Submitted:
13 June 2024
Posted:
14 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
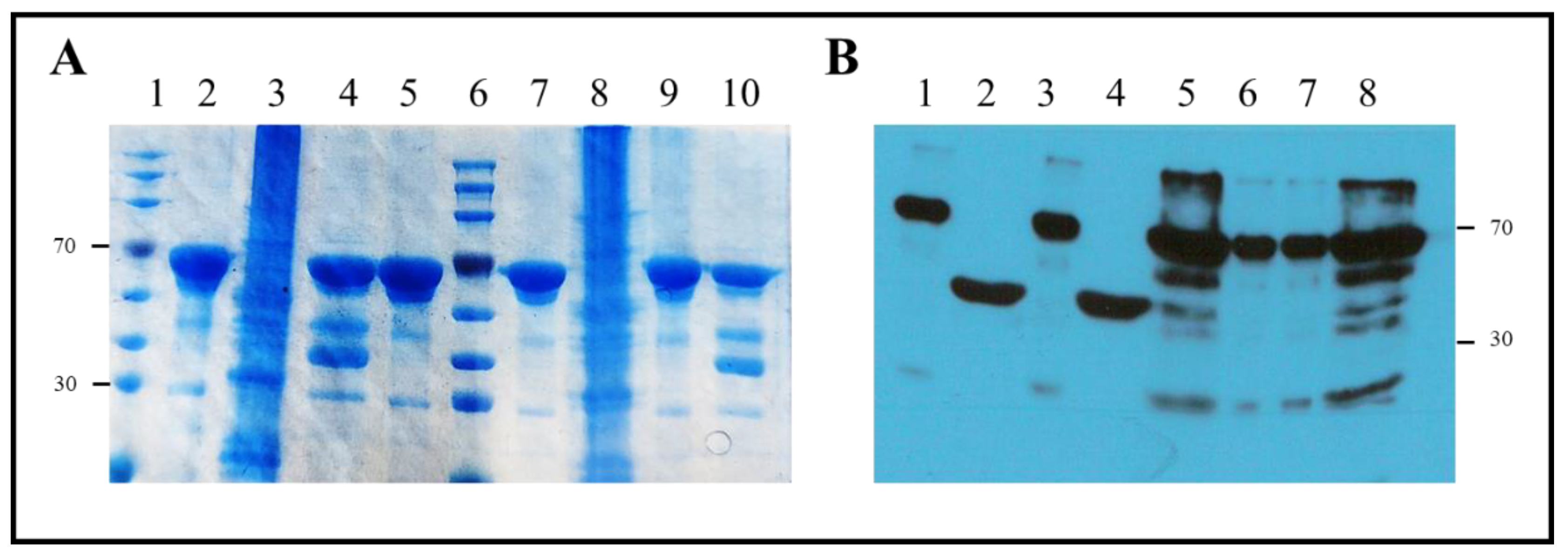
2.1. Proteins and Plasmids
2.2. Protein Expression and Purification
2.3. Protein Modeling
2.4. Web-Based Analysis Tools
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
2.6. Mass Spectrometry Assay and Analysis
2.7. Size Exclusion Chromatography
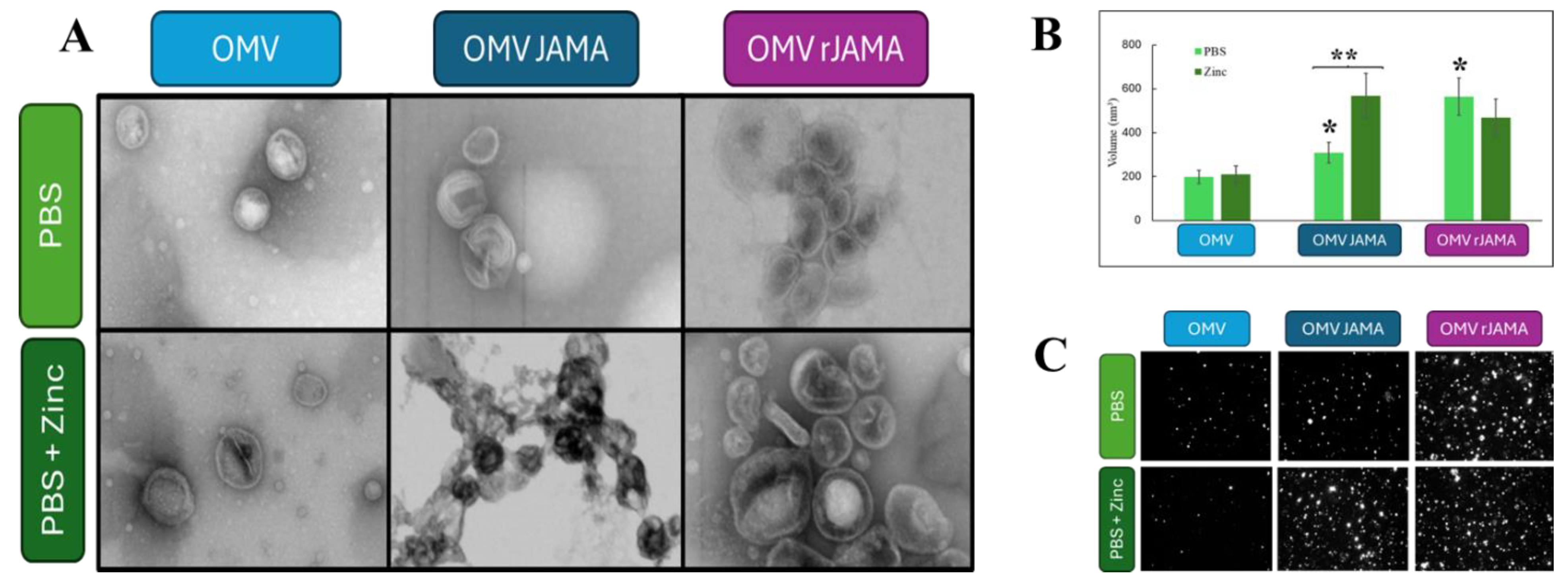
2.8. Outer Membrane Vesicle Production (OMV)
2.9. Cell Aggregation Assay Using FlowCytometry (FC)
2.10. Bacterial Outer Membrane Vesicles (OMVs) Analysis
2.11. Statistical Analysis
2.12. Negative Staining Electron Microscopy of a Preparation of Shuffle T7 Express E. coli OMVs
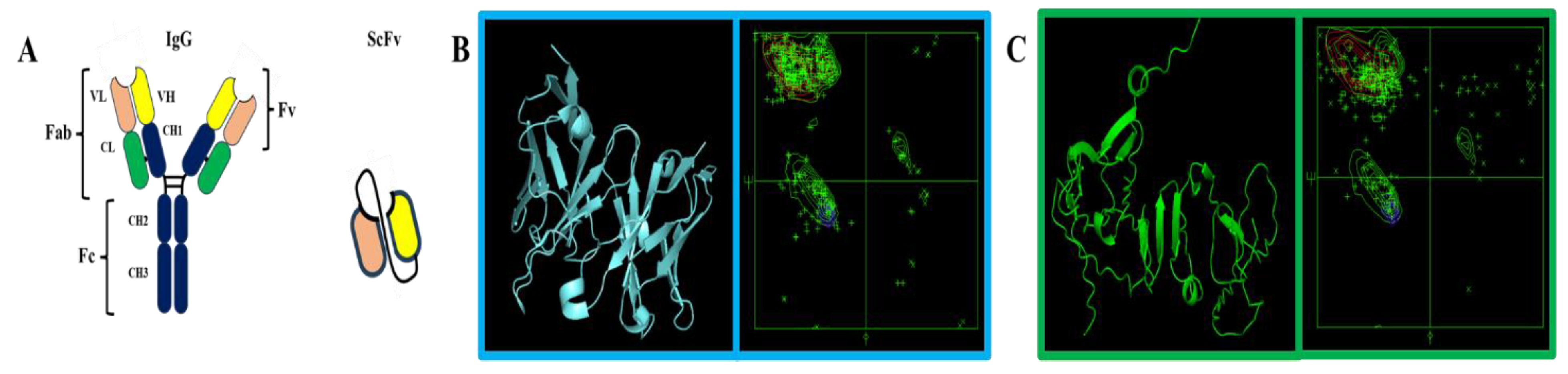
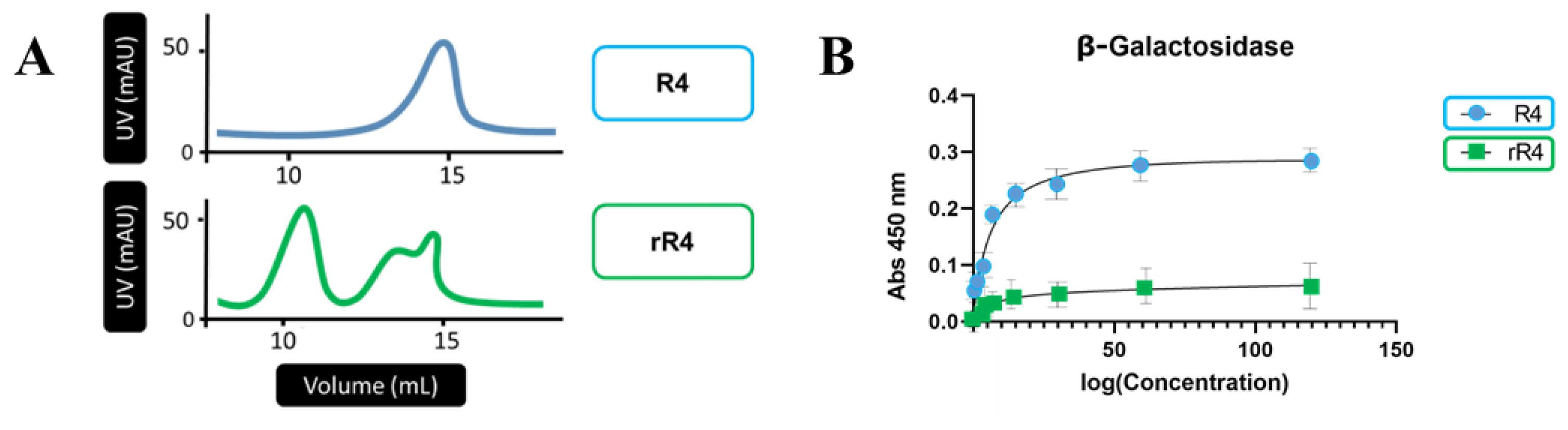
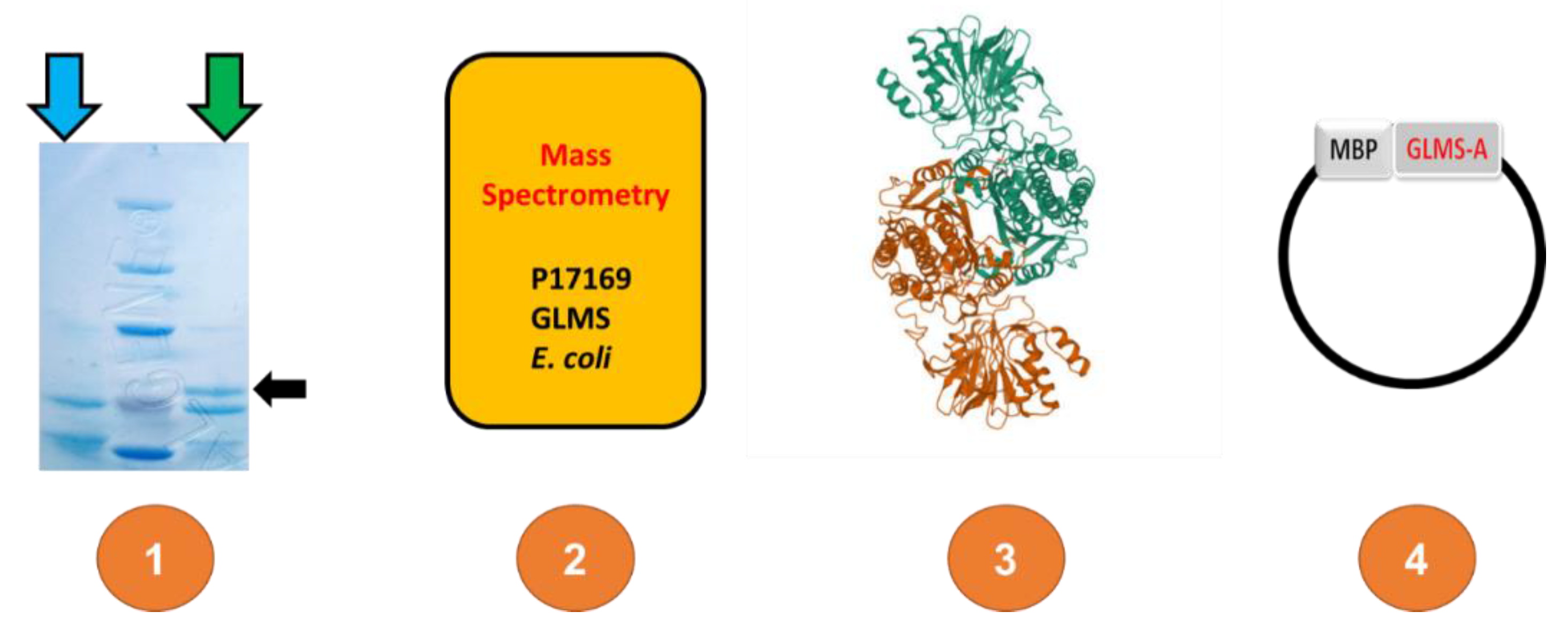
3. Results
3.1. Protein Sequences and Plasmids
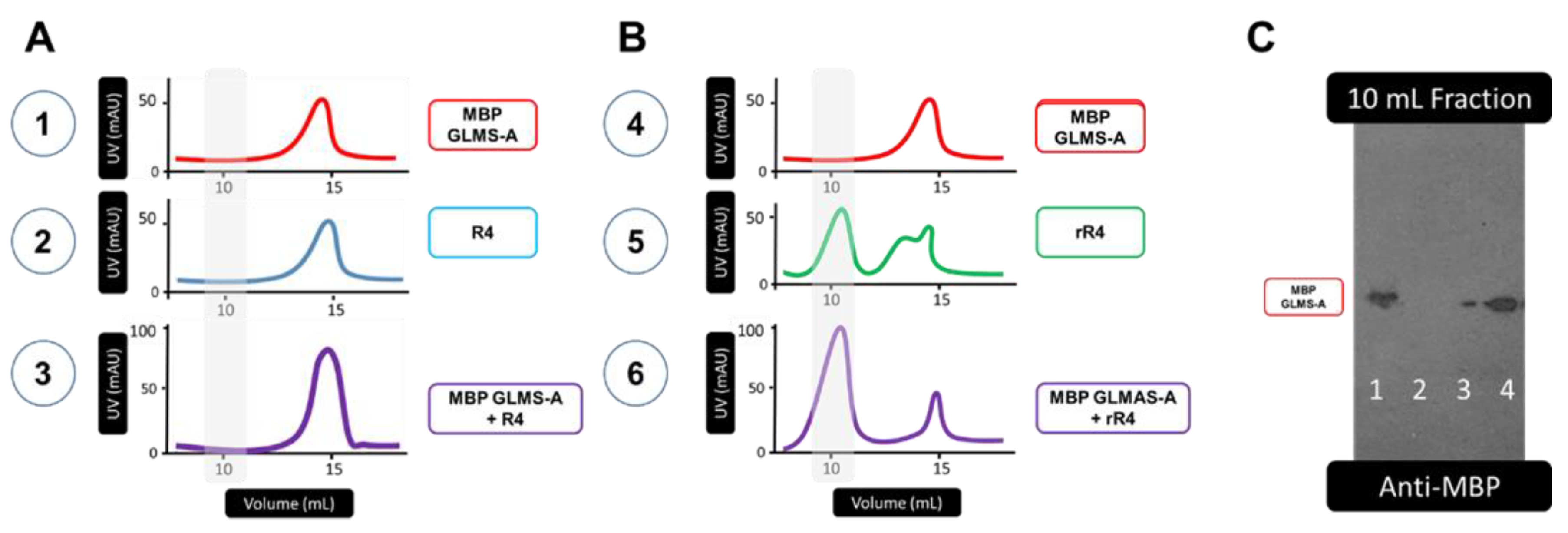
4. Discussion
5. Conclusions
Acknowledgments
References
- Adhav, V.A.; Saikrishnan, K. The Realm of Unconventional Noncovalent Interactions in Proteins: Their Significance in Structure and Function. ACS Omega 2023, 8, 22268–22284. [Google Scholar] [CrossRef]
- Anfinsen, C.B. Principles that govern the folding of protein chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef]
- Iorgulescu, J.B. Reporting of Immunotherapy and Biologic Therapy in the National Cancer Database. JAMA Oncol. 2023, 9, 1003. [Google Scholar] [CrossRef]
- Saeed, A.F.; et al. Antibody Engineering for Pursuing a Healthier Future. Front. Microbiol. 2017, 8, 495. [Google Scholar] [CrossRef]
- Aricescu, A.R.; Jones, E.Y. Immunoglobulin superfamily cell adhesion molecules: Zippers and signals. Curr. Opin. Cell Biol. 2007, 19, 543–550. [Google Scholar] [CrossRef]
- Berezin, V.A.; Walmod, P.S. Cell adhesion molecules: Implications in neurological diseases. Advances in neurobiology; Springer: New York, NY, USA, 2014; 410 pages. [Google Scholar]
- Alejandra, W.P.; et al. Production of monoclonal antibodies for therapeutic purposes: A review. Int. Immunopharmacol. 2023, 120, 110376. [Google Scholar] [CrossRef]
- Elgundi, Z.; et al. The state-of-play and future of antibody therapeutics. Adv. Drug Deliv. Rev. 2017, 122, 2–19. [Google Scholar] [CrossRef]
- Janeway, C. Immunobiology: The Immune System in Health and Disease. 2001: Garland Pub.
- Williams, A.F. and A. N. Barclay, The immunoglobulin superfamily--domains for cell surface recognition. Annu Rev Immunol 1988, 6, 381–405. [Google Scholar]
- Bemani, P., M. Mohammadi, and A. Hakakian, ScFv Improvement Approaches. Protein Pept Lett 2018, 25, 222–229. [Google Scholar] [CrossRef]
- Silva, T.A.; et al. Potential of an anti-bevacizumab idiotype scFv DNA-based immunization to elicit VEGF-binding antibody response. Gene Ther 2023, 30, 598–602. [Google Scholar] [CrossRef]
- Dreier, B. and A. Pluckthun, Rapid Selection of High-Affinity Antibody scFv Fragments Using Ribosome Display. Methods Mol Biol 2018, 1827, 235–268. [Google Scholar]
- Zhang, S., L. Wu, and M. Dang, Antibody mimetics: The next generation antibody engineering, a retrospective and prospective analysis. Biotechnol J 2024, 19, e2300532. [Google Scholar]
- Keri, D.; et al. Next generation of multispecific antibody engineering. Antib Ther 2024, 7, 37–52. [Google Scholar] [CrossRef]
- Qerqez, A.N., R. P. Silva, and J.A. Maynard, Outsmarting Pathogens with Antibody Engineering. Annu Rev Chem Biomol Eng 2023, 14, 217–241. [Google Scholar]
- Tsuchikama, K.; et al. Exploring the next generation of antibody-drug conjugates. Nat Rev Clin Oncol 2024, 21, 203–223. [Google Scholar] [CrossRef]
- Guichard, G.; et al. Antigenic mimicry of natural L-peptides with retro-inverso-peptidomimetics. Proc Natl Acad Sci U S A 1994, 91, 9765–9769. [Google Scholar] [CrossRef] [PubMed]
- Chorev, M. and M. Goodman, Recent developments in retro peptides and proteins--an ongoing topochemical exploration. Trends Biotechnol 1995, 13, 438–445. [Google Scholar]
- Lacroix, E., A. R. Viguera, and L. Serrano, Reading protein sequences backwards. Fold Des 1998, 3, 79–85. [Google Scholar]
- Olszewski, K.A., A. Kolinski, and J. Skolnick, Does a backwardly read protein sequence have a unique native state? Protein Eng 1996, 9, 5–14. [Google Scholar]
- Mittl, P.R.; et al. The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure. Proc Natl Acad Sci U S A 2000, 97, 2562–2566. [Google Scholar] [CrossRef]
- Jumper, J.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Martineau, P., P. Jones, and G. Winter, Expression of an antibody fragment at high levels in the bacterial cytoplasm. J Mol Biol 1998, 280, 117–127. [Google Scholar]
- Karlsson, A.J.; et al. Engineering antibody fitness and function using membrane-anchored display of correctly folded proteins. J Mol Biol 2012, 416, 94–107. [Google Scholar] [CrossRef]
- Pettersen, E.F.; et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Goldman, A.R.; et al. Proteome Analysis Using Gel-LC-MS/MS. Curr Protoc Protein Sci 2019, 96, e93. [Google Scholar] [CrossRef]
- Prachayasittikul, V.; et al. EDTA-induced membrane fluidization and destabilization: Biophysical studies on artificial lipid membranes. Acta Biochim Biophys Sin (Shanghai) 2007, 39, 901–913. [Google Scholar] [CrossRef]
- Rollins, J.; et al. Expression of Cell-Adhesion Molecules in E. coli: A High Throughput Screening to Identify Paracellular Modulators. Int J Mol Sci 2023, 24. [Google Scholar]
- Worn, A. and A. Pluckthun, An intrinsically stable antibody scFv fragment can tolerate the loss of both disulfide bonds and fold correctly. FEBS Lett 1998, 427, 357–361. [Google Scholar] [PubMed]
- Tassew, N.G.; et al. Sustained in vivo inhibition of protein domains using single-chain Fv recombinant antibodies and its application to dissect RGMa activity on axonal outgrowth. J Neurosci 2009, 29, 1126–1131. [Google Scholar] [CrossRef]
- Boldicke, T. , Single domain antibodies for the knockdown of cytosolic and nuclear proteins. Protein Sci 2017, 26, 925–945. [Google Scholar] [CrossRef]
- de Aguiar, R.B.; et al. Generation and functional characterization of a single-chain variable fragment (scFv) of the anti-FGF2 3F12E7 monoclonal antibody. Sci Rep 2021, 11, 1432. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, Z.A.; et al. scFv antibody: Principles and clinical application. Clin Dev Immunol 2012, 2012, 980250. [Google Scholar] [CrossRef] [PubMed]
- Steinbacher, T., D. Kummer, and K. Ebnet, Junctional adhesion molecule-A: Functional diversity through molecular promiscuity. Cell Mol Life Sci 2018, 75, 1393–1409. [Google Scholar] [PubMed]
- Mendoza, C.; et al. Calcium regulates the interplay between the tight junction and epithelial adherens junction at the plasma membrane. FEBS Lett 2022, 596, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Mendoza C., N. S.H., Peterson K., and Mizrachi D., Cations as Molecular Switches for Junctional Adhesion Molecule-A. Biomedical Journal of Scientific & Technical Research, 2022.
- Ebnet, K. Junctional Adhesion Molecules (JAMs): Cell Adhesion Receptors With Pleiotropic Functions in Cell Physiology and Development. Physiol Rev 2017, 97, 1529–1554. [Google Scholar] [CrossRef] [PubMed]
- Rollins, J.; et al. Expression of cell-adhesion molecules in E. coli: A high-throughput method to identify paracellular modulators. 2021, Cold Spring Harbor Laboratory.
- Duong-Ly, K.C.; Gabelli, S.B. Affinity Purification of a Recombinant Protein Expressed as a Fusion with the Maltose-Binding Protein (MBP) Tag. Methods Enzymol. 2015, 559, 17–26. [Google Scholar]
- Bachurski, D.; et al. Extracellular vesicle measurements with nanoparticle tracking analysis - An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J Extracell Vesicles 2019, 8, 1596016. [Google Scholar] [CrossRef] [PubMed]
- Daaboul, G.G.; et al. Digital Detection of Exosomes by Interferometric Imaging. Sci. Rep. 2016, 6, 37246. [Google Scholar] [CrossRef] [PubMed]
- Atzori, A.; et al. Effect of sequence and stereochemistry reversal on p53 peptide mimicry. PLoS ONE 2013, 8, e68723. [Google Scholar] [CrossRef]
- Ambroggio, E.E.; et al. Reversing the peptide sequence impacts on molecular surface behaviour. Colloids Surf B Biointerfaces 2016, 139, 25–32. [Google Scholar] [CrossRef]
- Li, C.; et al. Limitations of peptide retro-inverso isomerization in molecular mimicry. J Biol Chem 2010, 285, 19572–19581. [Google Scholar] [CrossRef] [PubMed]
- Carriero, M.V.; et al. Retro-inverso Urokinase Receptor Antagonists for the Treatment of Metastatic Sarcomas. Sci Rep 2017, 7, 1312. [Google Scholar] [CrossRef] [PubMed]
- Franco, O.L. , Development of antimicrobial peptides as a strategy for infectious disease control. Peptides 2022, 150, 170761. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; et al. Retro-protein XXA is a remarkable solubilizing fusion tag for inclusion bodies. Microb Cell Fact 2022, 21, 51. [Google Scholar] [CrossRef]
- Errante, F.; et al. Retro-Inverso Collagen Modulator Peptide Derived from Serpin A1 with Enhanced Stability and Activity In Vitro. J. Med. Chem. 2024, 67, 5053–5063. [Google Scholar] [CrossRef]
- Sercinoglu, O. and P. Ozbek, Sequence-structure-function relationships in class I MHC: A local frustration perspective. PLoS ONE 2020, 15, e0232849. [Google Scholar] [CrossRef]
- Pfanner, N. , Who chaperones nascent chains in bacteria? Curr. Biol. 1999, 9, R720–R724. [Google Scholar] [CrossRef]
- Lill, R.; et al. The “trigger factor cycle” includes ribosomes, presecretory proteins, and the plasma membrane. Cell 1988, 54, 1013–1018. [Google Scholar] [CrossRef]
- He, W.; et al. Chaperone Spy Protects Outer Membrane Proteins from Folding Stress via Dynamic Complex Formation. mBio 2021, 12, e0213021. [Google Scholar] [CrossRef]
- Castanie-Cornet, M.P., N. Bruel, and P. Genevaux, Chaperone networking facilitates protein targeting to the bacterial cytoplasmic membrane. Biochim. Biophys Acta 2014, 1843, 1442–1456. [Google Scholar] [CrossRef]
- Marchenkov, V.V. and G.V. Semisotnov, GroEL-assisted protein folding: Does it occur within the chaperonin inner cavity? Int. J. Mol. Sci. 2009, 10, 2066–2083. [Google Scholar] [CrossRef] [PubMed]
- Raran-Kurussi, S.; Keefe, K.; Waugh, D.S. Positional effects of fusion partners on the yield and solubility of MBP fusion proteins. Protein. Expr. Purif. 2015, 110, 159–164. [Google Scholar]
- Alvarez, A.B.; et al. Stitching together a nm thick peptide-based semiconductor sheet using UV light. Colloids Surf B Biointerfaces 2021, 203, 111734. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




