Submitted:
13 June 2024
Posted:
14 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Synuclein and Tau: ‘Seeding and Spreading’
3. Importance of Epigenetics
Epigenetics in Cognition
4. Epigenetics in PD
4.1. DNA Methylation in PD Cognitive Dysfunction
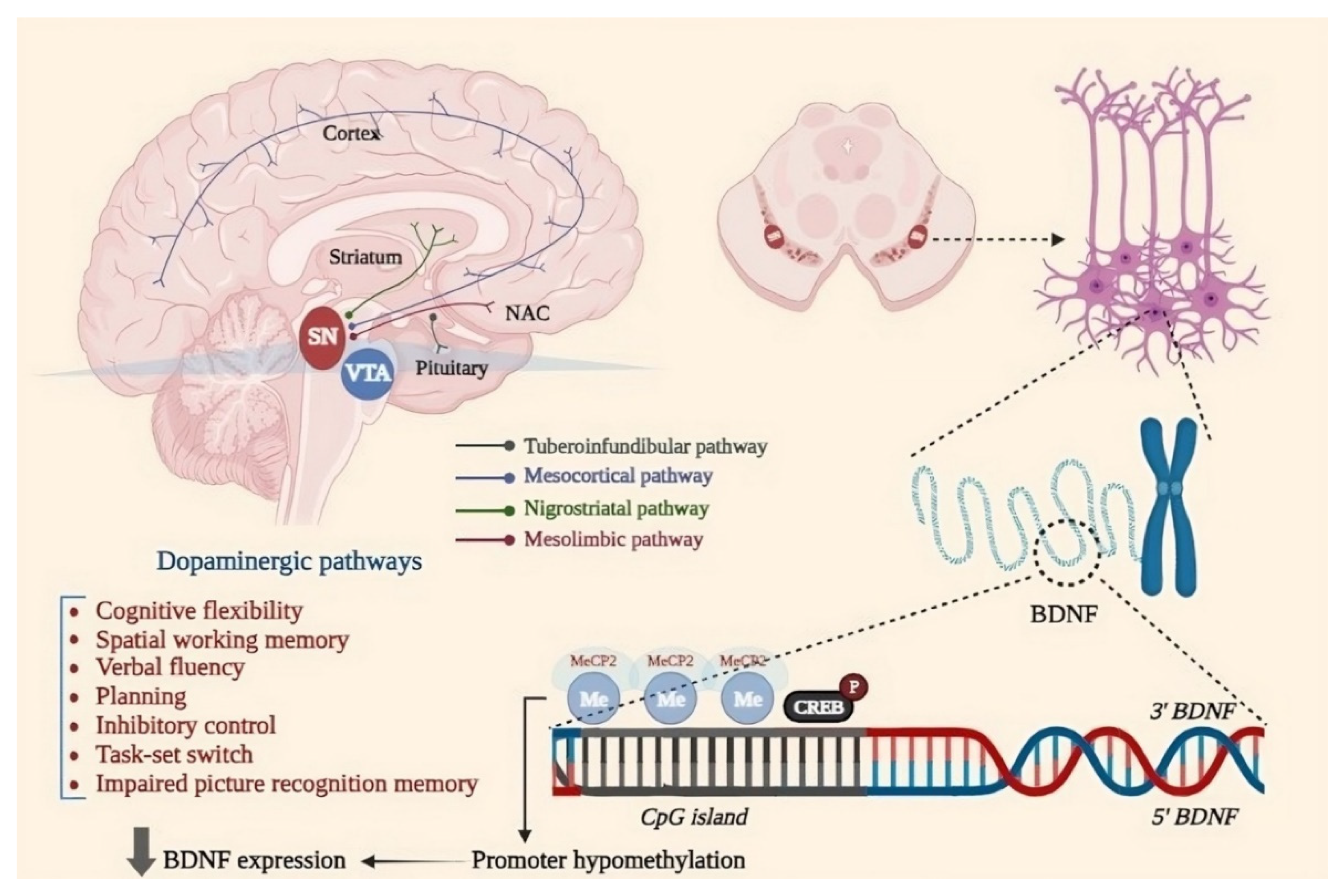
4.1.1. Brain-Derived Neurotrophic Factor (BDNF)
4.1.2. Peroxisome Proliferator-Activated Receptor Gamma Coactivator−1 α (PGC−1α)
4.1.3. Peroxisome Proliferator-Activated Receptor Gamma Coactivator−1 α (PGC−1α)
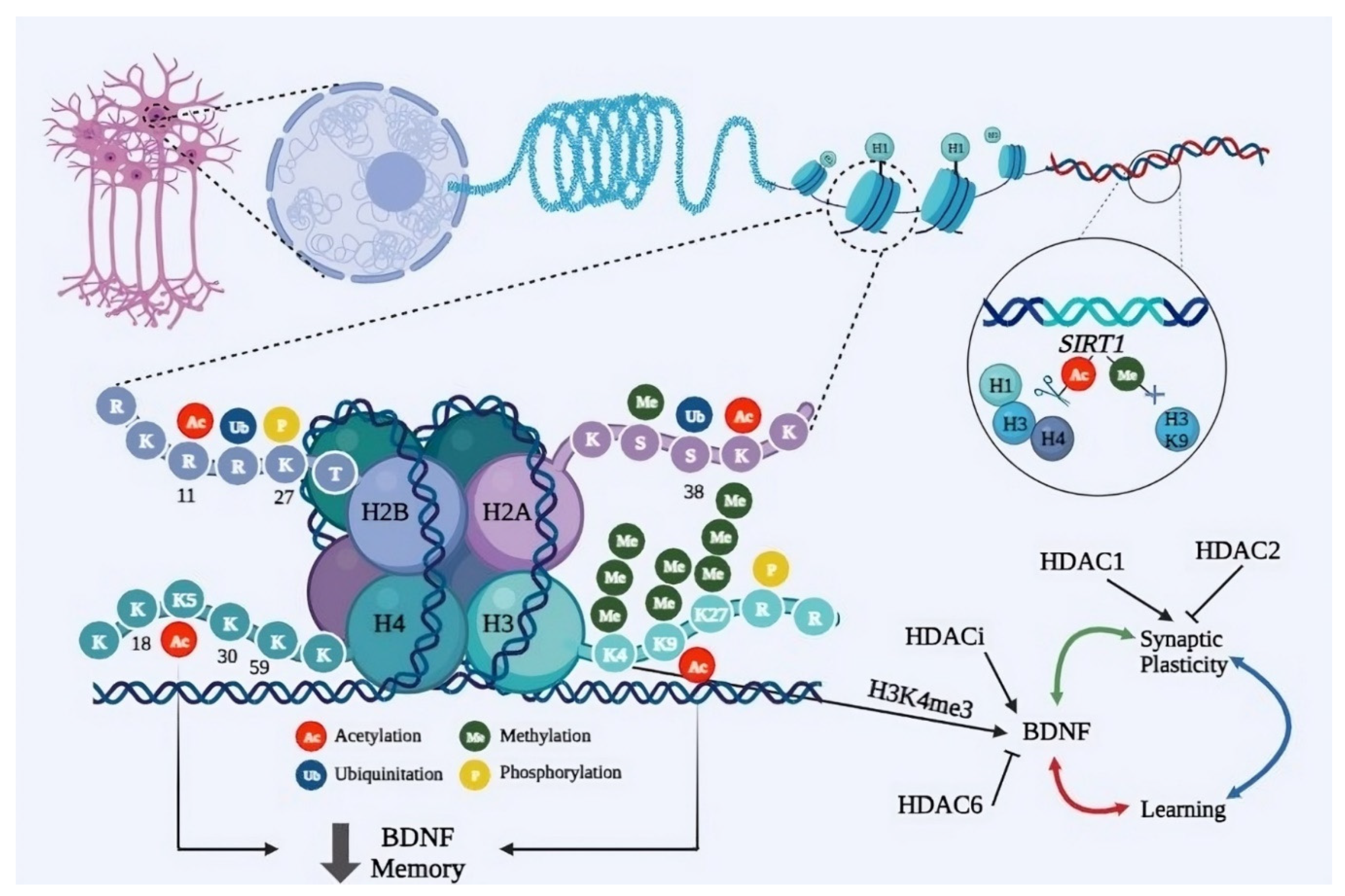
4.2. Histone Modification in PD Cognitive Dysfunction
4.2.1. H3K9me2
4.2.2. H3K4me3 and H3K27me3
4.2.3. K4me3
4.2.4. H3K27me3
4.2.5. H3K9ac
4.3. MicroRNAs in PD Cognitive Dysfunction
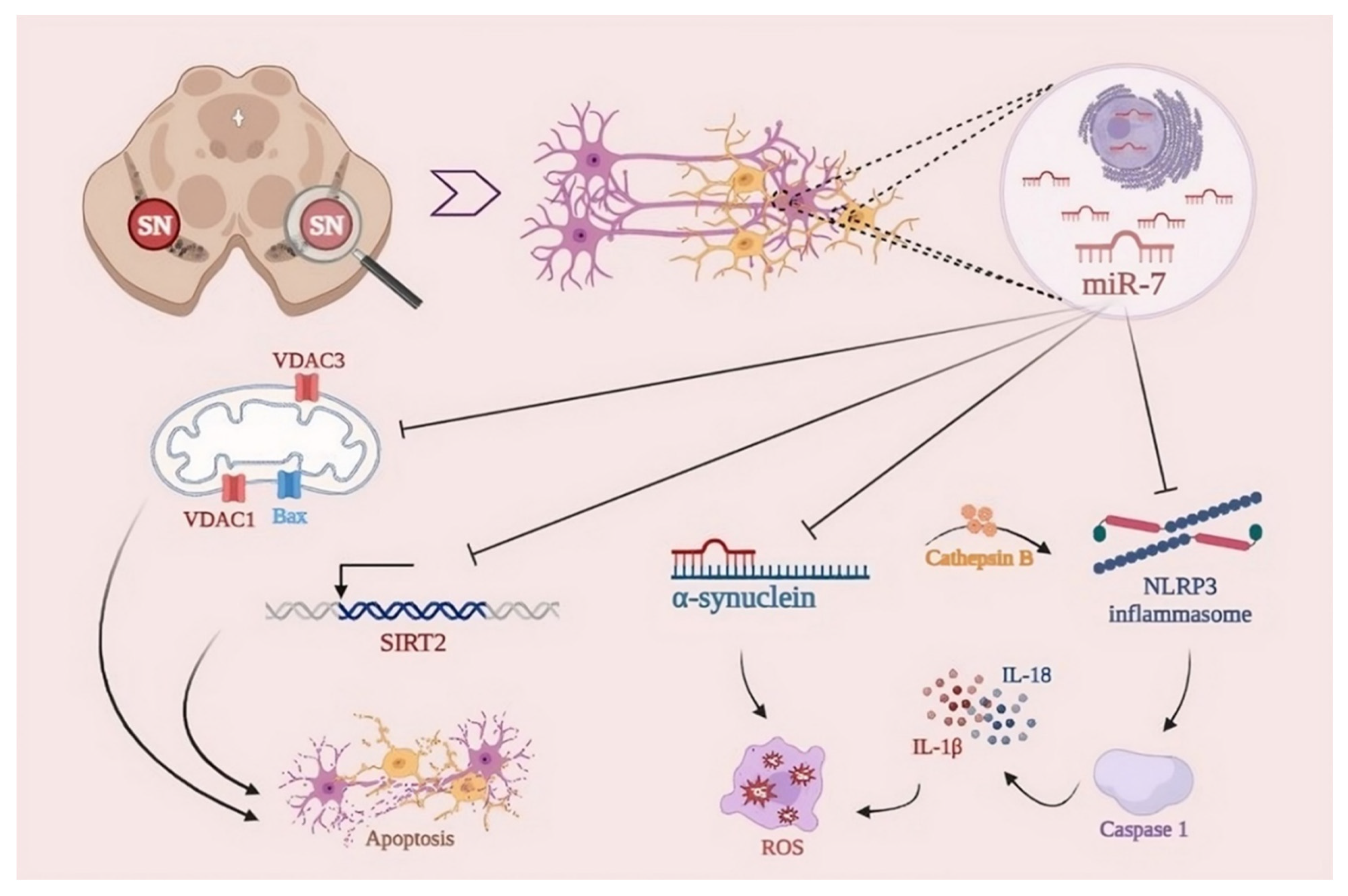
4.3.1. miR-7
4.3.2. miR-124-3p
4.3.3. miR-124
4.3.4. miR-195
4.3.5. miR-190
4.3.6. miR-146a
4.3.7. miR-155
4.3.8. miR-133b
4.3.9. miR-126
4.3.10. miR-144
4.3.11. miR-148b
4.3.12. miR-221
4.3.13. miR-199a
4.3.14. miR-132
5. Therapeutics Perspective of Epigenetics in PD Cognitive Dysfunction
Histone Deacetylases as Potential Epigenetic Targets
6. Conclusions
Abbreviations
References
- Moosavi A, Motevalizadeh Ardekani A. Role of Epigenetics in Biology and Human Diseases. Iran Biomed J. 2016;20(5):246-58. [CrossRef]
- Surguchov, A. α-Synuclein and Mechanisms of Epigenetic Regulation. Brain Sci. 2023, 13, 150. [CrossRef]
- Huynh, J.L.; Casaccia, P. Epigenetic mechanisms in multiple sclerosis: implications for pathogenesis and treatment. Lancet Neurol. 2013, 12, 195–206. [CrossRef]
- Doder, M.; Jahanshahi, M.; Turjanski, N.; Moseley, I.F.; Lees, A.J. Parkinson’s syndrome after closed head injury: a single case report. J. Neurol. Neurosurg. Psychiatry 1999, 66, 380–385. [CrossRef]
- Goetz, C.G. The History of Parkinson’s Disease: Early Clinical Descriptions and Neurological Therapies. Cold Spring Harb. Perspect. Med. 2011, 1, a008862. [CrossRef]
- Emamzadeh FN, Surguchov A. Parkinson’s Disease: Biomarkers, Treatment, and Risk Factors. Front Neurosci. 2018;12:612. [CrossRef]
- Nair, V.D.; Ge, Y. Alterations of miRNAs reveal a dysregulated molecular regulatory network in Parkinson’s disease striatum. Neurosci. Lett. 2016, 629, 99–104. [CrossRef]
- Cannon JR, Greenamyre JT. Gene-environment interactions in Parkinson’s disease: specific evidence in humans and mammalian models. Neurobiol Dis. 2013;57:38-46. [CrossRef]
- Zoghbi HY, Beaudet AL. Epigenetics and Human Disease. Cold Spring Harb Perspect Biol. 2016;8(2):a019497. [CrossRef]
- Vasili E, Dominguez-Meijide A, Outeiro TF. Spreading of α-Synuclein and Tau: A Systematic Comparison of the Mechanisms Involved. Front Mol Neurosci. 2019;12:107. [CrossRef]
- Chen X, de Silva HAR, Pettenati MJ, Rao PN, George-Hyslop PS, Roses AD, et al. The human NACP/α-synuclein gene: chromosome assignment to 4q21.3–q22 and TaqI RFLP analysis. Genomics. 1995;26(2):425-7. [CrossRef]
- Mehra, S.; Sahay, S.; Maji, S.K. α-Synuclein misfolding and aggregation: Implications in Parkinson’s disease pathogenesis. Biochim. et Biophys. Acta (BBA) - Proteins Proteom. 2019, 1867, 890–908. [CrossRef]
- Coppedè, F. Genetics and Epigenetics of Parkinson’s Disease. Sci. World J. 2012, 2012, 1–12. [CrossRef]
- Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364(9440):1167-9. [CrossRef]
- Deng, H.; Yuan, L. Genetic variants and animal models in SNCA and Parkinson disease. Ageing Res. Rev. 2014, 15, 161–176. [CrossRef]
- Myhre R, Toft M, Kachergus J, Hulihan M, Aasly J, Klungland H, et al. Multiple alpha-synuclein gene polymorphisms are associated with Parkinson’s disease in a Norwegian population. Acta Neurologica Scandinavica. 2008;118(5):320-7. [CrossRef]
- Mizuta, I.; Satake, W.; Nakabayashi, Y.; Ito, C.; Suzuki, S.; Momose, Y.; Nagai, Y.; Oka, A.; Inoko, H.; Fukae, J.; et al. Multiple candidate gene analysis identifies α-synuclein as a susceptibility gene for sporadic Parkinson’s disease. Hum. Mol. Genet. 2006, 15, 1151–1158. [CrossRef]
- Ross, O.A.; Gosal, D.; Stone, J.T.; Lincoln, S.J.; Heckman, M.G.; Irvine, G.B.; Johnston, J.A.; Gibson, J.M.; Farrer, M.J.; Lynch, T. Familial genes in sporadic disease: Common variants of α-synuclein gene associate with Parkinson’s disease. Mech. Ageing Dev. 2007, 128, 378–382. [CrossRef]
- Chung, S.J.; Armasu, S.M.; Biernacka, J.M.; Lesnick, T.G.; Rider, D.N.; Lincoln, S.J.; Ortolaza, A.I.; Farrer, M.J.; Cunningham, J.M.; Rocca, W.A.; et al. Common variants in PARK loci and related genes and Parkinson’s disease. Mov. Disord. 2010, 26, 280–288. [CrossRef]
- Trotta, L.; Guella, I.; Soldà, G.; Sironi, F.; Tesei, S.; Canesi, M.; Pezzoli, G.; Goldwurm, S.; Duga, S.; Asselta, R. SNCA and MAPT genes: Independent and joint effects in Parkinson disease in the Italian population. Park. Relat. Disord. 2012, 18, 257–262. [CrossRef]
- Wu-Chou, Y.-H.; Chen, Y.-T.; Yeh, T.-H.; Chang, H.-C.; Weng, Y.-H.; Lai, S.-C.; Huang, C.-L.; Chen, R.-S.; Huang, Y.-Z.; Chen, C.-C.; et al. Genetic variants of SNCA and LRRK2 genes are associated with sporadic PD susceptibility: A replication study in a Taiwanese cohort. Park. Relat. Disord. 2012, 19, 251–255. [CrossRef]
- Chen, Y.; Wei, Q.-Q.; Ou, R.; Cao, B.; Chen, X.; Zhao, B.; Guo, X.; Yang, Y.; Chen, K.; Wu, Y.; et al. Genetic Variants of SNCA Are Associated with Susceptibility to Parkinson’s Disease but Not Amyotrophic Lateral Sclerosis or Multiple System Atrophy in a Chinese Population. PLOS ONE 2015, 10, e0133776. [CrossRef]
- Davis, A.A.; Andruska, K.M.; Benitez, B.A.; Racette, B.A.; Perlmutter, J.S.; Cruchaga, C. Variants in GBA , SNCA , and MAPT influence Parkinson disease risk, age at onset, and progression. Neurobiol. Aging 2016, 37, 209.e1–209.e7. [CrossRef]
- Wei, Y.; Yang, N.; Xu, Q.; Sun, Q.; Guo, J.; Li, K.; Liu, Z.; Yan, X.; Zhu, X.; Tang, B. The rs3756063 polymorphism is associated with SNCA methylation in the Chinese Han population. J. Neurol. Sci. 2016, 367, 11–14. [CrossRef]
- Foo, J.N.; Tan, L.C.; Irwan, I.D.; Au, W.-L.; Low, H.Q.; Prakash, K.-M.; Ahmad-Annuar, A.; Bei, J.; Chan, A.Y.; Chen, C.M.; et al. Genome-wide association study of Parkinson’s disease in East Asians. Hum. Mol. Genet. 2016, 26, 226–232. [CrossRef]
- Nalls, M.A.; Pankratz, N.; Lill, C.M.; Do, C.B.; Hernandez, D.G.; Saad, M.; DeStefano, A.L.; Kara, E.; Bras, J.; Sharma, M.; et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014, 46, 989–993. [CrossRef]
- Zhang, Y.; Shu, L.; Sun, Q.; Pan, H.; Guo, J.; Tang, B. A Comprehensive Analysis of the Association Between SNCA Polymorphisms and the Risk of Parkinson’s Disease. Front. Mol. Neurosci. 2018, 11, 391. [CrossRef]
- Chartier-Harlin, M.-C.; Kachergus, J.; Roumier, C.; Mouroux, V.; Douay, X.; Lincoln, S.; Levecque, C.; Larvor, L.; Andrieux, J.; Hulihan, M.; et al. α-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet 2004, 364, 1167–1169. [CrossRef]
- Huang Y, Wang G, Rowe D, Wang Y, Kwok JB, Xiao Q, et al. SNCA gene, but not MAPT, influences onset age of Parkinson’s disease in Chinese and Australians. BioMed Research International. 2015;2015. [CrossRef]
- Cardo, L.F.; Coto, E.; De Mena, L.; Ribacoba, R.; Lorenzo-Betancor, O.; Pastor, P.; Samaranch, L.; Mata, I.F.; Díaz, M.; Morís, G.; et al. A Search for SNCA 3′ UTR Variants Identified SNP rs356165 as a Determinant of Disease Risk and Onset Age in Parkinson’s Disease. J. Mol. Neurosci. 2012, 47, 425–430. [CrossRef]
- Elbaz, A.; Ross, O.A.; Ioannidis, J.P.A.; Soto-Ortolaza, A.I.; Moisan, F.; Aasly, J.; Annesi, G.; Bozi, M.; Brighina, L.; Chartier-Harlin, M.; et al. Independent and joint effects of the MAPT and SNCA genes in Parkinson disease. Ann. Neurol. 2010, 69, 778–792. [CrossRef]
- Brockmann, K.; Schulte, C.; Hauser, A.; Lichtner, P.; Huber, H.; Maetzler, W.; Berg, D.; Gasser, T. SNCA: Major genetic modifier of age at onset of Parkinson’s disease. Mov. Disord. 2013, 28, 1217–1221. [CrossRef]
- Guhathakurta, S.; Bok, E.; Evangelista, B.A.; Kim, Y.-S. Deregulation of α-synuclein in Parkinson’s disease: Insight from epigenetic structure and transcriptional regulation of SNCA. Prog. Neurobiol. 2017, 154, 21–36. [CrossRef]
- Jowaed, A.; Schmitt, I.; Kaut, O.; Wüllner, U. Methylation Regulates Alpha-Synuclein Expression and Is Decreased in Parkinson’s Disease Patients’ Brains. J. Neurosci. 2010, 30, 6355–6359. [CrossRef]
- Matsumoto, L.; Takuma, H.; Tamaoka, A.; Kurisaki, H.; Date, H.; Tsuji, S.; Iwata, A. CpG Demethylation Enhances Alpha-Synuclein Expression and Affects the Pathogenesis of Parkinson’s Disease. PLOS ONE 2010, 5, e15522. [CrossRef]
- Wu, H.; D’alessio, A.C.; Ito, S.; Xia, K.; Wang, Z.; Cui, K.; Zhao, K.; Sun, Y.E.; Zhang, Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 2011, 473, 389–393. [CrossRef]
- Guhathakurta, S.; Song, M.K.; Basu, S.; Je, G.; Cristovao, A.C.; Kim, Y.-S. Regulation of Alpha-Synuclein Gene (SNCA) by Epigenetic Modifier TET1 in Parkinson Disease. Int. Neurourol. J. 2022, 26, S85–93. [CrossRef]
- Mueller JC, Fuchs J, Hofer A, Zimprich A, Lichtner P, Illig T, et al. Multiple regions of α-synuclein are associated with Parkinson’s disease. Annals of Neurology. 2005;57(4):535-41. [CrossRef]
- Kolarova, M.; García-Sierra, F.; Bartos, A.; Ricny, J.; Ripova, D. Structure and Pathology of Tau Protein in Alzheimer Disease. Int. J. Alzheimer’s Dis. 2012, 2012, 1–13. [CrossRef]
- Cleveland, D.W.; Hwo, S.-Y.; Kirschner, M.W. Physical and chemical properties of purified tau factor and the role of tau in microtubule assembly. J. Mol. Biol. 1977, 116, 227–247. [CrossRef]
- Aguzzi, A.; Montrasio, F.; Kaeser, P.S. Prions: health scare and biological challenge. Nat. Rev. Mol. Cell Biol. 2001, 2, 118–126. [CrossRef]
- Lewis, V.; Hooper, N.M. The role of lipid rafts in prion protein biology. Front. Biosci. 2011, 16, 151–168. [CrossRef]
- Costanzo, M.; Zurzolo, C. The cell biology of prion-like spread of protein aggregates: mechanisms and implication in neurodegeneration. Biochem. J. 2013, 452, 1–17. [CrossRef]
- Jucker, M.; Walker, L.C. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 2013, 501, 45–51. [CrossRef]
- Holmes, B.B.; Devos, S.L.; Kfoury, N.; Li, M.; Jacks, R.; Yanamandra, K.; Ouidja, M.O.; Brodsky, F.M.; Marasa, J.; Bagchi, D.P.; et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl. Acad. Sci. USA 2013, 110, E3138–E3147. [CrossRef]
- Weaver, I.C. Epigenetic Programming by Maternal Behavior and Pharmacological InterventionNature Versus Nurture: Let’s Call The Whole Thing Off. Epigenetics 2007, 2, 22–28. [CrossRef]
- Bredy, T.W.; Wu, H.; Crego, C.; Zellhoefer, J.; Sun, Y.E.; Barad, M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn. Mem. 2007, 14, 268–276. [CrossRef]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53(6):857-69. [CrossRef]
- Day, J.J.; Sweatt, J.D. Epigenetic Mechanisms in Cognition. Neuron 2011, 70, 813–829. [CrossRef]
- Parkinson disease 2022 [Available from: https://www.who.int/news-room/fact-sheets/detail/parkinson-disease.
- Hirsch, L.; Jette, N.; Frolkis, A.; Steeves, T.; Pringsheim, T. The Incidence of Parkinson’s Disease: A Systematic Review and Meta-Analysis. Neuroepidemiology 2016, 46, 292–300. [CrossRef]
- Cacabelos, R. Parkinson’s Disease: From Pathogenesis to Pharmacogenomics. Int. J. Mol. Sci. 2017, 18, 551. [CrossRef]
- Caillierez, R.; Bégard, S.; Lécolle, K.; Deramecourt, V.; Zommer, N.; Dujardin, S.; Loyens, A.; Dufour, N.; Aurégan, G.; Winderickx, J.; et al. Lentiviral Delivery of the Human Wild-type Tau Protein Mediates a Slow and Progressive Neurodegenerative Tau Pathology in the Rat Brain. Mol. Ther. 2013, 21, 1358–1368. [CrossRef]
- Clavaguera, F.; Akatsu, H.; Fraser, G.; Crowther, R.A.; Frank, S.; Hench, J.; Probst, A.; Winkler, D.T.; Reichwald, J.; Staufenbiel, M.; et al. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc. Natl. Acad. Sci. USA 2013, 110, 9535–9540. [CrossRef]
- Holmes, B.B.; Devos, S.L.; Kfoury, N.; Li, M.; Jacks, R.; Yanamandra, K.; Ouidja, M.O.; Brodsky, F.M.; Marasa, J.; Bagchi, D.P.; et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl. Acad. Sci. USA 2013, 110, E3138–E3147. [CrossRef]
- Levarska, L.; Zilka, N.; Jadhav, S.; Neradil, P.; Novak, M. Of Rodents and Men: The Mysterious Interneuronal Pilgrimage of Misfolded Protein Tau in Alzheimer’s Disease. J. Alzheimer’s Dis. 2013, 37, 569–577. [CrossRef]
- Masuda-Suzukake, M.; Nonaka, T.; Hosokawa, M.; Oikawa, T.; Arai, T.; Akiyama, H.; Mann, D.M.A.; Hasegawa, M. Prion-like spreading of pathological α-synuclein in brain. Brain 2013, 136, 1128–1138. [CrossRef]
- Oliveras-Salvá M, Van der Perren A, Casadei N, Stroobants S, Nuber S, D’Hooge R, et al. rAAV2/7 vector-mediated overexpression of alpha-synuclein in mouse substantia nigra induces protein aggregation and progressive dose-dependent neurodegeneration. Mol Neurodegener. 2013;8:44. [CrossRef]
- Pooler, A.M.; Phillips, E.C.; Lau, D.H.W.; Noble, W.; Hanger, D.P. Physiological release of endogenous tau is stimulated by neuronal activity. Embo Rep. 2013, 14, 389–394. [CrossRef]
- Sacino, A.N.; Brooks, M.; McGarvey, N.H.; McKinney, A.B.; A Thomas, M.; Levites, Y.; Ran, Y.; E Golde, T.; I Giasson, B. Induction of CNS α-synuclein pathology by fibrillar and non-amyloidogenic recombinant α-synuclein. Acta Neuropathol. Commun. 2013, 1, 38–38. [CrossRef]
- Ulusoy A, Rusconi R, Pérez-Revuelta BI, Musgrove RE, Helwig M, Winzen-Reichert B, et al. Caudo-rostral brain spreading of α-synuclein through vagal connections. EMBO Mol Med. 2013;5(7):1119-27. [CrossRef]
- Watts, J.C.; Giles, K.; Oehler, A.; Middleton, L.; Dexter, D.T.; Gentleman, S.M.; DeArmond, S.J.; Prusiner, S.B. Transmission of multiple system atrophy prions to transgenic mice. Proc. Natl. Acad. Sci. 2013, 110, 19555–19560. [CrossRef]
- Wu, J.W.; Herman, M.; Liu, L.; Simoes, S.; Acker, C.M.; Figueroa, H.; Steinberg, J.I.; Margittai, M.; Kayed, R.; Zurzolo, C.; et al. Small Misfolded Tau Species Are Internalized via Bulk Endocytosis and Anterogradely and Retrogradely Transported in Neurons. J. Biol. Chem. 2013, 288, 1856–1870. [CrossRef]
- Ahmed, Z.; Cooper, J.; Murray, T.K.; Garn, K.; McNaughton, E.; Clarke, H.; Parhizkar, S.; Ward, M.A.; Cavallini, A.; Jackson, S.; et al. A novel in vivo model of tau propagation with rapid and progressive neurofibrillary tangle pathology: the pattern of spread is determined by connectivity, not proximity. Acta Neuropathol. 2014, 127, 667–683. [CrossRef]
- Clavaguera, F.; Hench, J.; Lavenir, I.; Schweighauser, G.; Frank, S.; Goedert, M.; Tolnay, M. Peripheral administration of tau aggregates triggers intracerebral tauopathy in transgenic mice. Acta Neuropathol. 2013, 127, 299–301. [CrossRef]
- Dujardin, S.; Lécolle, K.; Caillierez, R.; Bégard, S.; Zommer, N.; Lachaud, C.; Carrier, S.; Dufour, N.; Aurégan, G.; Winderickx, J.; et al. Neuron-to-neuron wild-type Tau protein transfer through a trans-synaptic mechanism: relevance to sporadic tauopathies. Acta Neuropathol. Commun. 2013, 2, 14–14. [CrossRef]
- Holmes, B.B.; Furman, J.L.; Mahan, T.E.; Yamasaki, T.R.; Mirbaha, H.; Eades, W.C.; Belaygorod, L.; Cairns, N.J.; Holtzman, D.M.; Diamond, M.I. Proteopathic tau seeding predicts tauopathy in vivo. Proc. Natl. Acad. Sci. 2014, 111, E4376–85. [CrossRef]
- Holmqvist, S.; Chutna, O.; Bousset, L.; Aldrin-Kirk, P.; Li, W.; Björklund, T.; Wang, Z.-Y.; Roybon, L.; Melki, R.; Li, J.-Y. Direct evidence of Parkinson pathology spread from the gastrointestinal tract to the brain in rats. Acta Neuropathol. 2014, 128, 805–820. [CrossRef]
- Recasens A, Dehay B. Alpha-synuclein spreading in Parkinson’s disease. Front Neuroanat. 2014;8:159.
- Reyes, J.F.; Rey, N.L.; Bousset, L.; Melki, R.; Brundin, P.; Angot, E. Alpha-synuclein transfers from neurons to oligodendrocytes. Glia 2013, 62, 387–398. [CrossRef]
- Rotermund C, Truckenmüller FM, Schell H, Kahle PJ. Diet-induced obesity accelerates the onset of terminal phenotypes in α-synuclein transgenic mice. J Neurochem. 2014;131(6):848-58. [CrossRef]
- Sacino, A.N.; Brooks, M.; Thomas, M.A.; McKinney, A.B.; Lee, S.; Regenhardt, R.W.; McGarvey, N.H.; Ayers, J.I.; Notterpek, L.; Borchelt, D.R.; et al. Intramuscular injection of alpha-synuclein induces CNS alpha-synuclein pathology and a rapid-onset motor phenotype in transgenic mice. Proc. Natl. Acad. Sci. USA 2014, 111, 10732–10737. [CrossRef]
- Sacino AN, Brooks M, Thomas MA, McKinney AB, McGarvey NH, Rutherford NJ, et al. Amyloidogenic α-synuclein seeds do not invariably induce rapid, widespread pathology in mice. Acta Neuropathol. 2014;127(5):645-65. [CrossRef]
- Sanders, D.W.; Kaufman, S.K.; DeVos, S.L.; Sharma, A.M.; Mirbaha, H.; Li, A.; Barker, S.J.; Foley, A.C.; Thorpe, J.R.; Serpell, L.C.; et al. Distinct Tau Prion Strains Propagate in Cells and Mice and Define Different Tauopathies. Neuron 2014, 82, 1271–1288. [CrossRef]
- Asai, H.; Ikezu, S.; Tsunoda, S.; Medalla, M.; Luebke, J.; Haydar, T.; Wolozin, B.; Butovsky, O.; Kügler, S.; Ikezu, T. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat. Neurosci. 2015, 18, 1584–1593. [CrossRef]
- Bernis, M.E.; Babila, J.T.; Breid, S.; Wüsten, K.A.; Wüllner, U.; Tamgüney, G. Prion-like propagation of human brain-derived alpha-synuclein in transgenic mice expressing human wild-type alpha-synuclein. Acta Neuropathol. Commun. 2015, 3, 1–18. [CrossRef]
- Bourdenx M, Dovero S, Engeln M, Bido S, Bastide MF, Dutheil N, et al. Lack of additive role of ageing in nigrostriatal neurodegeneration triggered by α-synuclein overexpression. Acta Neuropathol Commun. 2015;3:46. [CrossRef]
- Daher, J.P.; Abdelmotilib, H.A.; Hu, X.; Volpicelli-Daley, L.A.; Moehle, M.S.; Fraser, K.B.; Needle, E.; Chen, Y.; Steyn, S.J.; Galatsis, P.; et al. Leucine-rich Repeat Kinase 2 (LRRK2) Pharmacological Inhibition Abates α-Synuclein Gene-induced Neurodegeneration. J. Biol. Chem. 2015, 290, 19433–19444. [CrossRef]
- Abounit, S.; Wu, J.W.; Duff, K.; Victoria, G.S.; Zurzolo, C. Tunneling nanotubes: A possible highway in the spreading of tau and other prion-like proteins in neurodegenerative diseases. Prion 2016, 10, 344–351. [CrossRef]
- D’Abramo, C.; Acker, C.M.; Schachter, J.B.; Terracina, G.; Wang, X.; Forest, S.K.; Davies, P. Detecting tau in serum of transgenic animal models after tau immunotherapy treatment. Neurobiol. Aging 2015, 37, 58–65. [CrossRef]
- Domert, J.; Sackmann, C.; Severinsson, E.; Agholme, L.; Bergström, J.; Ingelsson, M.; Hallbeck, M. Aggregated Alpha-Synuclein Transfer Efficiently between Cultured Human Neuron-Like Cells and Localize to Lysosomes. PLOS ONE 2016, 11, e0168700. [CrossRef]
- Helwig, M.; Klinkenberg, M.; Rusconi, R.; Musgrove, R.E.; Majbour, N.K.; El-Agnaf, O.M.A.; Ulusoy, A.; Di Monte, D.A. Brain propagation of transduced α-synuclein involves non-fibrillar protein species and is enhanced in α-synuclein null mice. Brain 2015, 139, 856–870. [CrossRef]
- Koprich JB, Johnston TH, Reyes G, Omana V, Brotchie JM. Towards a Non-Human Primate Model of Alpha-Synucleinopathy for Development of Therapeutics for Parkinson’s Disease: Optimization of AAV1/2 Delivery Parameters to Drive Sustained Expression of Alpha Synuclein and Dopaminergic Degeneration in Macaque. PLoS One. 2016;11(11):e0167235. [CrossRef]
- Aulić, S.; Masperone, L.; Narkiewicz, J.; Isopi, E.; Bistaffa, E.; Ambrosetti, E.; Pastore, B.; De Cecco, E.; Scaini, D.; Zago, P.; et al. α-Synuclein Amyloids Hijack Prion Protein to Gain Cell Entry, Facilitate Cell-to-Cell Spreading and Block Prion Replication. Sci. Rep. 2017, 7, 1–12. [CrossRef]
- Cavaliere, F.; Cerf, L.; Dehay, B.; Ramos-Gonzalez, P.; De Giorgi, F.; Bourdenx, M.; Bessede, A.; Obeso, J.A.; Matute, C.; Ichas, F.; et al. In vitro α-synuclein neurotoxicity and spreading among neurons and astrocytes using Lewy body extracts from Parkinson disease brains. Neurobiol. Dis. 2017, 103, 101–112. [CrossRef]
- DeVos, S.L.; Miller, R.L.; Schoch, K.M.; Holmes, B.B.; Kebodeaux, C.S.; Wegener, A.J.; Chen, G.; Shen, T.; Tran, H.; Nichols, B.; et al. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci. Transl. Med. 2017, 9. [CrossRef]
- Loria, F.; Vargas, J.Y.; Bousset, L.; Syan, S.; Salles, A.; Melki, R.; Zurzolo, C. α-Synuclein transfer between neurons and astrocytes indicates that astrocytes play a role in degradation rather than in spreading. Acta Neuropathol. 2017, 134, 789–808. [CrossRef]
- Ngolab, J.; Trinh, I.; Rockenstein, E.; Mante, M.; Florio, J.; Trejo, M.; Masliah, D.; Adame, A.; Masliah, E.; Rissman, R.A. Brain-derived exosomes from dementia with Lewy bodies propagate α-synuclein pathology. Acta Neuropathol. Commun. 2017, 5, 1–10. [CrossRef]
- Shimozawa, A.; Ono, M.; Takahara, D.; Tarutani, A.; Imura, S.; Masuda-Suzukake, M.; Higuchi, M.; Yanai, K.; Hisanaga, S.-I.; Hasegawa, M. Propagation of pathological α-synuclein in marmoset brain. Acta Neuropathol. Commun. 2017, 5, 12. [CrossRef]
- Ulusoy, A.; Phillips, R.J.; Helwig, M.; Klinkenberg, M.; Powley, T.L.; Di Monte, D.A. Brain-to-stomach transfer of α-synuclein via vagal preganglionic projections. Acta Neuropathol. 2016, 133, 381–393. [CrossRef]
- Wang, Y.; Balaji, V.; Kaniyappan, S.; Krüger, L.; Irsen, S.; Tepper, K.; Chandupatla, R.; Maetzler, W.; Schneider, A.; Mandelkow, E.; et al. The release and trans-synaptic transmission of Tau via exosomes. Mol. Neurodegener. 2017, 12, 1–25. [CrossRef]
- Polanco, J.C.; Li, C.; Durisic, N.; Sullivan, R.; Götz, J. Exosomes taken up by neurons hijack the endosomal pathway to spread to interconnected neurons. Acta Neuropathol. Commun. 2018, 6, 1–14. [CrossRef]
- Rusconi R, Ulusoy A, Aboutalebi H, Di Monte DA. Long-lasting pathological consequences of overexpression-induced α-synuclein spreading in the rat brain. Aging Cell. 2018;17(2). [CrossRef]
- Vitale, F.; Giliberto, L.; Ruiz, S.; Steslow, K.; Marambaud, P.; D’abramo, C. Anti-tau conformational scFv MC1 antibody efficiently reduces pathological tau species in adult JNPL3 mice. Acta Neuropathol. Commun. 2018, 6, 82. [CrossRef]
- Smolek, T.; Jadhav, S.; Brezovakova, V.; Cubinkova, V.; Valachova, B.; Novak, P.; Zilka, N. First-in-Rat Study of Human Alzheimer’s Disease Tau Propagation. Mol. Neurobiol. 2018, 56, 621–631. [CrossRef]
- Ferrer, I.; Zelaya, M.V.; García, M.A.; Carmona, M.; López-González, I.; Andrés-Benito, P.; Lidón, L.; Gavín, R.; Garcia-Esparcia, P.; del Rio, J.A. Relevance of host tau in tau seeding and spreading in tauopathies. Brain Pathol. 2020, 30, 298–318. [CrossRef]
- Ferrer, I.; García, M.A.; Carmona, M.; Andrés-Benito, P.; Torrejón-Escribano, B.; Garcia-Esparcia, P.; del Rio, J.A. Involvement of Oligodendrocytes in Tau Seeding and Spreading in Tauopathies. Front. Aging Neurosci. 2019, 11, 112. [CrossRef]
- Masuda-Suzukake, M.; Suzuki, G.; Hosokawa, M.; Nonaka, T.; Goedert, M.; Hasegawa, M. Dextran sulphate-induced tau assemblies cause endogenous tau aggregation and propagation in wild-type mice. Brain Commun. 2020, 2. [CrossRef]
- Mezias, C.; Rey, N.; Brundin, P.; Raj, A. Neural connectivity predicts spreading of alpha-synuclein pathology in fibril-injected mouse models: Involvement of retrograde and anterograde axonal propagation. Neurobiol. Dis. 2019, 134, 104623–104623. [CrossRef]
- Veys, L.; Van Houcke, J.; Aerts, J.; Van Pottelberge, S.; Mahieu, M.; Coens, A.; Melki, R.; Moechars, D.; De Muynck, L.; De Groef, L. Absence of Uptake and Prion-Like Spreading of Alpha-Synuclein and Tau After Intravitreal Injection of Preformed Fibrils. Front. Aging Neurosci. 2021, 12. [CrossRef]
- Courte, J.; Bousset, L.; Von Boxberg, Y.; Villard, C.; Melki, R.; Peyrin, J.-M. The expression level of alpha-synuclein in different neuronal populations is the primary determinant of its prion-like seeding. Sci. Rep. 2020, 10, 1–13. [CrossRef]
- Jimenez-Ferrer, I.; Bäckström, F.; Dueñas-Rey, A.; Jewett, M.; Boza-Serrano, A.; Luk, K.C.; Deierborg, T.; Swanberg, M. The MHC class II transactivator modulates seeded alpha-synuclein pathology and dopaminergic neurodegeneration in an in vivo rat model of Parkinson’s disease. Brain, Behav. Immun. 2020, 91, 369–382. [CrossRef]
- Thomsen, M.B.; Ferreira, S.A.; Schacht, A.C.; Jacobsen, J.; Simonsen, M.; Betzer, C.; Jensen, P.H.; Brooks, D.J.; Landau, A.M.; Romero-Ramos, M. PET imaging reveals early and progressive dopaminergic deficits after intra-striatal injection of preformed alpha-synuclein fibrils in rats. Neurobiol. Dis. 2020, 149, 105229. [CrossRef]
- Dutta D, Jana M, Majumder M, Mondal S, Roy A, Pahan K. Selective targeting of the TLR2/MyD88/NF-κB pathway reduces α-synuclein spreading in vitro and in vivo. Nat Commun. 2021;12(1):5382. [CrossRef]
- Bassil, F.; Meymand, E.S.; Brown, H.J.; Xu, H.; Cox, T.O.; Pattabhiraman, S.; Maghames, C.M.; Wu, Q.; Zhang, B.; Trojanowski, J.Q.; et al. α-Synuclein modulates tau spreading in mouse brains. J. Exp. Med. 2020, 218. [CrossRef]
- Matsuo, K.; Kawahata, I.; Melki, R.; Bousset, L.; Owada, Y.; Fukunaga, K. Suppression of α-synuclein propagation after intrastriatal injection in FABP3 null mice. Brain Res. 2021, 1760, 147383. [CrossRef]
- Pan L, Li C, Meng L, Tian Y, He M, Yuan X, et al. Tau accelerates α-synuclein aggregation and spreading in Parkinson’s disease. Brain. 2022;145(10):3454-71. [CrossRef]
- Dutta D, Paidi RK, Raha S, Roy A, Chandra S, Pahan K. Treadmill exercise reduces α-synuclein spreading via PPARα. Cell Rep. 2022;40(2):111058. [CrossRef]
- Garcia P, Jürgens-Wemheuer W, Uriarte Huarte O, Michelucci A, Masuch A, Brioschi S, et al. Neurodegeneration and neuroinflammation are linked, but independent of alpha-synuclein inclusions, in a seeding/spreading mouse model of Parkinson’s disease. Glia. 2022;70(5):935-60. [CrossRef]
- Vasili, E.; Dominguez-Meijide, A.; Flores-León, M.; Al-Azzani, M.; Kanellidi, A.; Melki, R.; Stefanis, L.; Outeiro, T.F. Endogenous Levels of Alpha-Synuclein Modulate Seeding and Aggregation in Cultured Cells. Mol. Neurobiol. 2022, 59, 1273–1284. [CrossRef]
- Sun, F.; Salinas, A.G.; Filser, S.; Blumenstock, S.; Medina-Luque, J.; Herms, J.; Sgobio, C. Impact of α-synuclein spreading on the nigrostriatal dopaminergic pathway depends on the onset of the pathology. Brain Pathol. 2021, 32, e13036. [CrossRef]
- Rahayel, S.; Mišić, B.; Zheng, Y.-Q.; Liu, Z.-Q.; Abdelgawad, A.; Abbasi, N.; Caputo, A.; Zhang, B.; Lo, A.; Kehm, V.; et al. Differentially targeted seeding reveals unique pathological alpha-synuclein propagation patterns. Brain 2021, 145, 1743–1756. [CrossRef]
- Lloyd, G.M.; Sorrentino, Z.A.; Quintin, S.; Gorion, K.-M.M.; Bell, B.M.; Paterno, G.; Long, B.; Prokop, S.; Giasson, B.I. Unique seeding profiles and prion-like propagation of synucleinopathies are highly dependent on the host in human α-synuclein transgenic mice. Acta Neuropathol. 2022, 143, 663–685. [CrossRef]
- Jain, N.; Lewis, C.A.; Ulrich, J.D.; Holtzman, D.M. Chronic TREM2 activation exacerbates Aβ-associated tau seeding and spreading. J. Exp. Med. 2022, 220. [CrossRef]
- De Gerando, A.M.; Welikovitch, L.A.; Khasnavis, A.; Commins, C.; Glynn, C.; Chun, J.E.; Perbet, R.; Hyman, B.T. Tau seeding and spreading in vivo is supported by both AD-derived fibrillar and oligomeric tau. Acta Neuropathol. 2023, 146, 191–210. [CrossRef]
- Deng H, Wang P, Jankovic J. The genetics of Parkinson disease. Ageing Res Rev. 2018;42:72-85. [CrossRef]
- Schulte C, Gasser T. Genetic basis of Parkinson’s disease: inheritance, penetrance, and expression. Appl Clin Genet. 2011;4:67-80. [CrossRef]
- Sidransky, E.; Lopez, G. The link between the GBA gene and parkinsonism. Lancet Neurol. 2012, 11, 986–998. [CrossRef]
- Nalls, M.A.; Pankratz, N.; Lill, C.M.; Do, C.B.; Hernandez, D.G.; Saad, M.; DeStefano, A.L.; Kara, E.; Bras, J.; Sharma, M.; et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014, 46, 989–993. [CrossRef]
- Simón-Sánchez, J.; Schulte, C.; Bras, J.M.; Sharma, M.; Gibbs, J.R.; Berg, D.; Paisan-Ruiz, C.; Lichtner, P.; Scholz, S.W.; Hernandez, D.G.; et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009, 41, 1308–1312. [CrossRef]
- Song, H.; Chen, J.; Huang, J.; Sun, P.; Liu, Y.; Xu, L.; Wei, C.; Mu, X.; Lu, X.; Wang, W.; et al. Epigenetic modification in Parkinson’s disease. Front. Cell Dev. Biol. 2023, 11, 1123621. [CrossRef]
- Goers, J.; Manning-Bog, A.B.; McCormack, A.L.; Millett, I.S.; Doniach, S.; Di Monte, D.A.; Uversky, V.N.; Fink, A.L. Nuclear Localization of alpha-Synuclein and Its Interaction with Histones. Biochemistry 2003, 42, 8465–8471. [CrossRef]
- Mouradian, M.M. MicroRNAs in Parkinson’s disease. Neurobiol. Dis. 2012, 46, 279–284. [CrossRef]
- McMillan, K.J.; Murray, T.K.; Bengoa-Vergniory, N.; Cordero-Llana, O.; Cooper, J.; Buckley, A.; Wade-Martins, R.; Uney, J.B.; O’neill, M.J.; Wong, L.F.; et al. Loss of MicroRNA-7 Regulation Leads to α-Synuclein Accumulation and Dopaminergic Neuronal Loss In Vivo. Mol. Ther. 2017, 25, 2404–2414. [CrossRef]
- Doxakis, E. Post-transcriptional Regulation of alpha-Synuclein Expression by mir-7 and mir-153. J. Biol. Chem. 2010, 285, 12726–12734. [CrossRef]
- Tan, Y.-Y.; Wu, L.; Zhao, Z.-B.; Wang, Y.; Xiao, Q.; Liu, J.; Wang, G.; Ma, J.-F.; Chen, S.-D. Methylation of α-synuclein and leucine-rich repeat kinase 2 in leukocyte DNA of Parkinson’s disease patients. Park. Relat. Disord. 2014, 20, 308–313. [CrossRef]
- de Boni, L.; Riedel, L.; Schmitt, I.; Kraus, T.F.; Kaut, O.; Piston, D.; Akbarian, S.; Wüllner, U. DNA methylation levels of α-synuclein intron 1 in the aging brain. Neurobiol. Aging 2015, 36, 3334.e7–3334.e11. [CrossRef]
- Feng, J.; Zhou, Y.; Campbell, S.L.; Le, T.; Li, E.; Sweatt, J.D.; Silva, A.J.; Fan, G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010, 13, 423–430. [CrossRef]
- Masliah E, Dumaop W, Galasko D, Desplats P. Distinctive patterns of DNA methylation associated with Parkinson disease: identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics. 2013;8(10):1030-8. [CrossRef]
- Wang, Y.; Liu, H.; Zhang, B.-S.; Soares, J.C.; Zhang, X.Y. Low BDNF is associated with cognitive impairments in patients with Parkinson’s disease. Park. Relat. Disord. 2016, 29, 66–71. [CrossRef]
- Habibi, E.; Masoudi-Nejad, A.; Abdolmaleky, H.M.; Haggarty, S.J. Emerging roles of epigenetic mechanisms in Parkinson’s disease. Funct. Integr. Genom. 2011, 11, 523–537. [CrossRef]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, et al. DNA Methylation-Related Chromatin Remodeling in Activity-Dependent Bdnf Gene Regulation. Science. 2003;302(5646):890-3. [CrossRef]
- Howells, D.; Porritt, M.; Wong, J.; Batchelor, P.; Kalnins, R.; Hughes, A.; Donnan, G. Reduced BDNF mRNA Expression in the Parkinson’s Disease Substantia Nigra. Exp. Neurol. 2000, 166, 127–135. [CrossRef]
- Muñoz, P.C.; A Aspé, M.; Contreras, L.S.; Palacios, A.G. Correlations of recognition memory performance with expression and methylation of brain-derived neurotrophic factor in rats. Biol. Res. 2010, 43, 251–258. [CrossRef]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28(42):10576-86. [CrossRef]
- Yang, X.; Xu, S.; Qian, Y.; He, X.; Chen, S.; Xiao, Q. Hypermethylation of the Gene Coding for PGC-1α in Peripheral Blood Leukocytes of Patients With Parkinson’s Disease. Front. Neurosci. 2020, 14, 97. [CrossRef]
- Su X, Chu Y, Kordower JH, Li B, Cao H, Huang L, et al. PGC-1α Promoter Methylation in Parkinson’s Disease. PLoS One. 2015;10(8):e0134087. [CrossRef]
- Gong, B.; Pan, Y.; Vempati, P.; Zhao, W.; Knable, L.; Ho, L.; Wang, J.; Sastre, M.; Ono, K.; Sauve, A.A.; et al. Nicotinamide riboside restores cognition through an upregulation of proliferator-activated receptor-γ coactivator 1α regulated β-secretase 1 degradation and mitochondrial gene expression in Alzheimer’s mouse models. Neurobiol. Aging 2013, 34, 1581–1588. [CrossRef]
- Zhao Y, Zhang J, Zheng Y, Zhang Y, Zhang XJ, Wang H, et al. NAD(+) improves cognitive function and reduces neuroinflammation by ameliorating mitochondrial damage and decreasing ROS production in chronic cerebral hypoperfusion models through Sirt1/PGC-1α pathway. J Neuroinflammation. 2021;18(1):207. [CrossRef]
- Han, B.; Jiang, W.; Liu, H.; Wang, J.; Zheng, K.; Cui, P.; Feng, Y.; Dang, C.; Bu, Y.; Wang, Q.M.; et al. Upregulation of neuronal PGC-1 α ameliorates cognitive impairment induced by chronic cerebral hypoperfusion. Theranostics 2020, 10, 2832–2848. [CrossRef]
- Coupland, K.G.; Mellick, G.D.; Silburn, P.A.; Mather, K.; Armstrong, N.J.; Sachdev, P.S.; Brodaty, H.; Huang, Y.; Halliday, G.M.; Hallupp, M.; et al. DNA methylation of the MAPT gene in Parkinson’s disease cohorts and modulation by vitamin E In Vitro. Mov. Disord. 2013, 29, 1606–1614. [CrossRef]
- Setó-Salvia N, Clarimón J, Pagonabarraga J, Pascual-Sedano B, Campolongo A, Combarros O, et al. Dementia risk in Parkinson disease: disentangling the role of MAPT haplotypes. Arch Neurol. 2011;68(3):359-64. [CrossRef]
- Winder-Rhodes, S.E.; Hampshire, A.; Rowe, J.B.; Peelle, J.E.; Robbins, T.W.; Owen, A.M.; Barker, R.A. Association between MAPT haplotype and memory function in patients with Parkinson’s disease and healthy aging individuals. Neurobiol. Aging 2014, 36, 1519–1528. [CrossRef]
- Tunold, J.-A.; Geut, H.; Rozemuller, J.M.A.; Henriksen, S.P.; Toft, M.; van de Berg, W.D.J.; Pihlstrøm, L. APOE and MAPT Are Associated With Dementia in Neuropathologically Confirmed Parkinson’s Disease. Front. Neurol. 2021, 12. [CrossRef]
- Jiskoot, L.C.; Panman, J.L.; van Asseldonk, L.; Franzen, S.; Meeter, L.H.H.; Kaat, L.D.; van der Ende, E.L.; Dopper, E.G.P.; Timman, R.; van Minkelen, R.; et al. Longitudinal cognitive biomarkers predicting symptom onset in presymptomatic frontotemporal dementia. J. Neurol. 2018, 265, 1381–1392. [CrossRef]
- Pal S, Tyler JK. Epigenetics and aging. Sci Adv. 2016;2(7):e1600584. [CrossRef]
- Levenson, J.M.; Sweatt, J.D. Epigenetic mechanisms in memory formation. Nat. Rev. Neurosci. 2005, 6, 108–118. [CrossRef]
- Hwang, J.-Y.; Aromolaran, K.A.; Zukin, R.S. The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat. Rev. Neurosci. 2017, 18, 347–361. [CrossRef]
- Federman N, de la Fuente V, Zalcman G, Corbi N, Onori A, Passananti C, et al. Nuclear factor κB-dependent histone acetylation is specifically involved in persistent forms of memory. J Neurosci. 2013;33(17):7603-14. [CrossRef]
- de la Fuente V, Federman N, Zalcman G, Salles A, Freudenthal R, Romano A. NF-κB transcription factor role in consolidation and reconsolidation of persistent memories. Front Mol Neurosci. 2015;8:50. [CrossRef]
- Basavarajappa, B.S.; Subbanna, S. Histone Methylation Regulation in Neurodegenerative Disorders. Int. J. Mol. Sci. 2021, 22, 4654. [CrossRef]
- Chatterjee, P.; Roy, D.; Bhattacharyya, M.; Bandyopadhyay, S. Biological networks in Parkinson’s disease: an insight into the epigenetic mechanisms associated with this disease. BMC Genom. 2017, 18, 721. [CrossRef]
- Morse, S.J.; Butler, A.A.; Davis, R.L.; Soller, I.J.; Lubin, F.D. Environmental Enrichment Reverses Histone Methylation Changes in the Aged Hippocampus and Restores Age-Related Memory Deficits. Biology 2015, 4, 298–313. [CrossRef]
- Gupta-Agarwal, S.; Jarome, T.J.; Fernandez, J.; Lubin, F.D. NMDA receptor- and ERK-dependent histone methylation changes in the lateral amygdala bidirectionally regulate fear memory formation. Learn. Mem. 2014, 21, 351–362. [CrossRef]
- Gupta-Agarwal, S.; Franklin, A.V.; DeRamus, T.; Wheelock, M.; Davis, R.L.; McMahon, L.L.; Lubin, F.D. G9a/GLP Histone Lysine Dimethyltransferase Complex Activity in the Hippocampus and the Entorhinal Cortex Is Required for Gene Activation and Silencing during Memory Consolidation. J. Neurosci. 2012, 32, 5440–5453. [CrossRef]
- Riddle, N.C.; Minoda, A.; Kharchenko, P.V.; Alekseyenko, A.A.; Schwartz, Y.B.; Tolstorukov, M.Y.; Gorchakov, A.A.; Jaffe, J.D.; Kennedy, C.; Linder-Basso, D.; et al. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 2010, 21, 147–163. [CrossRef]
- Satake, W.; Nakabayashi, Y.; Mizuta, I.; Hirota, Y.; Ito, C.; Kubo, M.; Kawaguchi, T.; Tsunoda, T.; Watanabe, M.; Takeda, A.; et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat. Genet. 2009, 41, 1303–1307. [CrossRef]
- Gupta, S.; Kim, S.Y.; Artis, S.; Molfese, D.L.; Schumacher, A.; Sweatt, J.D.; Paylor, R.E.; Lubin, F.D. Histone Methylation Regulates Memory Formation. J. Neurosci. 2010, 30, 3589–3599. [CrossRef]
- Södersten, E.; Feyder, M.; Lerdrup, M.; Gomes, A.-L.; Kryh, H.; Spigolon, G.; Caboche, J.; Fisone, G.; Hansen, K. Dopamine Signaling Leads to Loss of Polycomb Repression and Aberrant Gene Activation in Experimental Parkinsonism. PLOS Genet. 2014, 10, e1004574. [CrossRef]
- Gibney E, Nolan C. Epigenetics and gene expression. Heredity. 2010;105(1):4-13.
- Mu, M.-D.; Qian, Z.-M.; Yang, S.-X.; Rong, K.-L.; Yung, W.-H.; Ke, Y. Therapeutic effect of a histone demethylase inhibitor in Parkinson’s disease. Cell Death Dis. 2020, 11, 1–16. [CrossRef]
- Mullen, R.J.; Buck, C.R.; Smith, A.M. NeuN, a neuronal specific nuclear protein in vertebrates. Development 1992, 116, 201–211. [CrossRef]
- Kim, K.K.; Adelstein, R.S.; Kawamoto, S. Identification of Neuronal Nuclei (NeuN) as Fox-3, a New Member of the Fox-1 Gene Family of Splicing Factors. J. Biol. Chem. 2009, 284, 31052–31061. [CrossRef]
- Guhathakurta S, Kim J, Adams L, Basu S, Song MK, Adler E, et al. Targeted attenuation of elevated histone marks at SNCA alleviates α-synuclein in Parkinson’s disease. EMBO Molecular Medicine. 2021;13(2):e12188. [CrossRef]
- Miranda, M.; Morici, J.F.; Zanoni, M.B.; Bekinschtein, P. Brain-Derived Neurotrophic Factor: A Key Molecule for Memory in the Healthy and the Pathological Brain. Front. Cell. Neurosci. 2019, 13, 363. [CrossRef]
- Srivastav, S.; Walitza, S.; Grünblatt, E. Emerging role of miRNA in attention deficit hyperactivity disorder: a systematic review. Adhd Atten. Deficit Hyperact. Disord. 2017, 10, 49–63. [CrossRef]
- Tang, G.-B.; Zeng, Y.-Q.; Liu, P.-P.; Mi, T.-W.; Zhang, S.-F.; Dai, S.-K.; Tang, Q.-Y.; Yang, L.; Xu, Y.-J.; Yan, H.-L.; et al. The Histone H3K27 Demethylase UTX Regulates Synaptic Plasticity and Cognitive Behaviors in Mice. Front. Mol. Neurosci. 2017, 10, 267–267. [CrossRef]
- Harrison, I.F.; Smith, A.D.; Dexter, D.T. Pathological histone acetylation in Parkinson’s disease: Neuroprotection and inhibition of microglial activation through SIRT 2 inhibition. Neurosci. Lett. 2017, 666, 48–57. [CrossRef]
- Hait, N.C.; E Wise, L.; Allegood, J.C.; O’Brien, M.; Avni, D.; Reeves, T.M.; E Knapp, P.; Lu, J.; Luo, C.; Miles, M.F.; et al. Active, phosphorylated fingolimod inhibits histone deacetylases and facilitates fear extinction memory. Nat. Neurosci. 2014, 17, 971–980. [CrossRef]
- Peixoto, L.; Abel, T. The Role of Histone Acetylation in Memory Formation and Cognitive Impairments. Neuropsychopharmacology 2012, 38, 62–76. [CrossRef]
- Huang, M.; Malovic, E.; Ealy, A.; Jin, H.; Anantharam, V.; Kanthasamy, A.; Kanthasamy, A.G. Microglial immune regulation by epigenetic reprogramming through histone H3K27 acetylation in neuroinflammation. Front. Immunol. 2023, 14. [CrossRef]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522-31. [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [CrossRef]
- Duffy, C.P.; McCoy, C.E. The Role of MicroRNAs in Repair Processes in Multiple Sclerosis. Cells 2020, 9, 1711. [CrossRef]
- Zetterberg, H.; Burnham, S.C. Blood-based molecular biomarkers for Alzheimer’s disease. Mol. Brain 2019, 12, 1–7. [CrossRef]
- Prabhakar P, Chandra SR, Christopher R. Circulating microRNAs as potential biomarkers for the identification of vascular dementia due to cerebral small vessel disease. Age Ageing. 2017;46(5):861-4. [CrossRef]
- Nadim WD, Simion V, Benedetti H, Pichon C, Baril P, Morisset-Lopez S. MicroRNAs in Neurocognitive Dysfunctions: New Molecular Targets for Pharmacological Treatments? Curr Neuropharmacol. 2017;15(2):260-75.
- Miquel, S.; Champ, C.; Day, J.; Aarts, E.; Bahr, B.A.; Bakker, M.; Bánáti, D.; Calabrese, V.; Cederholm, T.; Cryan, J.; et al. Poor cognitive ageing: Vulnerabilities, mechanisms and the impact of nutritional interventions. Ageing Res. Rev. 2018, 42, 40–55. [CrossRef]
- Aarsland D, Creese B, Politis M, Chaudhuri KR, Ffytche DH, Weintraub D, et al. Cognitive decline in Parkinson disease. Nat Rev Neurol. 2017;13(4):217-31. [CrossRef]
- Junn, E.; Lee, K.-W.; Jeong, B.S.; Chan, T.W.; Im, J.-Y.; Mouradian, M.M. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. USA 2009, 106, 13052–13057. [CrossRef]
- Zhou, Y.; Lu, M.; Du, R.-H.; Qiao, C.; Jiang, C.-Y.; Zhang, K.-Z.; Ding, J.-H.; Hu, G. MicroRNA-7 targets Nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson’s disease. Mol. Neurodegener. 2016, 11, 1–15. [CrossRef]
- Yao, G.-Y.; Zhu, Q.; Xia, J.; Chen, F.-J.; Huang, M.; Liu, J.; Zhou, T.-T.; Wei, J.-F.; Cui, G.-Y.; Zheng, K.-Y.; et al. Ischemic postconditioning confers cerebroprotection by stabilizing VDACs after brain ischemia. Cell Death Dis. 2018, 9, 1–15. [CrossRef]
- Li, S.; Lv, X.; Zhai, K.; Xu, R.; Zhang, Y.; Zhao, S.; Qin, X.; Yin, L.; Lou, J. MicroRNA-7 inhibits neuronal apoptosis in a cellular Parkinson’s disease model by targeting Bax and Sirt2. Am J Transl Res. 2016;8(2):993-1004.
- Geng, L.; Liu, W.; Chen, Y. miR-124-3p attenuates MPP+-induced neuronal injury by targeting STAT3 in SH-SY5Y cells. Exp. Biol. Med. 2017, 242, 1757–1764. [CrossRef]
- Han, J.; Pu, C.-X.; Xiao, Q.-X.; Tang, L.-J.; Liu, T.; He, L.; Ren, Y.-B.; Liu, Q.; Zhang, Y. miRNA-124-3p targeting of LPIN1 attenuates inflammation and apoptosis in aged male rats cardiopulmonary bypass model of perioperative neurocognitive disorders. Exp. Gerontol. 2021, 155, 111578. [CrossRef]
- Gan, L.; Li, Z.; Lv, Q.; Huang, W. Rabies virus glycoprotein (RVG29)-linked microRNA-124-loaded polymeric nanoparticles inhibit neuroinflammation in a Parkinson’s disease model. Int. J. Pharm. 2019, 567, 118449. [CrossRef]
- Xing RX, Li LG, Liu XW, Tian BX, Cheng Y. Down regulation of miR-218, miR-124, and miR-144 relates to Parkinson’s disease via activating NF-κB signaling. Kaohsiung J Med Sci. 2020;36(10):786-92. [CrossRef]
- Pantano, L.; Friedländer, M.R.; Escaramís, G.; Lizano, E.; Pallarès-Albanell, J.; Ferrer, I.; Estivill, X.; Martí, E. Specific small-RNA signatures in the amygdala at premotor and motor stages of Parkinson’s disease revealed by deep sequencing analysis. Bioinformatics 2015, 32, 673–681. [CrossRef]
- Rajasethupathy, P.; Fiumara, F.; Sheridan, R.; Betel, D.; Puthanveettil, S.V.; Russo, J.J.; Sander, C.; Tuschl, T.; Kandel, E. Characterization of Small RNAs in Aplysia Reveals a Role for miR-124 in Constraining Synaptic Plasticity through CREB. Neuron 2009, 63, 803–817. [CrossRef]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27(3):435-48.
- Konopka, W.; Kiryk, A.; Novak, M.; Herwerth, M.; Parkitna, J.R.; Wawrzyniak, M.; Kowarsch, A.; Michaluk, P.; Dzwonek, J.; Arnsperger, T.; et al. MicroRNA Loss Enhances Learning and Memory in Mice. J. Neurosci. 2010, 30, 14835–14842. [CrossRef]
- Kang, Q.; Xiang, Y.; Li, D.; Liang, J.; Zhang, X.; Zhou, F.; Qiao, M.; Nie, Y.; He, Y.; Cheng, J.; et al. MiR-124-3p attenuates hyperphosphorylation of tau protein-induced apoptosis via caveolin-1-PI3K/Akt/GSK3β pathway in N2a/APP695swe cells. Oncotarget 2017, 8, 24314–24326. [CrossRef]
- Bartsch, D.; Casadio, A.; A Karl, K.; Serodio, P.; Kandel, E.R. CREB1 Encodes a Nuclear Activator, a Repressor, and a Cytoplasmic Modulator that Form a Regulatory Unit Critical for Long-Term Facilitation. Cell 1998, 95, 211–223. [CrossRef]
- Yang, Y.; Shu, X.; Liu, D.; Shang, Y.; Wu, Y.; Pei, L.; Xu, X.; Tian, Q.; Zhang, J.; Qian, K.; et al. EPAC Null Mutation Impairs Learning and Social Interactions via Aberrant Regulation of miR-124 and Zif268 Translation. Neuron 2012, 73, 774–788. [CrossRef]
- Blount, G.S.; Coursey, L.; Kocerha, J. MicroRNA Networks in Cognition and Dementia. Cells 2022, 11, 1882. [CrossRef]
- Ren Y, Li H, Xie W, Wei N, Liu M. MicroRNA-195 triggers neuroinflammation in Parkinson’s disease in a Rho-associated kinase 1-dependent manner. Mol Med Rep. 2019;19(6):5153-61. [CrossRef]
- Ai, J.; Sun, L.-H.; Che, H.; Zhang, R.; Zhang, T.-Z.; Wu, W.-C.; Su, X.-L.; Chen, X.; Yang, G.; Li, K.; et al. MicroRNA-195Protects Against Dementia Induced by Chronic Brain Hypoperfusion via Its Anti-Amyloidogenic Effect in Rats. J. Neurosci. 2013, 33, 3989–4001. [CrossRef]
- Sun LH, Ban T, Liu CD, Chen QX, Wang X, Yan ML, et al. Activation of Cdk5/p25 and Tau phosphorylation following chronic brain hypoperfusion in rats involves microRNA-195 down-regulation. J Neurochem. 2015;134(6):1139-51. [CrossRef]
- Fischer, A.; Sananbenesi, F.; Schrick, C.; Spiess, J.; Radulovic, J. Cyclin-Dependent Kinase 5 Is Required for Associative Learning. J. Neurosci. 2002, 22, 3700–3707. [CrossRef]
- Junker, A.; Krumbholz, M.; Eisele, S.; Mohan, H.; Augstein, F.; Bittner, R.; Lassmann, H.; Wekerle, H.; Hohlfeld, R.; Meinl, E. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 2009, 132, 3342–3352. [CrossRef]
- Sun, Q.; Wang, S.; Chen, J.; Cai, H.; Huang, W.; Zhang, Y.; Wang, L.; Xing, Y. MicroRNA-190 alleviates neuronal damage and inhibits neuroinflammation via Nlrp3 in MPTP-induced Parkinson’s disease mouse model. J. Cell. Physiol. 2019, 234, 23379–23387. [CrossRef]
- Liu, Q.; Hou, A.; Zhang, Y.; Guo, Y.; Li, J.; Yao, Y.; Niu, K.; Li, H.; Ma, Y.; Cao, J. MiR-190a potentially ameliorates postoperative cognitive dysfunction by regulating Tiam1. BMC Genom. 2019, 20, 1–12. [CrossRef]
- Jiang, C.; Dong, N.; Feng, J.; Hao, M. MiRNA-190 exerts neuroprotective effects against ischemic stroke through Rho/Rho-kinase pathway. Pfl?gers Arch. Eur. J. Physiol. 2020, 473, 121–130. [CrossRef]
- Iyer, A.; Zurolo, E.; Prabowo, A.; Fluiter, K.; Spliet, W.G.M.; van Rijen, P.C.; Gorter, J.A.; Aronica, E. MicroRNA-146a: A Key Regulator of Astrocyte-Mediated Inflammatory Response. PLOS ONE 2012, 7, e44789. [CrossRef]
- Lukiw WJ, Dua P, Pogue AI, Eicken C, Hill JM. Upregulation of micro RNA-146a (miRNA-146a), a marker for inflammatory neurodegeneration, in sporadic Creutzfeldt-Jakob disease (sCJD) and Gerstmann-Straussler-Scheinker (GSS) syndrome. J Toxicol Environ Health A. 2011;74(22-24):1460-8. [CrossRef]
- Alexandrov, P.N.; Dua, P.; Hill, J.M.; Bhattacharjee, S.; Zhao, Y.; Lukiw, W.J. microRNA (miRNA) speciation in Alzheimer’s disease (AD) cerebrospinal fluid (CSF) and extracellular fluid (ECF). Int. J. Biochem. Mol. Biol. 2012, 3, 365–373.
- Kim, J.-H.; Jou, I.; Joe, I.J.A.E.-H. Suppression of miR-155 Expression in IFN-γ-Treated Astrocytes and Microglia by DJ-1: A Possible Mechanism for Maintaining SOCS1 Expression. Exp. Neurobiol. 2014, 23, 148–154. [CrossRef]
- Cardoso AL, Guedes JR, Pereira de Almeida L, Pedroso de Lima MC. miR-155 modulates microglia-mediated immune response by down-regulating SOCS-1 and promoting cytokine and nitric oxide production. Immunology. 2012;135(1):73-88. [CrossRef]
- Kim, J.; Inoue, K.; Ishii, J.; Vanti, W.B.; Voronov, S.V.; Murchison, E.; Hannon, G.; Abeliovich, A. A MicroRNA Feedback Circuit in Midbrain Dopamine Neurons. Science 2007, 317, 1220–1224. [CrossRef]
- Zhang, Y.; Liu, J.; Xie, C.; Wu, P. Overexpression of miR-133b protects against isoflurane-induced learning and memory impairment. Exp. Ther. Med. 2021, 22, 1–7. [CrossRef]
- Yang, Q.; Zhao, Q.; Yin, Y. miR-133b is a potential diagnostic biomarker for Alzheimer’s disease and has a neuroprotective role. Exp. Ther. Med. 2019, 18, 2711–2718. [CrossRef]
- Schulz J, Takousis P, Wohlers I, Itua IOG, Dobricic V, Rücker G, et al. Meta-analyses identify differentially expressed micrornas in Parkinson’s disease. Ann Neurol. 2019;85(6):835-51. [CrossRef]
- Schlaudraff, F.; Gründemann, J.; Fauler, M.; Dragicevic, E.; Hardy, J.; Liss, B. Orchestrated increase of dopamine and PARK mRNAs but not miR-133b in dopamine neurons in Parkinson’s disease. Neurobiol. Aging 2014, 35, 2302–2315. [CrossRef]
- Villar-Menéndez, I.; Porta, S.; Buira, S.P.; Pereira-Veiga, T.; Díaz-Sánchez, S.; Albasanz, J.L.; Ferrer, I.; Martín, M.; Barrachina, M. Increased striatal adenosine A2A receptor levels is an early event in Parkinson’s disease-related pathology and it is potentially regulated by miR-34b. Neurobiol. Dis. 2014, 69, 206–214. [CrossRef]
- Kim, W.; Lee, Y.; McKenna, N.D.; Yi, M.; Simunovic, F.; Wang, Y.; Kong, B.; Rooney, R.J.; Seo, H.; Stephens, R.M.; et al. miR-126 contributes to Parkinson’s disease by dysregulating the insulin-like growth factor/phosphoinositide 3-kinase signaling. Neurobiol. Aging 2014, 35, 1712–1721. [CrossRef]
- Briggs, C.E.; Wang, Y.; Kong, B.; Woo, T.-U.W.; Iyer, L.K.; Sonntag, K.C. Midbrain dopamine neurons in Parkinson׳s disease exhibit a dysregulated miRNA and target-gene network. Brain Res. 2015, 1618, 111–121. [CrossRef]
- Kong, F.; Zhou, J.; Zhou, W.; Guo, Y.; Li, G.; Yang, L. Protective role of microRNA-126 in intracerebral hemorrhage. Mol. Med. Rep. 2017, 15, 1419–1425. [CrossRef]
- Yu, P.; Venkat, P.; Chopp, M.; Zacharek, A.; Shen, Y.; Ning, R.; Liang, L.; Li, W.; Zhang, L.; Landschoot-Ward, J.; et al. Role of microRNA-126 in vascular cognitive impairment in mice. J. Cereb. Blood Flow Metab. 2018, 39, 2497–2511. [CrossRef]
- Hoss, A.G.; Labadorf, A.; Beach, T.G.; Latourelle, J.C.; Myers, R.H. microRNA Profiles in Parkinson’s Disease Prefrontal Cortex. Front. Aging Neurosci. 2016, 8, 36. [CrossRef]
- Tatura, R.; Kraus, T.; Giese, A.; Arzberger, T.; Buchholz, M.; Höglinger, G.; Müller, U. Parkinson’s disease: SNCA-, PARK2-, and LRRK2- targeting microRNAs elevated in cingulate gyrus. Park. Relat. Disord. 2016, 33, 115–121. [CrossRef]
- Sun, L.; Zhao, M.; Zhang, J.; Liu, A.; Ji, W.; Li, Y.; Yang, X.; Wu, Z. MiR-144 promotes β-amyloid accumulation-induced cognitive impairments by targeting ADAM10 following traumatic brain injury. Oncotarget 2017, 8, 59181–59203. [CrossRef]
- Wang, J.; Chen, T.; Shan, G. miR-148b Regulates Proliferation and Differentiation of Neural Stem Cells via Wnt/β-Catenin Signaling in Rat Ischemic Stroke Model. Front. Cell. Neurosci. 2017, 11, 329–329. [CrossRef]
- Yang, H.; Zhang, Y.; Chen, H.; Zhu, Y.; Li, Y.; Ouyang, F.; Chu, L.; Liu, D. Mir-184 Contributes to Brain Injury Through Targeting PPAP2B Following Ischemic Stroke in Male Rats. Front. Mol. Neurosci. 2021, 14. [CrossRef]
- Mendes-Silva, A.P.; Fujimura, P.T.; Silva, J.R.d.C.; Teixeira, A.L.; Vieira, E.M.; Guedes, P.H.G.; Barroso, L.S.S.; Nicolau, M.d.S.; Ferreira, J.D.R.; Bertola, L.; et al. Brain-enriched MicroRNA-184 is downregulated in older adults with major depressive disorder: A translational study. J. Psychiatr. Res. 2019, 111, 110–120. [CrossRef]
- McKiernan, R.C.; Jimenez-Mateos, E.M.; Sano, T.; Bray, I.; Stallings, R.L.; Simon, R.P.; Henshall, D.C. Expression profiling the microRNA response to epileptic preconditioning identifies miR-184 as a modulator of seizure-induced neuronal death. Exp. Neurol. 2012, 237, 346–354. [CrossRef]
- Shan, Y.; Hu, J.; Lv, H.; Cui, X.; Di, W. miR-221 Exerts Neuroprotective Effects in Ischemic Stroke by Inhibiting the Proinflammatory Response. J. Stroke Cerebrovasc. Dis. 2020, 30, 105489. [CrossRef]
- Song, D.; Li, G.; Hong, Y.; Zhang, P.; Zhu, J.; Yang, L.; Huang, J. miR-199a decreases Neuritin expression involved in the development of Alzheimer’s disease in APP/PS1 mice. Int. J. Mol. Med. 2020, 46, 384–396. [CrossRef]
- Tolosa E, Botta-Orfila T, Morató X, Calatayud C, Ferrer-Lorente R, Martí MJ, et al. MicroRNA alterations in iPSC-derived dopaminergic neurons from Parkinson disease patients. Neurobiol Aging. 2018;69:283-91. [CrossRef]
- Nakahama T, Hanieh H, Nguyen NT, Chinen I, Ripley B, Millrine D, et al. Aryl hydrocarbon receptor-mediated induction of the microRNA-132/212 cluster promotes interleukin-17–producing T-helper cell differentiation. Proceedings of the National Academy of Sciences. 2013;110(29):11964-9. [CrossRef]
- Wayman, G.A.; Davare, M.; Ando, H.; Fortin, D.; Varlamova, O.; Cheng, H.-Y.M.; Marks, D.; Obrietan, K.; Soderling, T.R.; Goodman, R.H.; et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl. Acad. Sci. USA 2008, 105, 9093–9098. [CrossRef]
- Cong, L.; Cong, Y.; Feng, N.; Liang, W.; Wu, Y. Up-regulated microRNA-132 reduces the cognition-damaging effect of sevoflurane on Alzheimer’s disease rats by inhibiting FOXA1. Genomics 2021, 113, 3644–3652. [CrossRef]
- Deng, Y.; Zhang, J.; Sun, X.; Ma, G.; Luo, G.; Miao, Z.; Song, L. miR-132 improves the cognitive function of rats with Alzheimer’s disease by inhibiting the MAPK1 signal pathway. Exp. Ther. Med. 2020, 20, 1–1. [CrossRef]
- Hansen, K.F.; Karelina, K.; Sakamoto, K.; Wayman, G.A.; Impey, S.; Obrietan, K. miRNA-132: a dynamic regulator of cognitive capacity. Brain Structure and Function. 2013;218(3):817-31. [CrossRef]
- Wang RY, Phang RZ, Hsu PH, Wang WH, Huang HT, Liu IY. In vivo knockdown of hippocampal miR-132 expression impairs memory acquisition of trace fear conditioning. Hippocampus. 2013;23(7):625-33.
- Gong, X.; Huang, M.; Chen, L. Mechanism of miR-132-3p Promoting Neuroinflammation and Dopaminergic Neurodegeneration in Parkinson’s Disease. eneuro 2021, 9, 1–17. [CrossRef]
- Clovis, Y.M.; Enard, W.; Marinaro, F.; Huttner, W.B.; De Pietri Tonelli, D. Convergent repression of Foxp2 3′UTR by miR-9 and miR-132 in embryonic mouse neocortex: implications for radial migration of neurons. Development 2012, 139, 3332–3342. [CrossRef]
- Gascon, E.; Lynch, K.; Ruan, H.; Almeida, S.; Verheyden, J.M.; Seeley, W.W.; Dickson, D.W.; Petrucelli, L.; Sun, D.; Jiao, J.; et al. Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nat. Med. 2014, 20, 1444–1451. [CrossRef]
- Pan, S.; Feng, W.; Li, Y.; Huang, J.; Chen, S.; Cui, Y.; Tian, B.; Tan, S.; Wang, Z.; Yao, S.; et al. The microRNA-195 - BDNF pathway and cognitive deficits in schizophrenia patients with minimal antipsychotic medication exposure. Transl. Psychiatry 2021, 11, 1–8. [CrossRef]
- Kondratyev, A.; Gale, K. Intracerebral injection of caspase-3 inhibitor prevents neuronal apoptosis after kainic acid-evoked status epilepticus. Mol. Brain Res. 2000, 75, 216–224. [CrossRef]
- Caggiu, E.; Paulus, K.; Mameli, G.; Arru, G.; Sechi, G.P.; Sechi, L.A. Differential expression of miRNA 155 and miRNA 146a in Parkinson’s disease patients. eNeurologicalSci 2018, 13, 1–4. [CrossRef]
- Chen, L.; Dong, R.; Lu, Y.; Zhou, Y.; Li, K.; Zhang, Z.; Peng, M. MicroRNA-146a protects against cognitive decline induced by surgical trauma by suppressing hippocampal neuroinflammation in mice. Brain, Behav. Immun. 2019, 78, 188–201. [CrossRef]
- Zhan-Qiang H, Hai-Hua Q, Chi Z, Miao W, Cui Z, Zi-Yin L, et al. miR-146a aggravates cognitive impairment and Alzheimer disease-like pathology by triggering oxidative stress through MAPK signaling. Neurologia (Engl Ed). 2021. [CrossRef]
- Liu, D.; Zhao, D.; Zhao, Y.; Wang, Y.; Zhao, Y.; Wen, C. Inhibition of microRNA-155 Alleviates Cognitive Impairment in Alzheimer’s Disease and Involvement of Neuroinflammation. Curr. Alzheimer Res. 2019, 16, 473–482. [CrossRef]
- Zago E, Dal Molin A, Dimitri G, Xumerle L, Pirazzini C, Bacalini MG, et al. Early downregulation of hsa-miR-144-3p in serum from drug-naïve Parkinson’s disease patients. Scientific reports. 2022;12:1330. [CrossRef]
- Li K, Zhang J, Ji C, Wang L. MiR-144-3p and Its Target Gene β-Amyloid Precursor Protein Regulate 1-Methyl-4-Phenyl-1,2-3,6-Tetrahydropyridine-Induced Mitochondrial Dysfunction. Mol Cells. 2016;39(7):543-9. [CrossRef]
- Murphy CP, Li X, Maurer V, Oberhauser M, Gstir R, Wearick-Silva LE, et al. MicroRNA-Mediated Rescue of Fear Extinction Memory by miR-144-3p in Extinction-Impaired Mice. Biological Psychiatry. 2017;81(12):979-89. [CrossRef]
- da Silva FC, Iop RdR, Vietta GG, Kair DA, Gutierres Filho PJB, de Alvarenga JGS, et al. microRNAs involved in Parkinson’s disease: A systematic review. Mol Med Rep. 2016;14(5):4015-22. [CrossRef]
- Zamanian, M.Y.; Ivraghi, M.S.; Gupta, R.; Prasad, K.D.V.; Alsaab, H.O.; Hussien, B.M.; Ahmed, H.; Ramadan, M.F.; Golmohammadi, M.; Nikbakht, N.; et al. miR-221 and Parkinson’s disease: A biomarker with therapeutic potential. Eur. J. Neurosci. 2023, 59, 283–297. [CrossRef]
- Ma, W.; Li, Y.; Wang, C.; Xu, F.; Wang, M.; Liu, Y. Serum miR-221 serves as a biomarker for Parkinson’s disease. Cell Biochem. Funct. 2016, 34, 511–515. [CrossRef]
- Li, L.; Xu, J.; Wu, M.; Hu, J.M. Protective role of microRNA-221 in Parkinson’s disease. Bratisl. Med J. 2018, 119, 22–27. [CrossRef]
- Ishihara, Y.; Yamada, H.; Munetuna, E.; Hagiwara, C.; Fujii, R.; Yamazaki, M.; Hatta, T.; Iwahara, A.; Ohashi, K.; Ishikawa, H.; et al. 486Serum levels of miRNA-221 and -222 may be predictive biomarkers for cognitive decline. International Journal of Epidemiology. 2021;50. [CrossRef]
- Gullett, J.M.; Chen, Z.; O’shea, A.; Akbar, M.; Bian, J.; Rani, A.; Porges, E.C.; Foster, T.C.; Woods, A.J.; Modave, F.; et al. MicroRNA predicts cognitive performance in healthy older adults. Neurobiol. Aging 2020, 95, 186–194. [CrossRef]
- Ba R-Q, Liu J, Fan X-J, Jin G-L, Huang B-G, Liu M-W, et al. Effects of miR-199a on autophagy by targeting glycogen synthase kinase 3β to activate PTEN/AKT/mTOR signaling in an MPP+ in vitro model of Parkinson’s disease. Neurological Research. 2020;42(4):308-18. [CrossRef]
- Nakashima, H.; Tsujimura, K.; Irie, K.; Imamura, T.; Trujillo, C.A.; Ishizu, M.; Uesaka, M.; Pan, M.; Noguchi, H.; Okada, K.; et al. MeCP2 controls neural stem cell fate specification through miR-199a-mediated inhibition of BMP-Smad signaling. Cell Rep. 2021, 35, 109124. [CrossRef]
- Wang RY, Phang RZ, Hsu PH, Wang WH, Huang HT, Liu IY. In vivo knockdown of hippocampal miR-132 expression impairs memory acquisition of trace fear conditioning. Hippocampus. 2013;23(7):625-33. [CrossRef]
- Harvey J, Smith AR, Weymouth LS, Smith RG, Castanho I, Hubbard L, et al. Epigenetic insights into neuropsychiatric and cognitive symptoms in Parkinson’s disease: A DNA co-methylation network analysis. Res Sq. 2023. [CrossRef]
- Chuang, Y.-H.; Lu, A.T.; Paul, K.C.; Folle, A.D.; Bronstein, J.M.; Bordelon, Y.; Horvath, S.; Ritz, B. Longitudinal Epigenome-Wide Methylation Study of Cognitive Decline and Motor Progression in Parkinson’s Disease. J. Park. Dis. 2019, 9, 389–400. [CrossRef]
- Outeiro, T.F.; Kontopoulos, E.; Altmann, S.M.; Kufareva, I.; Strathearn, K.E.; Amore, A.M.; Volk, C.B.; Maxwell, M.M.; Rochet, J.-C.; McLean, P.J.; et al. Sirtuin 2 Inhibitors Rescue α-Synuclein-Mediated Toxicity in Models of Parkinson’s Disease. Science 2007, 317, 516–519. [CrossRef]
- Rane, P.; Shields, J.; Heffernan, M.; Guo, Y.; Akbarian, S.; King, J.A. The histone deacetylase inhibitor, sodium butyrate, alleviates cognitive deficits in pre-motor stage PD. Neuropharmacology 2012, 62, 2409–2412. [CrossRef]
- Song, C.; Anantharam, V.; Sun, F.; Kanthasamy, A.G. Environmental Neurotoxic Pesticide Increases Histone Acetylation to Promote Apoptosis in Dopaminergic Neuronal Cells: Relevance to Epigenetic Mechanisms of Neurodegeneration. Mol. Pharmacol. 2010, 77, 621–632. [CrossRef]
| Study | Protein | Findings | Reference |
|---|---|---|---|
| Caillierez et al. 2013 | α-Syn | AT8 immunoreactivity spans from the CA1 to the cortex in rats receiving LV-hTau46WT injections, while, in rats injected with LV-hTau46P301L, it is restricted to the hippocampal formation. | [53] |
| Clavaguera et al. 2013 | Tau | Self-spreading of Tau inclusions in a manner like prions, not influenced by other pathological mechanisms | [54] |
| Holmes et al. 2013 | Tau and α-Syn | Tau fibrils enter cells via macropinocytosis; HSPGs act as receptors for binding and uptake of Tau, inhibition of HSPG blocks aggregate propagations, HSPGs mediate α-Syn uptake | [55] |
| Levarska et al. 2013 | Tau | Different spreading capacities of disease-modified Tau strains as Spreading only of the SHR72 4R Tau variant | [56] |
| Masuda-Suzukake et al. 2013 | α-Syn | Intracerebral injection of insoluble α-Syn induces aggregation of endogenous mouse α-Syn through a prion-like spreading. | [57] |
| Oliveras-Salvá et al. 2013 | α-Syn | Following viral vector-mediated α-Syn overexpression, dose-dependent dopaminergic neuron loss in SNpc 8 months post-injection occurred, also motor issues, and protein aggregates in remaining surviving neurons. | [58] |
| Pooler et al. 2013 | Tau | AMPA-stimulated Tau release is dependent on pre-synaptic vesicle secretion instead of exosome extrusion | [59] |
| Sacino et al. 2013 | α-Syn | Injection of amyloidogenic and non-amyloidogenic human α-Syn induced limited α-Syn inclusions. Delayed Strong induction of neuroinflammation with or without α-Syn inclusions | [60] |
| Ulusoy et al. 2013 | α-Syn | Propagation from medulla oblongata to rostral brain regions | [61] |
| Watts et al. 2013 | α-Syn | Lethality upon transmission to animals resembles how kuru, CJD, and similar diseases spread in nonhuman primates | [62] |
| Wu et al. 2013 | Tau | Transferring through the cell via anterograde and retrograde endocytosis of Small Misfolded Tau species | [63] |
| Ahmed et al. 2014 | Tau | Spreading relied on synaptic connectivity | [64] |
| Clavaguera et al. 2014 | Tau | Seeded Tau aggregates can spread to the CNS, peripherally | [65] |
| Dujardin et al. 2014 | Tau | Indication of trans-synaptic protein transfer due to Wild-type human Tau protein could transferred via the axons from ventral hippocampus neurons to connected secondary neurons e.g., olfactory and limbic systems | [66] |
| Holmes et al. 2014 | Tau | The seeding activity of mutant mice rises with age, interaction with synuclein | [67] |
| Holmqvist et al. 2014 | α-Syn | Spreading of α-Syn from intestine to the brain via the vagal nerve | [68] |
| Recasens and Dehay 2014 | α-Syn | Regardless of whether it was injected into the striatum or the SNpc. LB-induced degeneration manifested earlier and more extensively in the striatal dopaminergic axon terminals rather than the cell bodies in the SNpc, | [69] |
| Reyes et al. 2014 | α-Syn | Reuptake of α-Syn monomers, oligomers, and fibrils by oligodendrocytes, internalization of α-Syn by in vivo Oligodendrocyte, Transfer of α-Syn from host brain to grafted oligodendroglial cells | [70] |
| Rotermund et al. 2014 | α-Syn | Nutritional factors can have a significant impact on α-Synucleinopathy. Possibility of diet-induced obesity as a risk factor for α-Synucleinopathy | [71] |
| Sacino et al. 2014 | α-Syn | Fibrillar α-Syn intramuscular injection led to CNS inclusion pathology | [72] |
| Sacino et al. 2014 | α-Syn | Spreading of inclusion pathology only in M83 mice after 4 months | [73] |
| Sanders et al. 2014 | Tau | Spreading to the ipsilateral and contralateral entorhinal cortex, retrosplenial cortex, and contralateral hippocampus from the left hippocampus after 5 weeks | [74] |
| Asai et al. 2015 | Tau | Microglia’s significant role in Tau protein spreading and neurotoxicity | [75] |
| Bernis et al. 2015 | α-Syn | Spreading of α-Syn after intrastriatal injection to the contralateral side | [76] |
| Bourdenx et al. 2015 | α-Syn | Spreading into the striatal area and throughout the whole mesencephalon in rats 16 weeks after surgery. Absence of phosphorylation levels of α-Syn and neurodegeneration in rats and marmosets | [77] |
| Daher et al. 2015 | α-Syn | Neurodegeneration in the contralateral side | [78] |
| Abounit et al. 2016 | Tau | Tau fibrils capability to enter cells and induce TNT (tunneling nano tubules) | [79] |
| d’Abramo et al. 2016 | Tau | Tau detection in serum | [80] |
| Domert et al. 2016 | α-Syn | Cell-to-cell transfer in of α-Syn between neuron-like cell model |
[81] |
| Helwig et al. 2016 | α-Syn | Spreading from the medulla oblongata to rostral brain regions | [82] |
| Koprich et al. 2016 | α-Syn | Accumulation of A53T shows an age-related progressive rise, higher levels, and a broader extent of degeneration with A53T | [83] |
| Aulić et al. 2017 | α-Syn | Spreading and the entrance of α-Syn amyloid in N2a cells is facilitated by cells expressing PrPc | [84] |
| Cavaliere et al. 2017 | α-Syn | Quantification of α-Syn uptake in neurons and astrocytes | [85] |
| DeVos et al. 2017 | Tau | Reduction and prevention of Tau seeding capability by human Tau | [86] |
| Loria et al. 2017 | α-Syn | Transfer of α-Syn between primary cortical astrocytes and neurons | [87] |
| Ngolab et al. 2017 | α-Syn | Intracellular aggregation of α-Syn is correlated with internalization of exosomes via endocytosis | [88] |
| Shimozawa et al. 2017 | α-Syn | Retrograde spreading of abnormal α-Syn and neurotoxicity following intracerebral injection of synthetic α-Syn fibrils, Clearance of α-Syn inclusions by microglial cells | [89] |
| Ulusoy et al. 2017 | α-Syn | The Vagus nerve acts as a conduit for α-Syn to reach from the brain to peripheral tissues e.g., stomach | [90] |
| Wang et al. 2017 | Tau | Exomes from CSF have the capability of stimulating Tau aggregation within cultured cells | [91] |
| Polanco et al. 2018 | Tau | When interconnected axons extend in proximity, they exchange exomes | [92] |
| Rusconi et al. 2018 | α-Syn | Sustained overexpression of α-Syn is crucial for continuous propagation. Increased spreading of α-Syn through the brain following increased level α-Syn in the medulla oblongata | [93] |
| Vitale et al. 2018 | Tau | Diffusion to distant areas | [94] |
| Smolek et al. 2019 | Tau | Tau pathology induction following bilateral hippocampus injection | [95] |
| Ferrer et al. 2020 | Tau | Potential role of oligodendropathy and neuronopathy in Tauopathies progression. 3RTau and 4RTau production and deposition and activation specific Tau kinases following injection of human Tau inoculations leads to modification of Tau metabolism | [96] |
| Ferrer et al. 2019 | Tau | Role of oligodendrocytes in seeding and spreading of Tauopathies in white matter | [97] |
| Masuda-Suzukake et al. 2020 | Tau | Tau spreading after dextran sulphate-induced Tau assemblies, propagation of Tau-assemblies does not depend on Tau to be either mutated or overexpressed. | [98] |
| Mezias et al. 2020 | α-Syn | After injection of protein fibrils, the α-Syn spreading follows neural networks | [99] |
| Veys et al. 2020 | Tau, α-Syn | α-Syn PFFs similar to the Tau K18 PFFs fail to enter the retina after IVT injection | [100] |
| Courte et al. 2020 | α-Syn | The ability of exogenous α-Syn seeding is dependent on the level of α-Syn expression within the regional neurons | [101] |
| Jimenez-Ferrer et al. 2021 | α-Syn | The significant role of Mhc2a as the key regulator of MHCII expression for regulation of seeding, spreading, and toxicity of α-Syn in vivo | [102] |
| Thomsen et al. 2021) | α-Syn | Striatal injection of α-Syn fibrils causes gradual impairment of synaptic function before cell death, detectable by PET scan, α-Syn striatal injection creates progressive α-Syn pathology as found in human PD | [103] |
| Dutta et al. 2021 | α-Syn | Decrease in α-Syn spreading by Intranasal administration of wtTIDM peptide, NEMO-binding domain (wtNBD) peptide, or genetic deletion of TLR2 | [104] |
| Bassil et al. 2021 | Tau, α-Syn | Tau spreading is reduced by endogenous α-Syn in mice while α-Syn seeding and spreading are not affected by endogenous Tau | [105] |
| Matsuo et al. 2021 | α -Syn | Deletion of Fabp3 results in the prevention of exogenous α-Syn spreading into SNpc | [106] |
| Pan et al. 2022 | α-Syn | Striatal injection of Tau-modified α-Syn fibrils results in more severe α-Syn pathology and motor and cognitive symptoms comparing pure α-Syn injection | [107] |
| Dutta et al. 2022 | α-Syn | Treadmill exercise reduces α-Syn spreading | [108] |
| Garcia et al. 2022 | α-Syn | Microgliosis occurs as a part of response independent from α-Syn inclusion formation, neurodegeneration could occur even if α-Syn inclusion is not present. | [109] |
| Vasili et al. 2022 | α-Syn | Seeding and accumulation of pS129A-Syn is induced by adding α-Syn PFFs, endogenous α-Syn aggregates are dependent on α-Syn levels | [110] |
| Sun et al. 2022 | α-Syn | Pathological mechanisms induced by the spreading of misfolded α-Syn throughout the nigrostriatal pathway vary based on the age of the dopamine network, resulting in a striatal dopamine release decline specifically observed in adult mice. | [111] |
| Rahayel et al. 2022 | α-Syn | Differentially targeted seeding of pathological α-Syn led to distinct spreading patterns observed over 24 months indicating most brain regions were susceptible to this pathology | [112] |
| Lloyd et al. 2022 | α-Syn | The phenotypic and pathological progression of the disease.is significantly influenced by initial inoculation | [113] |
| Jain et al. 2023 | Tau | Chronic administration of activatingTREM2 antibody enhances the activation of microglia around plaques. This amplification correlates with an increase in peri-plaque NP-Tau pathology and neuritic dystrophy, while the presence of Aβ plaques remains unaffected. | [114] |
| Mate De Gerando et al. 2023 | Tau | Soluble HMW Tau has similar seeding potential comparing fibrillar sarkosyl-insoluble Tau but could be more bioactive in terms of spreading across neural systems | [115] |
| miRNA | miR Expression |
Studied Population | Cognition Impaired | Studied Species/Specimen | References |
|---|---|---|---|---|---|
| miR-7 | ↑ | PD patients/ MPTP mice model/ MPP+-SH-SY5Y cell model |
Acute ischemic stroke | Human/blood | [180]/ [182] |
| ↑ | MPTP mouse model | Neuroprotection | Human/brain | [181]/ [183] | |
| miR-124-3p | ↓ | MPP+-SH-SY5Y cell model | [184] | ||
| ↑ | Ease cognitive function, inflammation and apoptosis in PND |
Rat/ brain | [185] | ||
| miR-124 | ↓ | LPS-BV-2 cell models MPTP mice model, MPP+-SH-SY5Y cell model |
AD/suppress memory and learning; Suppress CREB and GluA2 |
Mouse /brain /hepatopancreas, muscle, heart, ovotestis, and central nervous system | [186]/ [189,190,191,192,193] |
| ↓ | prefrontal cortex of the left cerebral hemisphere | Impaired LTP and learning and memory, spatial learning, and social interactions |
Mouse/brain | [187]/ [194] | |
| ↑ | amygdala | FTD | Mouse/ | [188]/ [195] | |
| miR-195 | ↓ | LPS-BV-2 cell model | Impaired spatial memory | Rat/brain | [196]/ [197] |
| ↓ | AD | Rat/brain | [198,199,200] | ||
| miR-190 | ↓ | LPS-BV-2 cell model, MPTP mice model | Upreg.ameliorates POCD | Mice/brain | [201] /[202] |
| ↑ | Neuroprotective | Rat | [203] | ||
| miR-146a | ↑ | human glial cell lines | AD | human, mouse/archived tissue, or total RNA extract sources | [204]/ [205] |
| ↑ | AD | human/CSF & ECF | [206] | ||
| ↓ | AD | human/plasma and CSF | [175] | ||
| miR-155 | ↑ | microglia and astrocytes cultured from DJ-1-knockout mouse brain | neuroprotective effect | mouse/brain | [207]/ [208] |
| miR-133b | ↓ | aphakia mice 6OHDA-treated mice |
[209] | ||
| ↑ | protects against isoflurane-induced learning and memory impairment | rat/brain | [210] | ||
| ↑ | neuroprotective in AD | AD vs. HC /serum | [211] | ||
| ↓ | midbrain | [209,212,213] | |||
| miR-34b | ↓ | putamen / FC / amygdala / SN / cerebellum | [214] | ||
| miR-126 | ↑ | DA neurons / amygdala | improved hemorrhagic lesion and the number of apoptotic cortical neurons | rat/brain | [188,215,216]/ [217] |
| ↓ | induces cognitive impairment and neuroinflammation | mice/serum | [218] | ||
| miR-204 | ↑ | putamen | [7] | ||
| ↓ | amygdala | [188] | |||
| miR-144 | ↑ | the prefrontal cortex anterior cingulate gyrus |
promoted cognitive impairments induced by β-amyloid accumulation post-TBI via suppres- sing ADAM10 expression (spatial learning and memory). | human, rat/brain | [219,220] / [221] |
| ↓ | the prefrontal cortex of the left cerebral hemisphere | [187] | |||
| miR-148b | ↓ | the prefrontal cortex/amygdala | [188,219] | ||
| ↑ | attenuated neuroprotection by inhibiting Wnt/ β-catenin signaling | rat | [222] | ||
| miR-184 | ↑ | DA neurons and amygdala | increased viability and reduced apoptosis | human, rat/brain | [188,216] / [223] |
| ↓ | worse learning and memory capacity | MDD vs. HC/blood | [224] | ||
| ↓ | Neuroprotective | mice/brain | [225] | ||
| miR-221 | ↑ | putamen / anterior cingulate gyrus, and amygdala | [7,188,220] | ||
| ↓ | neuroprotection | human/plasma | [226] | ||
| miR-199a | ↑ | amygdala | spatial memory ability began to decrease at 4 months and was significantly decreased at 7 months | mice/brain | [188]/ [227] |
| ↓ | iPSC-derived DNs from PD patients | [228] | |||
| miR-132 | ↓ | prefrontal cortex, and in a meta-analysis of different PD brain specimens | [212,219] | ||
| ↑ | MS | human/ | [229] | ||
| regulates the process of cognitive impairment after stress | rat/ | [230] | |||
| reduces the cognition-damaging effect of sevoflurane | rat/brain | [231,232] | |||
| spatial memory mission | /brain | [233] | |||
| significantly damage the cognition of spatial memory | mice/brain | [234] | |||
| midbrain | axon growth, neural migration, and plasticity | mice/dorsal root ganglion | [235]/ [236] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





