutant strains to keep the antibacterial activity at the level of P482 wt.
Disruption of the genes within another P482 cluster identified, 'cluster 17', had a diverse impact on the inhibition of pathogen growth by P482, also dependent on the carbon source present in the tested medium and the type of pathogen used in the antibiosis assays [
14]. Mutants in this cluster maintained antibacterial activity when glucose was present in the assay medium, but the activity was substantially diminished in the presence of glycerol.
This study aimed to evaluate the effects of the carbon source on the production of 7-HT by P. donghuensis P482 strain. Metabolomic analyses revealed the profiles of the secondary metabolites produced by P482 wt and its selected mutants, grown in nutrient-diluted (10% tryptone soy broth, TSB) or nutrient-restricted (M9 with either glucose or glycerol as a sole carbon source) media.
2. Results and Discussion
Under
in vitro conditions, the secondary metabolism of
Pseudomonas spp. has been repeatedly shown to be influenced by nutrients and carbon sources in culture media [
20]. Although the diverse effects of glucose and glycerol utilization on producing valuable secondary metabolites by
Pseudomonas spp. are indisputable, they have not been explored broadly for
P. donghuensis.
2.1. Complex Nutrient Availability Supports the Synthesis of Metabolites with Antimicrobial Activity
Our earlier analyses concerning nutritional dependencies of the secondary metabolism of
P. donghuensis P482 showed that, when tested in a minimal M9 medium supplemented with a single carbon source (restricted nutritional conditions), the carbon sources impact the antibacterial activity of P482 [
14]. In the presence of glucose, mutants affected in the synthesis of 7-HT exhibited a significantly decreased ability to inhibit pathogen growth. However, when glycerol served as a single carbon source, the 7-HT mutants efficiently suppressed the pathogens' growth, reflecting the activity of the P482 wild-type strain.
Interestingly, we found that on undefined nutrient-diluted medium (agar-solidified 0.1 TSB), the tested mutants showed only traces (KN4705, KN4706) or no (KN4709) activity towards
P. syringae pv.
syringae Pss762, and none of them inhibited the growth of
D. solani IFB0102 (
Supplementary Figure S1). The 'cluster 17' mutants (KN4240 and KN4243), on the other hand, showed comparable or slightly reduced antibacterial activity (as in the case of strain KN4240), or heavily reduced activity (up to -60% as in the case of strain KN4243) compared to the wt strain against both tested pathogens (
Supplementary Figure S1). We observed similar levels of antibacterial activity of these two mutants on the minimal M9 medium supplemented with glucose, but they were very little active when glycerol was the sole carbon source [
14].
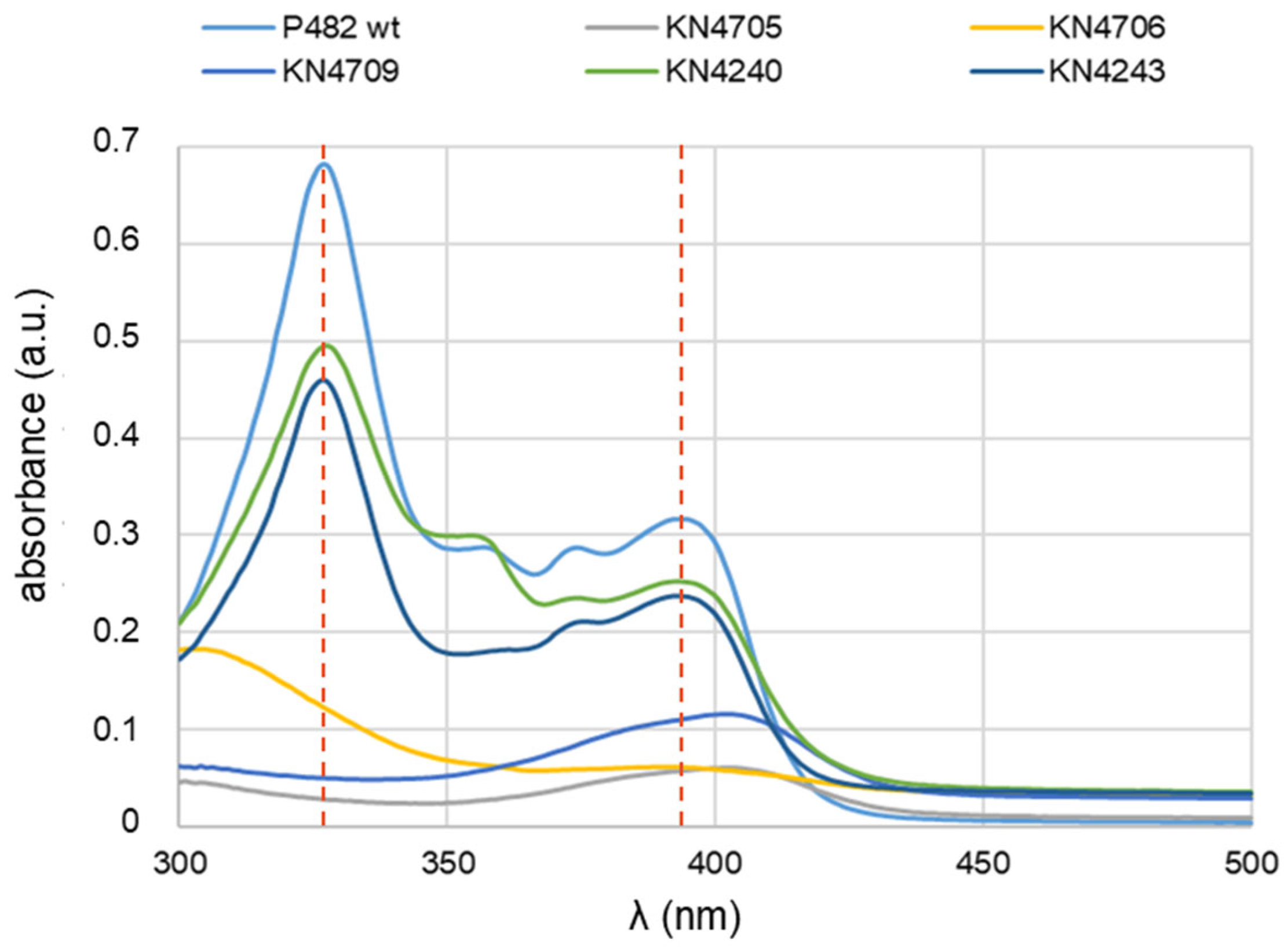
2.2. UV-Vis Spectra of Post-Culture P482 Strains Supernatants Confirmed the Absence of 7-HT in KN4705, KN4706 and KN4709 Strains Grown in 0.1 TSB
The UV-Vis spectra of post-culture supernatants reveal metabolic differences between
P. donghuensis strains. The study by Chen et al. (2018a) [
18] demonstrated the presence of two peaks that correspond to the iron scavenger 7-HT: at 330 nm and 392 nm, in the UV-Vis absorption spectrum of the supernatant obtained from
P. donghuensis HYS
T culture in King's B medium [
16]. To verify the presence of the 7-HT in the nutrient-diluted medium, the UV-Vis absorption spectra in the wavelength range 300-500 nm were obtained from post-culture supernatants of the P482 strains, both wt strain and its mutants, grown in 0.1 TSB for 24 h. The expected spectra for 7-HT were obtained only for P482 wt and two mutants, KN4240 and KN4243 (
Figure 1).
They contain two peaks at wavelengths 327 nm and 393 nm (a slightly shifted spectrum of 7-HT), pointing to the lack of influence of these mutations on the production of 7-HT under the tested conditions. The spectrophotometry outcome for KN4240 and KN4243 in 0.1 TSB (
Figure 1) differed substantially from that in the minimal M9 medium [
14], where no peaks at 330 or 327 nm were recorded, regardless of the carbon source used. Thus, no 7-HT biosynthesis was detected in the minimal medium. The gene in the BV82_4243 locus might be linked to the 7-HT secretion [
14], since it encodes part of an efflux pump – a homologue of the EmrA protein. It was shown for
Salmonella typhimurium subjected to several antibacterial compounds that the activity of efflux pumps involved in expelling antibiotics from the cells was affected by the presence of different carbon sources in the growth media [
21]. A similar regulation mechanism might also play a role in
P. donghuensis strain P482 when secondary metabolites need to be secreted outside the cells through an efflux pump.
As expected, the mutants KN4705, KN4706, and KN4709 did not produce 7-HT in 0.1 TSB medium at a detectable level, and no peaks characteristic for 7-HT were observed. In the case of these mutants, the results also agree with those obtained for direct antibiosis tests towards bacterial plant pathogens
D. solani IFB0102 and
P. syringae pv.
syringae Pss762 described in the earlier paragraph. The antibacterial activity of these mutants was nearly or entirely abolished (
Supplementary Figure S1). The lack of growth inhibition zones on the agar-solidified 0.1 TSB medium might, therefore, be attributed to the lack of production of 7-HT by KN4705, KN4706, and KN4709 mutants.
2.3. LC-MS Analyses Confirmed the Lack of 7-HT Biosynthesis by P482 in the Nutrient-Restricted Medium
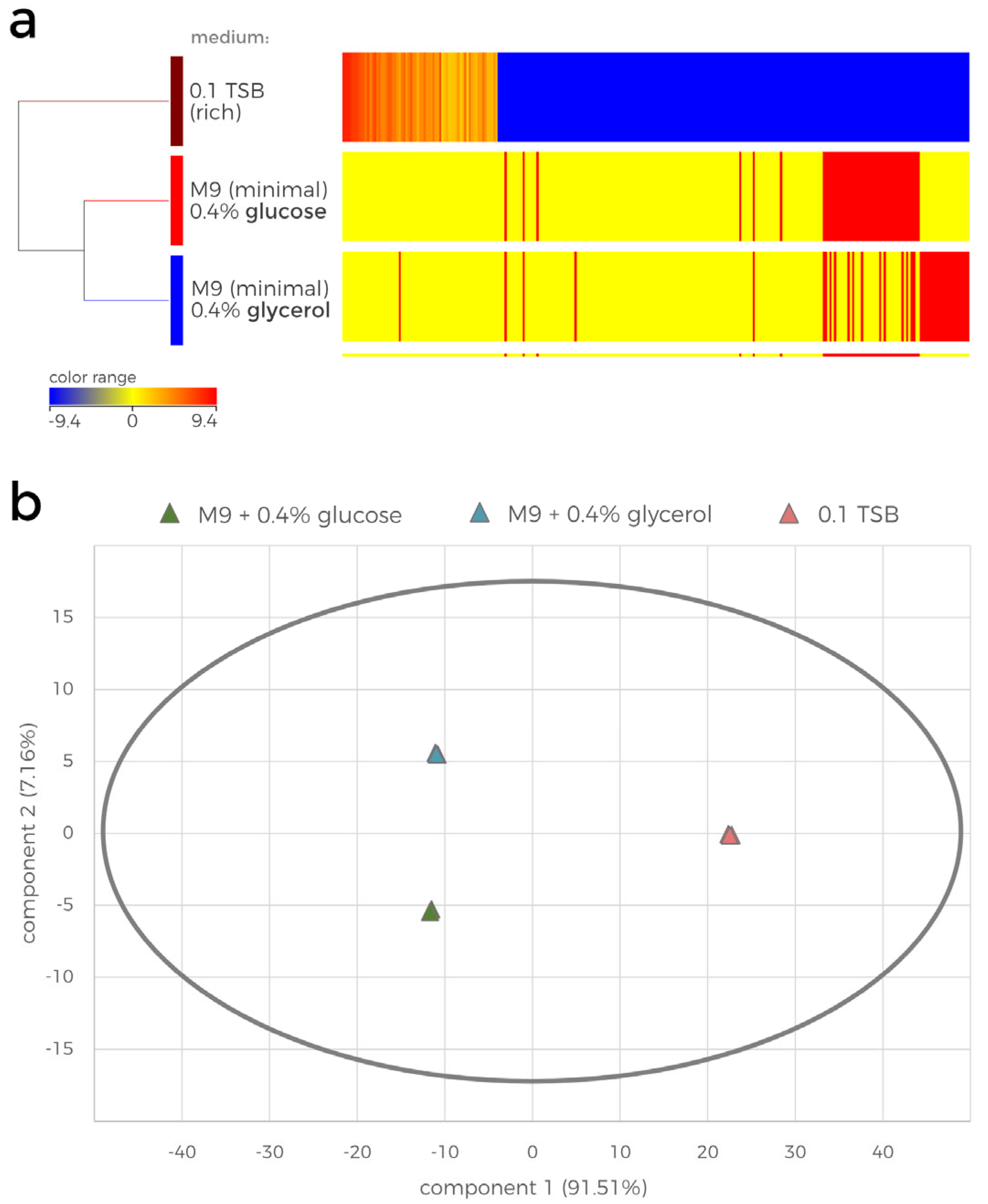
Post-culture supernatants of P482 wt obtained in undefined nutrient-diluted (0.1 TSB) or in nutrient-restricted (M9 with either 0.4% glucose or 0.4% glycerol as a sole carbon source) media were subjected to LC-MS analysis. The 0.1 TSB medium was chosen here as the production of the crucial metabolite, 7-HT, was most efficient and reflected the highest antibacterial activity of P482 strain. Overall, the LC-MS analysis of the extracts showed that the growth media influenced the P482 metabolome (
Supplementary Figure S2). The number and retention time of the peaks varied depending on whether P482 was cultured in a nutrient-diluted undefined 0.1 TSB medium or nutrient-restricted M9 minimal medium. In contrast, the carbon source (glucose or glycerol) in the M9 medium showed a minor impact on the P482 metabolome (
Supplementary Figure S2).
The LC-MS data were further subjected to statistical analysis (
Figure 2). The hierarchically clustered heatmap shows the results of the ANOVA analysis, revealing the growth medium-dependent log2 fold change (FC) in the normalized abundances of the compounds detected in the samples (
Figure 2a). The heatmap visualizes the striking differences between the contents of the tested extracts. Overall, 205 compounds were detected in the 0.1 TSB-cultured P482 extract, which were also present in the minimal media samples. Furthermore, 22 and 30 unique compounds were detected in the glycerol and glucose culture extracts, respectively, which did not appear to be present in the 0.1 TSB extracts (Supplementary dataset_1). The principal component analysis (PCA) additionally confirmed the statistical differences between the overall metabolite profiles of the samples (
Figure 2b). The first component (x-axis, 91.51% of total variance) distinguishes the effect of the rich medium from the impact of minimal media on the metabolome of P482. The second component (y-axis, 7.16% of total variance) further separates the minimal media extracts by the difference in the P482 metabolome caused by the carbon source (glucose/glycerol). The variation between the biological replicates on the PCA plot is insignificant, suggesting there is no observable effect of other, possibly uncontrollable conditions of the experiment.
The automated and manual database search could structurally match very few detected compounds. Additionally, most automatically matched bioactive compounds have never been reported in
Pseudomonas spp. Of these automatically annotated metabolites, only two compounds have been previously described as antimicrobials of pseudomonads: 7-HT (
m/
z 139) [
16] and zafrin (
m/
z 213) [
22]. 7-HT is known to be a metabolite of
P. donghuensis and can be additionally detected with UV-Vis spectrophotometry. Only 7-HT, which was hypothesised to contribute significantly to P482 antimicrobial activity, was evaluated in the following analyses.
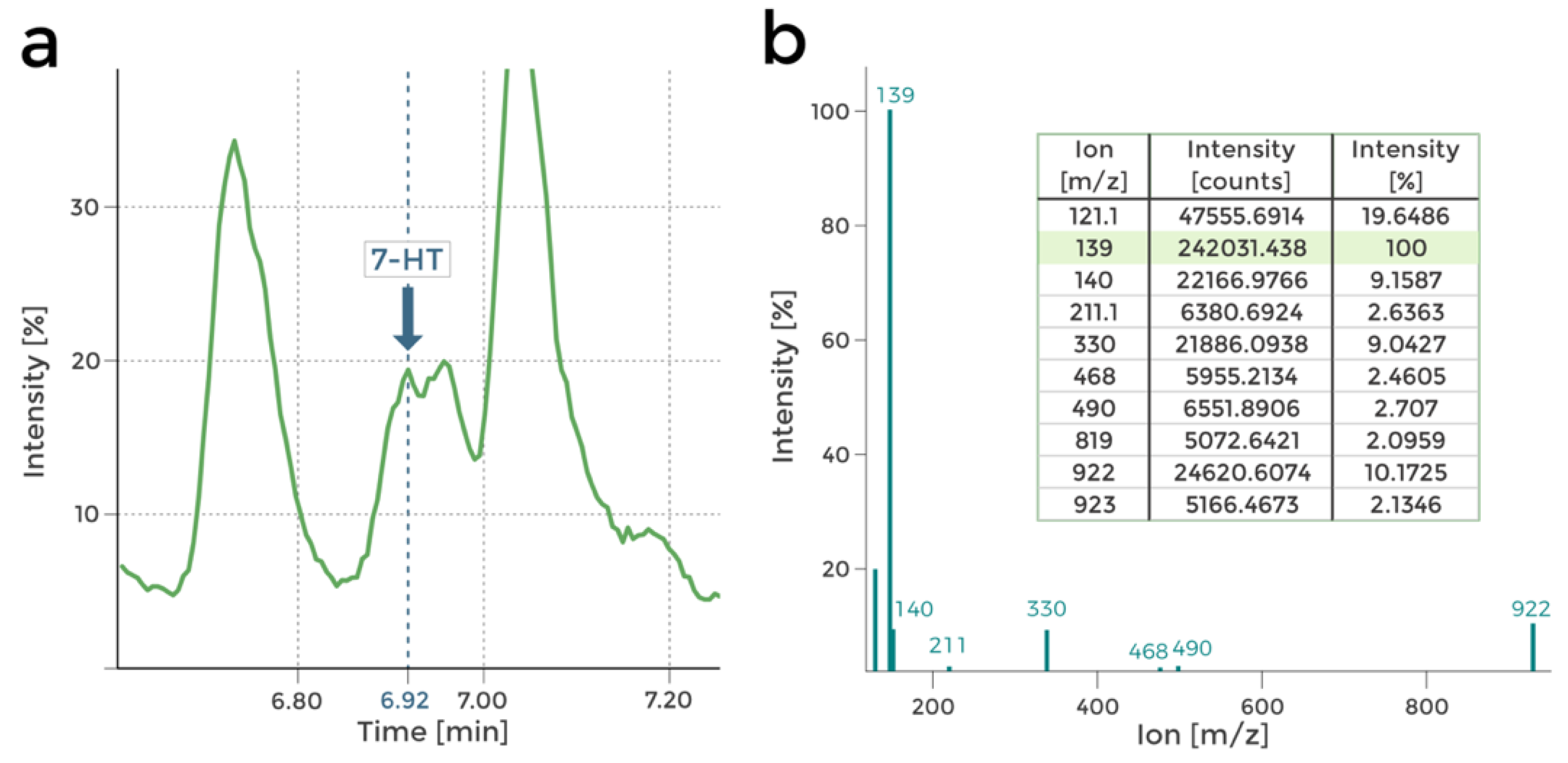
The presence of 7-HT in the tested P482 wt post-culture extracts was assessed using LC-MS analyses. The UV-Vis results obtained for the culture supernatants of P482 wt suggested that while 7-HT is indeed produced under rich or nutrient-diluted medium conditions (
Figure 1), its biosynthesis is inhibited in minimal media, regardless of the carbon source used (glucose or glycerol) [
14]. The LC-MS investigation confirmed this hypothesis. Peaks in the mass spectra corresponding to 7-HT were found in the chromatograms obtained in 0.1 TSB-grown P482 wt post-culture extracts (nutrient-diluted medium), as reported in
Figure 3.
Moreover, no 7-HT presence was detected in the extracts from P482 wt grown in M9 minimal media with either glucose or glycerol as the carbon source. This outcome reinforces the assumption that P482 requires nutrients other than those included in the M9 medium formulation or a specific molecular signalization to biosynthesize 7-HT.
To summarize, the analysis established the important effect of the nutritional environment on the metabolome of P482 strain, while the single carbon source as a component of a minimal medium did not prove to be as contributory. Moreover, 7-HT was found to be produced by P482 only in the rich (LB, TSB and nutrient-diluted, 0.1 TSB), undefined media (as found here and in [
14]).
The metabolomic analysis of P482 wt extracts produced in three distinct media (0.1 TSB, M9 supplemented with glucose or M9 supplemented with glycerol) revealed substantial disparities in secondary microbial metabolism. This visualization underscores the stark contrast between the metabolic profiles observed in environments abundant in nutrients and those characterized by nutritional limitations. These results show that P482 produces numerous compounds in a nutrient-rich environment, where all the essential precursors are present, and the energy supply is sufficient for more than the primary metabolism [
23,
24,
25]. The analysis also reveals the difficulties of investigating the effects of a single carbon source on the bacterial metabolome. While minimal media offer limited nutrients, hindering the activation of robust secondary metabolism, the complexity of media complicates direct comparisons of carbon source effects on microbial phenotype. Nevertheless, the fact that the difference between glucose and glycerol as a sole carbon source does not significantly affect the P482 metabolome builds on the comprehensive analysis. Van der Werf (2008) [
26] study on
P. putida S12 showed that the metabolic profiles after S12 growth on various carbon sources are very similar, mainly differing in the concentration of several detected compounds. Moreover, a study on the metabolome of
P. aeruginosa showed that the growth under minimal nutritional conditions engages the metabolism of bacteria mostly in pathways directly related to the utilisation of the given carbon source, while the secondary metabolism is reduced [
24].
2.4. Mutations in Four of P482 key Antimicrobial Loci Have an Unpredicted Influence on the Strain Metabolome
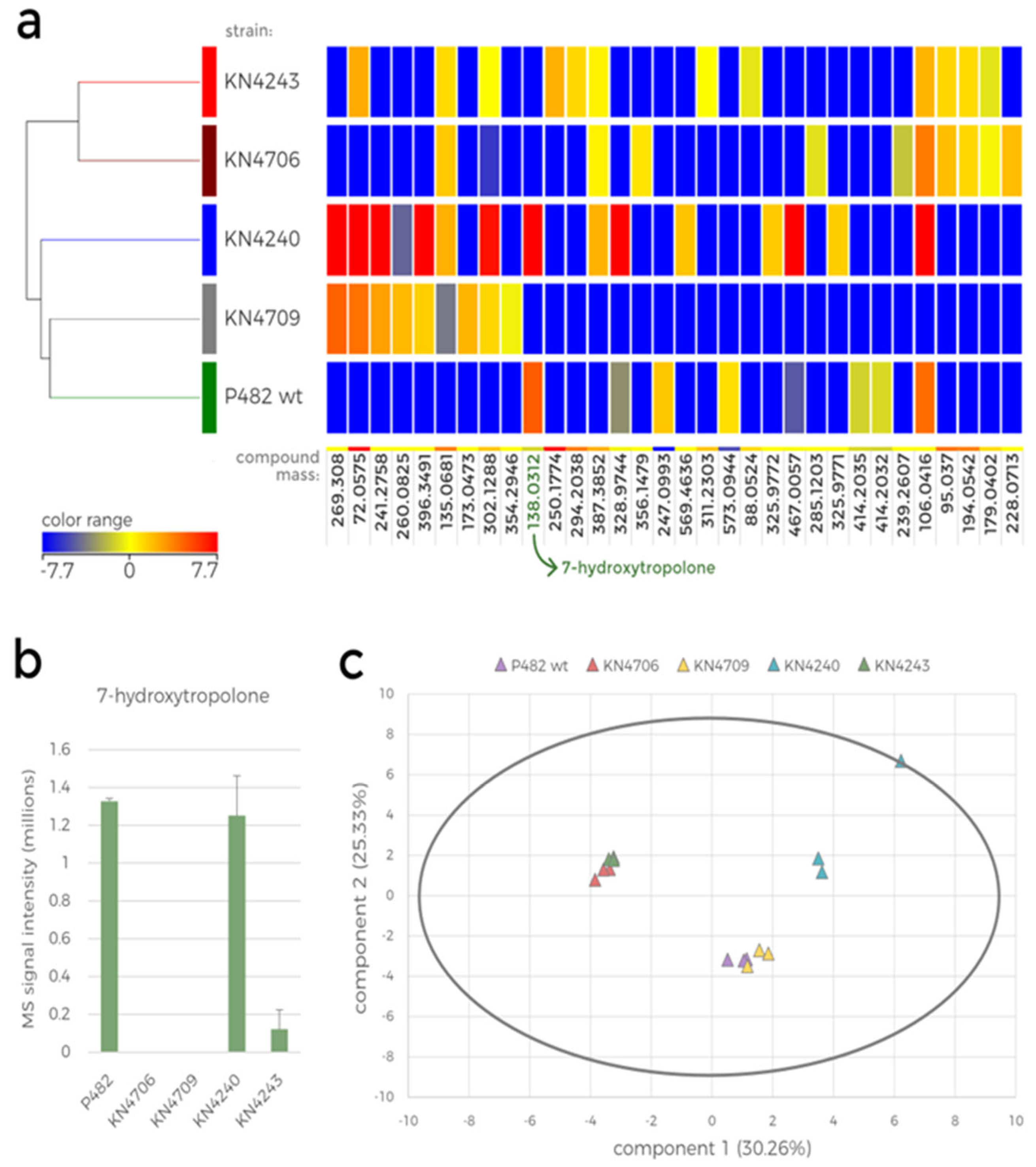
To test whether and how the mutations in the 7-HT biosynthesis cluster and 'cluster 17' genes affect the metabolome of P482, the extracts from the 0.1 TSB post-culture supernatants of P482 wt, and of its mutants (KN4706, KN4709, KN4240, and KN4243) were analysed by LC-MS.
Firstly, metabolic differences between the mutants can be observed by comparing their HPLC chromatograms (
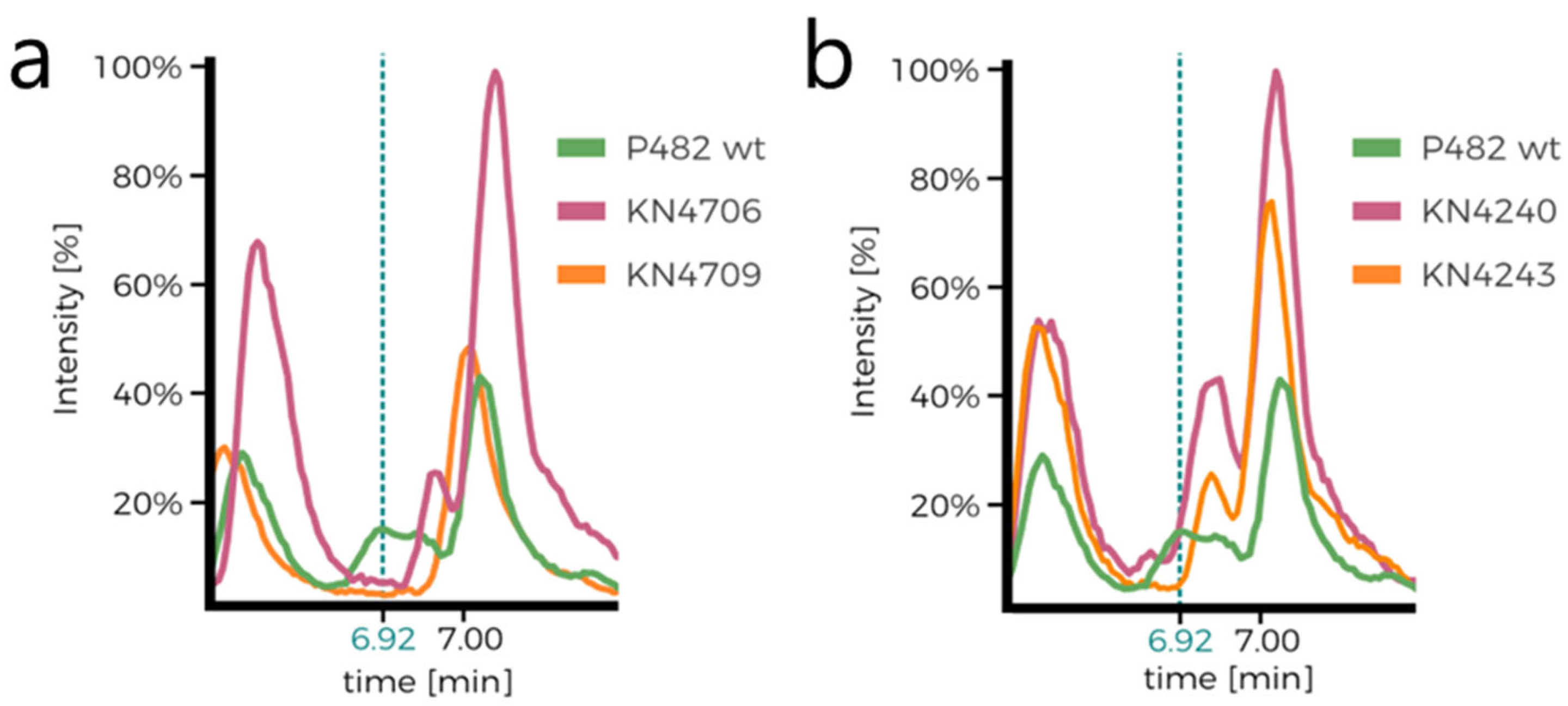
Supplementary Figure S3a,b). Comparison of the chromatograms of P482 wt and the 7-HT mutants (KN4706, KN4709) is the first confirmation of 7-HT biosynthesis ability loss in the mutants (no peak at approx. 6.92 min;
Figure 4a,b). The only other difference between the mutants seems to lie in the ratios of normalised concentrations of the numerous detected compounds, although it is not cleanly visible in the chromatograms. Nevertheless, it suggests that the introduced mutations influence P482 metabolome.
Statistical analyses of the metabolomic data were performed as described in the previous section (
Figure 5). The hierarchically clustered heatmap represents the mutant-dependent difference in the abundance of the ions detected in the extracts (
Figure 5a). Only the compounds for which at least a 2-fold change in abundance between the mutants was observed were included in this analysis, resulting in only 32 compounds. Among these, only eight were present in the extracts from P482 wt cultures, four of which were not detected in any mutant culture. None of the 32 compounds, except 7-HT, could be fully identified based on the database search; however, the mass was used to calculate the formula of these metabolites. The metabolites present only in the wild-type strain culture were: C
17H
13NO (mass: 247.0993), C
19H
15N
11O
11 (mass: 573.0944), C
19H
39ClO
3S
2 (mass: 414.2035) and C
19H
31ClN
4O
4 (mass: 414.2032) (Supplementary Data_set_1).
Furthermore, three metabolites were only present in P482 wt and KN4240. These metabolites were putatively identified as: C
27H
49N (mass: 387.3852), C
27H
15S
4 (mass: 467.0057), and the antimicrobial iron chelator, 7-HT (mass: 138.0312). Unsurprisingly, 7-HT was not produced by the P482 mutants designed to be defective in its biosynthesis, namely, KN4706 and KN4709 (
Figure 5b). This result suggests that the gene in the
BV82_4243 locus is linked to 7-HT biosynthesis and/or secretion [
14], since it encodes a part of an efflux pump (EmrA). This might also explain the detection of 7-HT in one of the replicates of KN4243 extract, as it could originate from the cells disrupted during centrifugation when the bacteria might still synthesize the compound but is not released outside the cells.
The PCA of the obtained data is presented in a biplot showing the two first principal component scores for each sample (
Figure 5c). These two principal components are responsible for only ca. 55.5% of the total variance in the analysed data, meaning that the other two components, omitted in the biplot, might also be significant for understanding the correlations (
Supplementary Table S2). Since each biological replicate was scored separately, the plot presents three data points for each strain. First, it shows the variance in metabolome between the mutants and the variance between biological replicates. The correlations between the mutants were similar to those observed in the hierarchical clustering (
Figure 5a): the KN4706 and KN4243 mutants' metabolomes resemble each other, and the metabolome of KN4709 was similar to that of P482 wt.
Figure 5.
Statistical analysis of the LC-MS data obtained for the metabolite extracts of P482 wt and its mutants cultured in 0.1 TSB. a. Heatmap with hierarchical clustering showing the diverse metabolic profile of P482 wt and its selected mutants (KN4706, KN4709, KN4240 and KN4243). The colour range represents log2 fold change (FC) of the normalised intensity values for compounds detected in the samples, a calculated mass of the compared compound is below each column (compounds with FC > 2, ANOVA, p < 0.01 were included in the analysis). b. Bar plot showing the mean raw MS signal intensity obtained for the m/z 139 ion (7-HT) for P482 wt and each investigated mutant. The error bars represent standard deviation. c. Principal component analysis (PCA) biplot representing the similarity between all the samples included in the analysis (three biological replicates for each mutant).
Figure 5.
Statistical analysis of the LC-MS data obtained for the metabolite extracts of P482 wt and its mutants cultured in 0.1 TSB. a. Heatmap with hierarchical clustering showing the diverse metabolic profile of P482 wt and its selected mutants (KN4706, KN4709, KN4240 and KN4243). The colour range represents log2 fold change (FC) of the normalised intensity values for compounds detected in the samples, a calculated mass of the compared compound is below each column (compounds with FC > 2, ANOVA, p < 0.01 were included in the analysis). b. Bar plot showing the mean raw MS signal intensity obtained for the m/z 139 ion (7-HT) for P482 wt and each investigated mutant. The error bars represent standard deviation. c. Principal component analysis (PCA) biplot representing the similarity between all the samples included in the analysis (three biological replicates for each mutant).
However, the metabolic profile of the KN4240 mutant is substantially different from other tested strains, including KN4243. Although the genes inactivated in these two mutants were classified into one biosynthetic cluster [
14], they are the members of the separate operons and thus, the lack of correlation between their metabolomes (as well as the antibacterial activity) confirms their various functions in P482 metabolism. The metabolic profiles of KN4706 and KN4709, while both missing 7-HT, differ significantly, which implies the involvement of the 7-HT cluster genes in distinct additional processes, yet unidentified. The product of the BV82_4240 gene might be involved in various cellular processes since the gene encodes an SDR family NAD(P)-dependent oxidoreductase, including the synthesis of secondary metabolites [
27].
Taken together, the results of the selected P482 mutants' metabolomic analysis do not comply with the predictions which would have clustered the mutants in the neighbouring genes together. The KN4243 mutant from 'cluster 17' shares the metabolomic similarity with KN4706, a 7-HT biosynthesis cluster mutant, although their carbon source-dependent antagonism towards bacterial plant pathogens differs substantially (
Table 1).
2.5. 7-HT Biosynthesis Is Not Stimulated by the Substances Produced by the Pathogen
The production of 7-HT could not be confirmed for P482 in nutrient-restricted media. However, the 7-HT biosynthesis cluster mutants (KN4705, KN4706, KN4709) showed reduced antimicrobial abilities in the direct antagonism tests performed on the same, albeit solidified, media with added glucose ([
14] and summarized in
Table 2). This suggests the contribution of this gene cluster's product to P482 antibacterial activity against plant pathogens. Furthermore, since the direct antagonism tests are performed to allow the contact between the antagonist and the pathogen (primarily indirect, through secreted compounds), we hypothesized that pathogen signalling might be responsible for the 7-HT biosynthesis by P482 under minimal media conditions.
To prepare the ethyl acetate (EtOAc) extract, P482 was cultured for 96 hours in minimal M9 medium with either glucose or glycerol as the only carbon source and supplemented with the 48-hour culture filtrate of
D. solani IFB0102 (DsFIL) in the corresponding minimal medium (M9 supplemented with 0.4% glucose or 0.4% glycerol). Before extracting secondary metabolites, the post-culture supernatants were sampled for the preliminary UV-Vis spectrophotometry test. The obtained spectra of the supernatants of the P482 cultures with added DsFIL exhibited substantial differences that reasonably depend on the carbon source present in the medium (
Supplementary Figure S4). When glycerol was the carbon source for P482 in the DsFIL-supplemented M9 medium, the peaks corresponding to the produced compounds absorbing 350-425 nm light wavelengths were much higher than those recorded for the conditions with glucose, suggesting higher levels of synthesis of the metabolites. Furthermore, the absorption peaks found in the glycerol/DsFIL P482 culture supernatant were analogous to those detected in our previous study where P482 supernatant was obtained from the culture without DsFIL [
14]. Thus, as no detectable peak was found at ~330 nm, we assume that P482 did not produce 7-HT in presence of glycerol and DsFIL. However, a vague shape of the typical 7-HT 330 nm peak was observed in the glucose/DsFIL culture supernatant of P482 (
Supplementary Figure S4). This contrasts with the absorption spectrum obtained earlier [
14] for P482 cultured under corresponding conditions without DsFIL supplementation, where no peak at 330 nm was detected. Although the method does not allow for quantitative analysis, the flat shape of the P482 glucose/DsFIL supernatant spectrum was unexpected.
Moreover, the organic extracts from the described P482 culture supernatants were subjected to LC-MS analysis. This part of the study was performed to ultimately determine whether the addition of DsFIL stimulates the 7-HT production by P482 under nutrient-restricted conditions. The comprehensive analysis of the spectra directed towards the detection of ions with
m/
z in the range 138.5-139.5 showed that 7-HT (
m/
z = 139; [M+H]
+) was present in trace amounts in the tested extracts regardless of the carbon source being glucose (
Supplementary Figure S5) or glycerol (
Supplementary Figure S6). For all the MS peaks with possible 7-HT ion mass detected,
m/
z 139 was one of the minor ions in the spectrum, and the fragmentation did not show the previously obtained pattern. Moreover, the
m/
z value of several detected ions found in the query range was 139.1. This disagrees with the calculated 7-HT
m/
z (139.038) and does not indicate the presence of this compound in the analysed extracts. These results suggest that the DsFIL supplementation of M9 minimal media with either glucose or glycerol does not cause any noteworthy changes in 7-HT biosynthesis by P482.
Together, the analyses do not support the hypothesis that the compounds synthesised by the pathogen might stimulate the production of 7-HT by P482 and thus the antibacterial activity of this strain under minimal medium conditions.
The postulate that compounds secreted by IFB0102 might modulate antimicrobials production by P482 is not novel and came from the published reports of such interspecies interactions between various microorganisms [
28,
29,
30,
31,
32], although it was not confirmed in the case of P482 and IFB0102. The puzzling lack of 7-HT in P482 secondary metabolites extracts might have several causes. The addition of IFB0102 supernatant does not take the other direction of signalisation into account – P482 might be releasing compounds that trigger IFB0102 to produce modulators of P482 activity. A co-culture would be an optimal way to test this idea [
28], it is problematic due to the fact that P482 substantially inhibits the growth of IFB0102. It is, nevertheless, an intriguing topic for further studies on metabolic pathways of P482. It is also plausible that the 7-HT production might be induced only on the solid media, as the regulation of bacterial metabolism might be significantly rearranged during colony formation [
33]. Another explanation of the 7-HT absence in the described extracts is that M9 minimal medium might not contain the necessary precursors for 7-HT biosynthesis, thus the gene expression alone would not equal the synthesis of the final product [
34,
35]. However, in the case of P482 it is unclear as the biochemical pathway of 7-HT biosynthesis in
P. donghuensis is not well understood and needs further research. Future studies could be based on the recently elucidated dihydroxytropolone biosynthetic pathway in
Streptomyces spp. [
36]. Interestingly, glucose is shown to be a substrate in this process, which supports the conclusion of glucose rather than glycerol promoting the 7-HT genes' expression and their importance in antimicrobial activity of P482.
3. Conclusions
While 7-HT appears to be the primary metabolite involved in the antimicrobial activity of P. donghuensis P482, this study suggests that other metabolites may also contribute to its antagonistic effects against bacterial plant pathogens. Investigation into the influence carbon sources on the secondary metabolism of P482 revealed that under minimal media supplemented with either glucose or glycerol (nutrient-restricted conditions) 7-HT was notably absent in post-culture extracts, This absence persisted even when the media were supplemented with cell-free culture supernatant from the pathogens. Intriguingly, both wild-type and mutant strains of P482 retained antibacterial activities under nutrient these conditions, indicating the involvement of alternative mechanisms of antagonism or biosynthetic pathways.
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
The bacterial strains used in the study are listed in
Table 2.
Unless otherwise stated, all bacterial strains were grown in Miller's Lysogeny Broth (LB, Novagen, Merck Group, Germany) or on LB solidified with 1.5% agar (LB-agar, Novagen, Merck Group, Germany). The bacteria were cultured at 28 °C, and the liquid cultures were grown with shaking. The medium was supplemented with kanamycin (30 μg ml
-1) when necessary. 10% (0.1) tryptic soy broth (TSB, Oxoid, USA) stands for tenfold diluted TSB medium and was used as an optimal liquid medium to produce antimicrobials by P482 in metabolomic analyses (Matuszewska M, IFB UG and MUG, University of Gdańsk, Gdańsk, Master thesis, 2016). The minimal M9 medium [
41] was supplemented with a single carbon source: glucose (final concentration 0.4%, Sigma-Aldrich) or glycerol (final concentration 0.4%, Sigma-Aldrich).
4.2. Extraction of Secondary Metabolites
Bacteria (P482 strain or its mutants) were cultured in 250 ml or 500 ml (M9 nutrient-restricted medium) of a given liquid medium at 28 °C with shaking (120 rpm). Culture flasks with baffled bottoms and vented caps with filters (Duran, Germany) were used to keep the environment well-oxygenated. The cultures were finished after 96 hours, long after bacteria entered the stationary growth phase, regardless of the nutritional conditions. Subsequently, they were centrifuged for 30 min in 10000×
g, and the supernatants were collected and subjected to the extraction of secondary metabolites with ethyl acetate (EtOAc) (Merck, USA). The organic residues were collected and traces of water were removed with anhydrous sodium sulphate [
42]
The dry extract was then dissolved in HPLC-grade methanol (MeOH) (100 µl MeOH per 500 ml of the initial culture) (Merck, USA) and stored at -20 °C until further analysis.
4.3. UV-Vis Absorption Spectra
The selected culture supernatants of P482 wt and its mutants were tested for 7-HT using UV and visible light (UV-Vis) spectrophotometry [
43]. The bacteria were cultured in a liquid 0.1 TSB medium at 28 °C with shaking. The samples of the cultures were collected after overnight incubation. The samples of bacterial suspensions were centrifuged, and the supernatants were collected and subsequently subjected to filtration to exclude the presence of cells that may interfere with the absorption read. The supernatant samples were then placed in a 96-well plate with a transparent bottom, and their absorption spectra in the wavelength range from 200 to 900 nm were measured using Epoch2 Microplate Spectrophotometer (BioTek, USA).
4.4. HPLC-MS Analyses
4.4.1. Metabolomics
The extracts of P482 and its mutants' secondary metabolites obtained in this study were subjected to high-performance liquid chromatography coupled with mass spectrometry (HPLC-MS). The extract samples were prepared by evaporating the solvent, weighing the crude extract, and dissolving it in 40 µl of EtOAc:MeOH mixture (1:1) per 1 mg of the extract (all used solvents were of HPLC-grade purity (Merck, USA). All samples were filtered with a 0.22 µm regenerated cellulose syringe filter and diluted tenfold before chromatographic procedures.
HPLC-MS was carried out using the Agilent 1260 Infinity Series chromatograph coupled with Ultra High Definition (UHD) quadrupole-time of flight (Q-TOF) mass spectrometer (model G6540B) with a dual electrospray ionization (Dual ESI) source (Agilent Technologies, USA). Agilent Mass Hunter software (Agilent Technologies, USA) was used for instrument control and data acquisition. The sample injection volume was 5 µl. The reverse-phase (RP) HPLC separation utilized a Zorbax Eclipse Plus C-18 column (4.6 × 100 mm, with 3.5 µm particles, Agilent Technologies) and the linear gradient elution program with H
2O and acetonitrile (both acidified with 0.1% formic acid (FA)) as the solvents (
Supplementary Table S5). The flow was set at 0.6 ml/min. The UV detection on the diode array detector (DAD) was set for 254 nm, 300 nm and 420 nm wavelengths. The MS equipment operated in positive mode. Mass spectra were recorded as centroid spectra in the m/z range of 100-1700. The spectra were analysed using MassHunter Qualitative Analysis Software and subjected to MassHunter Profinder (Agilent Technologies, USA) batch feature analysis using the bacterial compound database created at IPSP-CNR. Further data analyses were carried out also with the use of OpenChrom software version 1.4.x (Lablicate GmbH, Germany) and using publicly available online databases: ChemSpider (
www.chemspider.com, Royal Society of Chemistry), MetaCyc (
www.metacyc.org [
44] PubChem (
https://pubchem.ncbi.nlm.nih.gov, [
45] and KEGG COMPOUND (
www.kegg.jp/kegg/compound/, [
46].
The m/z data obtained in the study were transformed into mass and compound formulas and used as queries against microbial metabolic databases.
4.4.2. Detection of 7-Hydroxytropolone
The extracts obtained from the P482 wild type (referred to as P482 wt) cultures in M9 media containing cell-free filter sterilised D. solani IFB0102 filtrate were subjected to HPLC-MS analysis to determine whether these conditions promote 7-HT biosynthesis by P482. The crude extracts (from an initial 200 ml of culture each) were dissolved in 50 µl of HPLC grade MeOH (Merck, USA) per sample. HPLC-MS was carried out using the micro-HPLC LC200 (Eksigent, Dublin, CA, USA) system with autosampler CTC PAL (CTC Analytics AG, Zwinger, Switzerland). The chromatograph was paired with QTRAP 6500 mass spectrometer (AB Sciex LLC, Framingham, MA, USA) with Sciex Analyst software (version 1.6.2) for data acquisition and analysis. The sample injection volume was 1 µl. The RP-HPLC separation utilized a ChromXP C18 CL column (0.3 × 150 mm, 3 µm particles, 120 Å, AB Sciex LLC). The elution program was a linear gradient (A: 2% → 98%; 10 minutes) of buffers A: H2O + 0.1% FA and B: AcN + 0.1% FA. The MS equipment operated in the positive mode with the scan type Q1. Mass spectra were recorded as centroid spectra in the m/z range of 30-500.
4.5. Statistical Analyses
Statistical metabolomic data analyses were conducted using Mass Profiler Professional software (Agilent Technologies, USA). First, the signal values of the entities detected in the samples included in a given analysis were normalised and subjected to ANOVA analysis with Tukey's posthoc test (p < 0.01). Then, they were further filtered using a fold change (FC) threshold. Only samples with FC ≥ 2 were included in the analyses. Finally, these results were subjected to hierarchical clustering and principal component analysis (PCA).
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Table S1. HPLC elution program for the extract samples. Table S2. Principal component analysis of the LC-MS results of P482 wt and mutants 10% TSB culture extract samples. Figure S1. Antibiosis assay for P482 wt and its mutants against Pseudomonas syringae pv. syringae Pss762 and Dickeya solani IFB102 strain on 0.1 TSB-agar medium. Figure S2. The overlay of the representative LFiC-MS chromatograms of the P482 wt extracts. Figure S3. The overlay of the representative LC-MS chromatograms of the studied mutant extracts. Figure S4. UV-Vis absorption spectra of culture supernatants of P482 wt cultured with D. solani IFB0102 culture filtrate. Figure S5. The results of 7-HT detection in the metabolite extracts from P482 cultured in M9 + 0.4% glucose supplemented with D. solani culture filtrate. Figure S6. The results of 7-HT detection in the metabolite extracts from P482 cultured in M9 + 0.4% glycerol supplemented with D. solani culture filtrate.
Author Contributions
Conceptualization, M.M., F.V., and S.J.; Methodology, M.M., M.R., F.V., A.S., M.P., S.J.; Software, M.M., R.M., A.S.; Validation, M.M., A.S., and R.M.; Formal Analysis, M.M.; Investigation, M.M., M.R., A.S., M.P., Resources, S.J., F.V., M.L.; Data Curation, M.M., A.S.; Writing – Original Draft Preparation, M.M., M.R., S.J., R.C.; Writing – Review & Editing, M.R., S.J., R.C., A.S., R.M., Visualization, M.M., M.R.; Supervision, S.J., A.S., F.V., M.L.; Project Administration, S.J., F.V., M.L.; Funding Acquisition, S.J., F.V., M.L.
Funding
This study was supported by the Polish Ministry of Education and Science (Ministerstwo Edukacji i Nauki) 531-N105-D786-24 (S.J.), by the Polish National Science Centre (Narodowe Centrum Nauki, Polska) research grant OPUS4 no. 2012/07/B/NZ9/01623 (S.J.), by the Agritech National Research Center and received funding from the European Union Next-Generation EU (Piano Nazionale Di Ripresa e Resilienza (PNRR)—Missione 4 Componente 2, Investimento 1.4—D.D. 1032 17/06/2022, CN00000022), the MISE CRESO (grant number Protection no. F/050421/01-03/X32 - Viabio no. F/200095/01-03/X45), Regione Campania (Project Dioniso, n. B98H19005010009) and Regione Calabria (project FILO, n. 84250071887). This manuscript reflects only the authors’ views and opinions neither the European Union nor the European Commission can be considered responsible for them.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data generated or analysed during this study are included in this published article and its supplementary materials. The data that support the findings of this study are available from the corresponding author upon reasonable request. The original data (Raw metabolomic, Compounds analyses, Hierarchical Condition tress and PCA Scores) presented in the study are openly available in
https://doi.org/10.5281/zenodo.11220997 repository.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sanchez, S.; Chavez, A.; Forero, A.; Garcia-Huante, Y.; Romero, A.; Sanchez, M.; Rocha, D.; Sanchez, B.; Avalos, M.; Guzman-Trampe, S.; et al. Carbon source regulation of antibiotic production. J Antibiot 2010, 63, 442–459. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Sanz, D.; Redondo-Nieto, M.; Martin, M.; Rivilla, R. Comparative genomics of the Pseudomonas corrugata subgroup reveals high species diversity and allows the description of Pseudomonas ogarae sp. nov. Microb Genomics 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, P.; O'Sullivan, D.J.; Simpson, P.; Glennon, J.D.; O'Gara, F. Isolation of 2,4-Diacetylphloroglucinol from a Fluorescent Pseudomonad and Investigation of Physiological Parameters Influencing Its Production. Applied and Environmental Microbiology 1992, 58, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Duffy, B.K.; Defago, G. Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Applied and Environmental Microbiology 1999, 65, 2429–2438. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, H.; Xu, Y.; Zhang, X. Optimization of nutrient components for enhanced phenazine-1-carboxylic acid production by gacA-inactivated Pseudomonas sp. M18G using response surface method. Applied Microbiology and Biotechnology 2008, 77, 1207–1217. [Google Scholar] [CrossRef]
- Park, J.; Oh, S.; Anderson, A.; Neiswender, J.; Kim, J.C.; Kim, Y. Production of the antifungal compounds phenazine and pyrrolnitrin from Pseudomonas chlororaphis O6 is differentially regulated by glucose. Letters in Applied Microbiology 2011, 52, 532–537. [Google Scholar] [CrossRef] [PubMed]
- van Rij, E.T.; Wesselink, M.; Chin-A-Woeng, T.F.; Bloemberg, G.V.; Lugtenberg, B.J. Influence of environmental conditions on the production of phenazine-1-carboxamide by Pseudomonas chlororaphis PCL1391. Molecular Plant-Microbe Interactions 2004, 17, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Vindeirinho, J.M.; Soares, H.M.V.M.; Soares, E.V. Modulation of Siderophore Production by Pseudomonas fluorescens Through the Manipulation of the Culture Medium Composition. Applied Biochemistry and Biotechnology 2021, 193, 607–618. [Google Scholar] [CrossRef]
- Dolan, S.K.; Kohlstedt, M.; Trigg, S.; Vallejo Ramirez, P.; Kaminski, C.F.; Wittmann, C.; Welch, M. Contextual flexibility in Pseudomonas aeruginosa central carbon metabolism during growth in single carbon sources. MBio 2020, 11, e02684-19. [Google Scholar] [CrossRef]
- Poblete-Castro, I.; Wittmann, C.; Nikel, P.I. Biochemistry, genetics and biotechnology of glycerol utilization in Pseudomonas species. Microb Biotechnol 2020, 13, 32–53. [Google Scholar] [CrossRef] [PubMed]
- Rojo, F. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiology Reviews 2010, 34, 658–684. [Google Scholar] [CrossRef] [PubMed]
- Nikel, P.I.; Kim, J.; de Lorenzo, V. Metabolic and regulatory rearrangements underlying glycerol metabolism in Pseudomonas putida KT2440. Environmental Microbiology 2014, 16, 239–254. [Google Scholar] [CrossRef] [PubMed]
- Belda, E.; Van Heck, R.G.; José Lopez-Sanchez, M.; Cruveiller, S.; Barbe, V.; Fraser, C.; Klenk, H.P.; Petersen, J.; Morgat, A.; Nikel, P.I. The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environmental Microbiology 2016, 18, 3403–3424. [Google Scholar] [CrossRef] [PubMed]
- Matuszewska, M.; Maciag, T.; Rajewska, M.; Wierzbicka, A.; Jafra, S. The carbon source-dependent pattern of antimicrobial activity and gene expression in Pseudomonas donghuensis P482. Sci Rep 2021, 11, 10994. [Google Scholar] [CrossRef] [PubMed]
- Gui, Z.; You, J.; Xie, G.; Qin, Y.; Wu, T.; Xie, Z. Pseudomonas donghuensis HYS 7-hydroxytropolone contributes to pathogenicity toward Caenorhabditis elegans and is influenced by pantothenic acid. Biochem Biophys Res Commun 2020, 533, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Chen, M.; Yu, X.; Xie, Z. 7-Hydroxytropolone produced and utilized as an iron-scavenger by Pseudomonas donghuensis. Biometals 2016, 29, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Krzyzanowska, D.M.; Ossowicki, A.; Rajewska, M.; Maciag, T.; Jablonska, M.; Obuchowski, M.; Heeb, S.; Jafra, S. When genome-based approach meets the "Old but Good": revealing genes involved in the antibacterial activity of Pseudomonas sp. P482 against soft rot pathogens. Front Microbiol 2016, 7, 782. [Google Scholar] [CrossRef]
- Chen, M.; Wang, P.; Xie, Z. A Complex Mechanism Involving LysR and TetR/AcrR That Regulates Iron Scavenger Biosynthesis in Pseudomonas donghuensis HYS. J Bacteriol 2018, 200, e00087-18. [Google Scholar] [CrossRef] [PubMed]
- Agaras, B.C.; Iriarte, A.; Valverde, C.F. Genomic insights into the broad antifungal activity, plant-probiotic properties, and their regulation, in Pseudomonas donghuensis strain SVBP6. PLoS One 2018, 13, e0194088. [Google Scholar] [CrossRef]
- Udaondo, Z.; Ramos, J.L.; Segura, A.; Krell, T.; Daddaoua, A. Regulation of carbohydrate degradation pathways in Pseudomonas involves a versatile set of transcriptional regulators. Microbial biotechnology 2018, 11, 442–454. [Google Scholar] [CrossRef]
- Villagra, N.A.; Fuentes, J.A.; Jofré, M.R.; Hidalgo, A.A.; García, P.; Mora, G.C. The carbon source influences the efflux pump-mediated antimicrobial resistance in clinically important Gram-negative bacteria. Journal of Antimicrobial Chemotherapy 2012, 67, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Uzair, B.; Ahmed, N.; Ahmad, V.U.; Mohammad, F.V.; Edwards, D.H. The isolation, purification and biological activity of a novel antibacterial compound produced by Pseudomonas stutzeri. FEMS Microbiol Lett 2008, 279, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Breitling, R.; Ceniceros, A.; Jankevics, A.; Takano, E. Metabolomics for secondary metabolite research. Metabolites 2013, 3, 1076–1083. [Google Scholar] [CrossRef] [PubMed]
- Frimmersdorf, E.; Horatzek, S.; Pelnikevich, A.; Wiehlmann, L.; Schomburg, D. How Pseudomonas aeruginosa adapts to various environments: a metabolomic approach. Environmental microbiology 2010, 12, 1734–1747. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, B.; Chavez, A.; Forero, A.; Garcia-Huante, Y.; Romero, A.; Sanchez, M.; Rocha, D.; Sanchez, B.; Rodriguez-Sanoja, R.; Sanchez, S.; et al. Production of microbial secondary metabolites: regulation by the carbon source. Crit Rev Microbiol 2010, 36, 146–167. [Google Scholar] [CrossRef] [PubMed]
- van der Werf, M.J.; Overkamp, K.M.; Muilwijk, B.; Koek, M.M.; van der Werff-van, B.J.; Jellema, R.H.; Coulier, L.; Hankemeier, T. Comprehensive analysis of the metabolome of Pseudomonas putida S12 grown on different carbon sources. Molecular BioSystems 2008, 4, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Kavanagh, K.; Jörnvall, H.; Persson, B.; Oppermann, U. Medium-and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cellular and Molecular Life Sciences 2008, 65, 3895–3906. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.-L. Metabolite induction via microorganism co-culture: a potential way to enhance chemical diversity for drug discovery. Biotechnology advances 2014, 32, 1180–1204. [Google Scholar] [CrossRef] [PubMed]
- Ezaki, M.; Shigematsu, N.; Yamashita, M.; Komori, T.; Umehara, K.; Imanaka, H. Biphenomycin C, a precursor of biphenomycin A in mixed culture. The Journal of Antibiotics 1993, 46, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Traxler, M.F.; Watrous, J.D.; Alexandrov, T.; Dorrestein, P.C.; Kolter, R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio 2013, 4, e00459-13. [Google Scholar] [CrossRef]
- Vinale, F.; Nicoletti, R.; Borrelli, F.; Mangoni, A.; Parisi, O.; Marra, R.; Lombardi, N.; Lacatena, F.; Grauso, L.; Finizio, S. Co-culture of plant beneficial microbes as source of bioactive metabolites. Scientific reports 2017, 7, 14330. [Google Scholar] [CrossRef]
- Vinale, F.; Nicoletti, R.; Lacatena, F.; Marra, R.; Sacco, A.; Lombardi, N.; d’Errico, G.; Digilio, M.; Lorito, M.; Woo, S. Secondary metabolites from the endophytic fungus Talaromyces pinophilus. Natural product research 2017, 31, 1778–1785. [Google Scholar] [CrossRef] [PubMed]
- Pasqua, M.; Grossi, M.; Zennaro, A.; Fanelli, G.; Micheli, G.; Barras, F.; Colonna, B.; Prosseda, G. The varied role of efflux pumps of the MFS family in the interplay of bacteria with animal and plant cells. Microorganisms 2019, 7, 285. [Google Scholar] [CrossRef] [PubMed]
- Iwai, Y.; Omura, S. Culture conditions for screening of new antibiotics. The Journal of antibiotics 1982, 35, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Oda, Y.; Lankford, P.K.; Zhang, B.; Samatova, N.F.; Pelletier, D.A.; Harwood, C.S.; Hettich, R.L. Characterization of anaerobic catabolism of p-coumarate in Rhodopseudomonas palustris by integrating transcriptomics and quantitative proteomics. Mol Cell Proteomics 2008, 7, 938–948. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, M.; Lü, J.; Xu, J.; Wang, Y.; Lin, S.; Deng, Z.; Tao, M. Biosynthesis of tropolones in Streptomyces spp.: interweaving biosynthesis and degradation of phenylacetic acid and hydroxylations on the tropone ring. Applied and Environmental Microbiology 2018, 84, e00349-18. [Google Scholar] [PubMed]
- Krzyzanowska, D.M.; Ossowicki, A.; Jafra, S. Genome sequence of Pseudomonas sp. strain P482, a tomato rhizosphere isolate with broad-spectrum antimicrobial activity. Genome Announc 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Tvrzova, L.; Schumann, P.; Sproer, C.; Sedlacek, I.; Pacova, Z.; Sedo, O.; Zdrahal, Z.; Steffen, M.; Lang, E. Pseudomonas moraviensis sp. nov. and Pseudomonas vranovensis sp. nov., soil bacteria isolated on nitroaromatic compounds, and emended description of Pseudomonas asplenii. International Journal of Systematic and Evolutionary Microbiology 2006, 56, 2657–2663. [Google Scholar] [CrossRef] [PubMed]
- Sławiak, M.; Łojkowska, E.; van der Wolf, J.M. First report of bacterial soft rot on potato caused by Dickeya sp. (syn. Erwinia chrysanthemi) in Poland. Plant Pathology 2009, 58, 794–794. [Google Scholar] [CrossRef]
- Kałużna, M.; Puławska, J.; Sobiczewski, P. The use of PCR melting profile for typing of Pseudomonas syringae isolates from stone fruit trees. European Journal of Plant Pathology 2009, 126, 437–443. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, 2001; pp. 3.17–3.32. [Google Scholar]
- Staropoli, A.; Iacomino, G.; De Cicco, P.; Woo, S.L.; Di Costanzo, L.; Vinale, F. Induced secondary metabolites of the beneficial fungus Trichoderma harzianum M10 through OSMAC approach. Chemical and Biological Technologies in Agriculture 2023, 10, 28. [Google Scholar] [CrossRef]
- Gao, J.; Xie, G.; Peng, F.; Xie, Z. Pseudomonas donghuensis sp. nov., exhibiting high-yields of siderophore. Antonie Van Leeuwenhoek 2015, 107, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K. The MetaCyc database of metabolic pathways and enzymes. Nucleic acids research 2018, 46, D633–D639. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B. PubChem in 2021: new data content and improved web interfaces. Nucleic acids research 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).