Submitted:
13 June 2024
Posted:
14 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
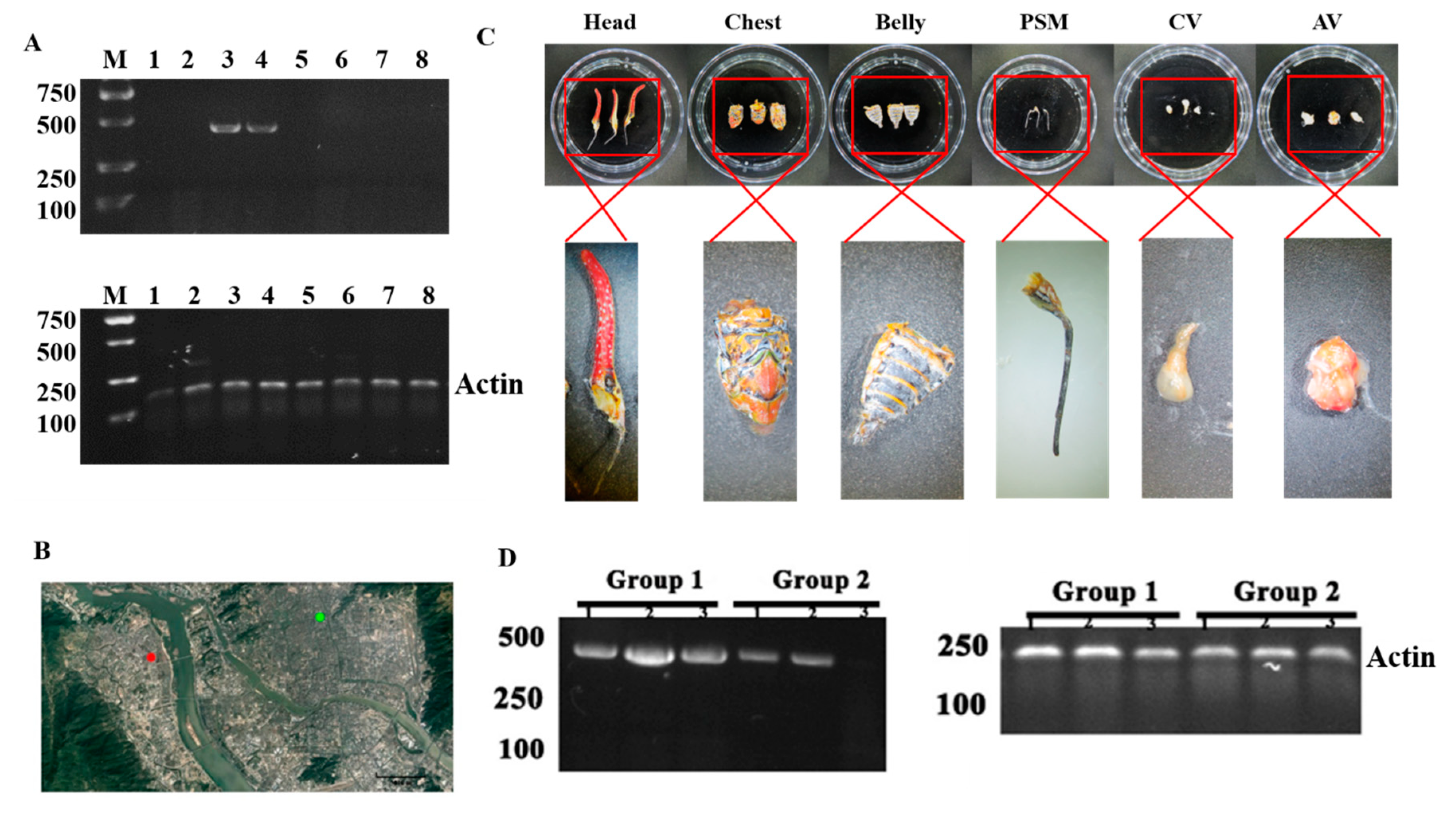
2.1. Insect Sample Collection
2.2. RNA Sequencing (RNA-seq) and Data Assembly
2.3. Rapid Amplification of cDNA Ends
2.4. Bioinformatics Analysis
2.5. RNA Extraction
2.7. DNA Purification
2.8. Virus Verification
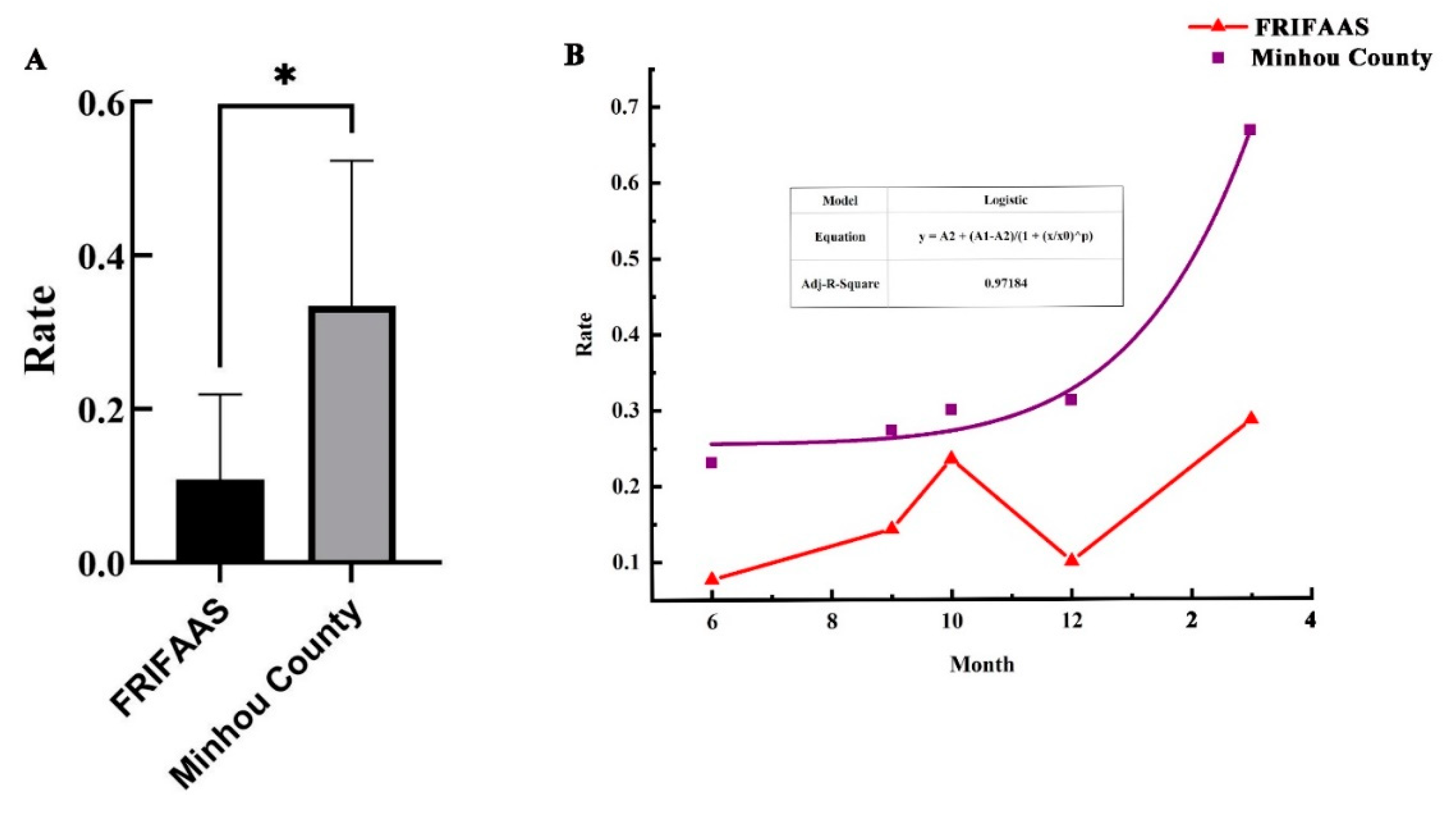
2.9. Statistics Analysis and Model Construction
3. Results
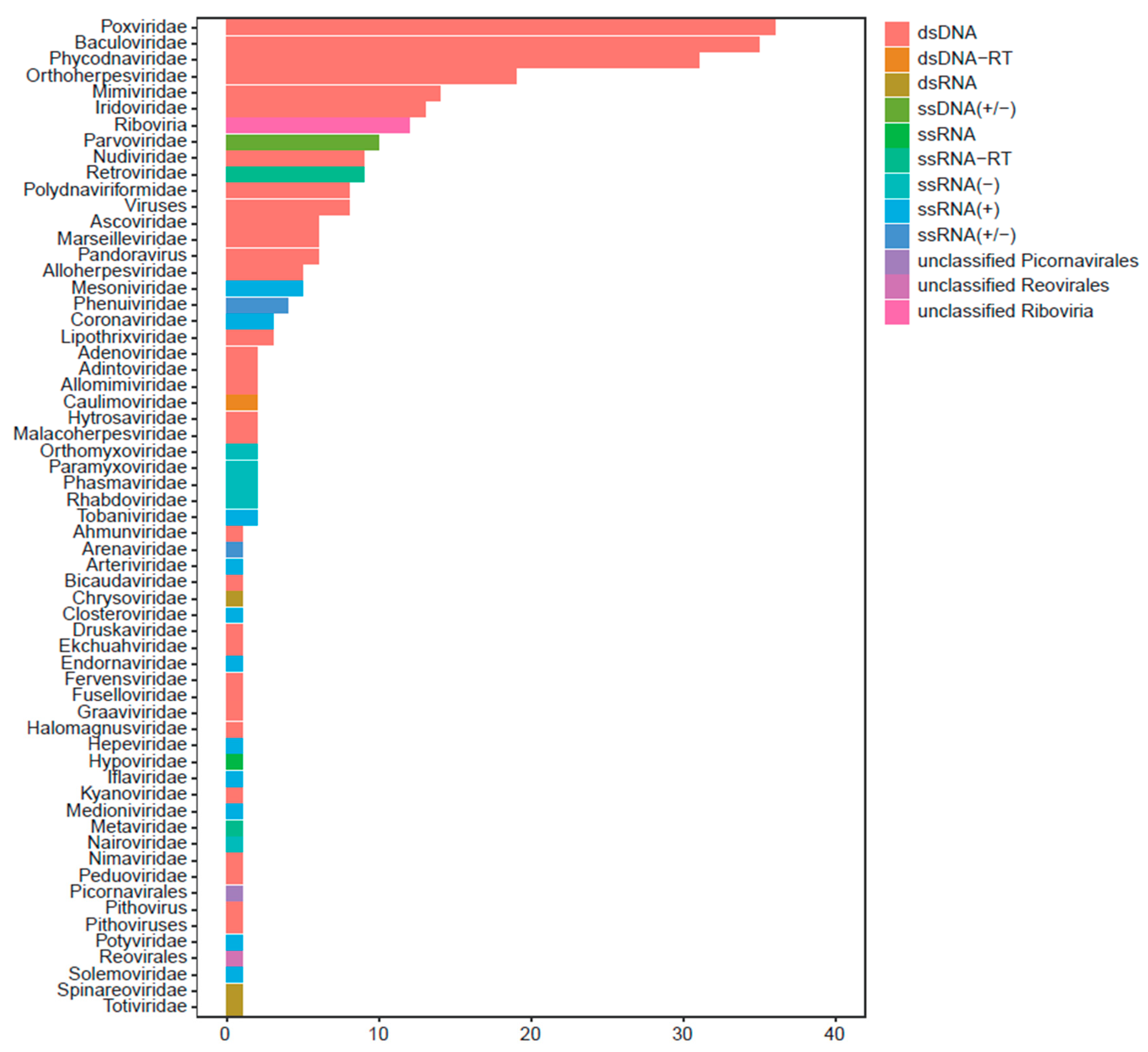
3.1. The Virus Screening from Pyrops Candelaria by RNA-seq
3.2. The Full-Length Sequencing and Related Bioinformatics Analysis Reflect PyCaV a New Virus
3.3. Construction of PyCaV Phylogenetic Tree
3.4. P. Candelaria Is One of the Important Carriers of the Virus PyCaV
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Valles, S.M.; Chen, Y.; Firth, A.E.; Guérin, D.M.A.; Hashimoto, Y.; Herrero, S.; de Miranda, J.R.; Ryabov, E.; Consortium, I.R. ICTV Virus Taxonomy Profile: Iflaviridae. J. Gen. Virol. 2017, 98, 527–528. [Google Scholar] [CrossRef] [PubMed]
- Ongus, J.R.; Roode, E.C.; Pleij, C.W.A.; Vlak, J.M.; van Oers, M.M. The 5' non-translated region of Varroa destructor virus 1 (genus Iflavirus): Structure prediction and IRES activity in Lymantria dispar cells. J Gen Virol 2006, 87, 3397–3407. [Google Scholar] [CrossRef]
- Lee, Y.F.; Nomoto, A.; Detjen, B.M.; Wimmer, E. A protein covalently linked to poliovirus genome RNA. Proc Natl Acad Sci USA 1977, 74, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.Y.; Wu, C.Y.; Chen, Y.J.; Chen, C.Y.; Wang, C.H. The 5' untranslated region of Perina nuda virus (PnV) possesses a strong internal translation activity in baculovirus-infected insect cells. Febs Lett 2007, 581, 3120–3126. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.O.; Groppelli, E. An atypical IRES within the 5' UTR of a dicistrovirus genome. Virus Res 2009, 139, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Lanzi, G.; de Miranda, J.R.; Boniotti, M.B.; Cameron, C.E.; Lavazza, A.; Capucci, L.; Camazine, S.M.; Rossi, C. Molecular and biological characterization of deformed wing virus of honeybees (Apis mellifera L.). J Virol 2006, 80, 4998–5009. [Google Scholar] [CrossRef] [PubMed]
- Isawa, H.; Asano, S.; Sahara, K.; Iizuka, T.; Bando, H. Analysis of genetic information of an insect picorna-like virus, infectious flacherie virus of silkworm: Evidence for evolutionary relationships among insect, mammalian and plant picorna(-like) viruses. Arch Virol 1998, 143, 127–143. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Lo, C.F.; Huang, C.J.; Yu, H.T.; Wang, C.H. The complete genome sequence of Perina nuda picorna-like virus, an insect-infecting RNA virus with a genome organization similar to that of the mammalian picornaviruses. Virology 2002, 294, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, J.; Lu, J.; Yi, F.; Liu, C.; Hu, Y. Sequence analysis and genomic organization of a new insect picorna-like virus, Ectropis obliqua picorna-like virus, isolated from Ectropis obliqua. J Gen Virol 2004, 85, 1145–1151. [Google Scholar] [CrossRef]
- Ongus, J.R.; Peters, D.; Bonmatin, J.M.; Bengsch, E.; Vlak, J.M.; van Oers, M.M. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J Gen Virol 2004, 85, 3747–3755. [Google Scholar] [CrossRef]
- Choe, S.E.; Nguyen, T.T.; Hyun, B.H.; Noh, J.H.; Lee, H.S.; Lee, C.H.; Kang, S.W. Genetic and phylogenetic analysis of South Korean sacbrood virus isolates from infected honey bees (Apis cerana). Vet Microbiol 2012, 157, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, S.; Bonning, B.C. Genome Sequence of a Novel Iflavirus from the Leafhopper Graminella nigrifrons. Genome Announc 2015, 3. [Google Scholar] [CrossRef]
- Wu, N.; Zhang, P.; Liu, W.; Cao, M.; Wang, X. Sequence analysis and genomic organization of a new insect iflavirus, Sogatella furcifera honeydew virus 1. Arch Virol 2018, 163, 2001–2003. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, Y.; Liu, W.; Cao, M.; Wang, X. Full genome sequence of a novel iflavirus from the leafhopper Psammotettix alienus. Arch Virol 2019, 164, 309–311. [Google Scholar] [CrossRef] [PubMed]
- de Miranda, J.R.; Genersch, E. Deformed wing virus. J Invertebr Pathol 2010, 103 (Suppl. 1), S48–S61. [Google Scholar] [CrossRef] [PubMed]
- Ryabov, E.V.; Wood, G.R.; Fannon, J.M.; Moore, J.D.; Bull, J.C.; Chandler, D.; Mead, A.; Burroughs, N.; Evans, D.J. A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog 2014, 10, e1004230. [Google Scholar] [CrossRef] [PubMed]
- Carballo, A.; Williams, T.; Murillo, R.; Caballero, P. Iflavirus Covert Infection Increases Susceptibility to Nucleopolyhedrovirus Disease in Spodoptera exigua. Viruses 2020, 12, 509. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Xu, P.; Yang, X.; Graham, R.I.; Wilson, K.; Wu, K. Characterization of a novel member of genus Iflavirus in Helicoverpa armigera. J Invertebr Pathol 2017, 144, 65–73. [Google Scholar] [CrossRef]
- Bitondi, M.M.; Nascimento, A.M.; Cunha, A.D.; Guidugli, K.R.; Nunes, F.M.; Simoes, Z.L. Characterization and expression of the Hex 110 gene encoding a glutamine-rich hexamerin in the honey bee, Apis mellifera. Arch Insect Biochem Physiol 2006, 63, 57–72. [Google Scholar] [CrossRef]
- Park, C.; Kang, H.S.; Jeong, J.; Kang, I.; Choi, K.; Yoo, M.S.; Kim, Y.H.; Kang, S.W.; Lim, H.Y.; Yoon, B.S.; et al. In-situ Hybridization for the Detection of Sacbrood Virus in Infected Larvae of the Honey Bee (Apis cerana). J Comp Pathol 2016, 154, 258–262. [Google Scholar] [CrossRef]
- Lin, Y.S.; Liao, J.R.; Shiao, S.F.; Ko, C.C. Origin and Potential Expansion of the Invasive Longan Lanternfly, Pyrops candelaria (Hemiptera: Fulgoridae) in Taiwan. Biology 2021, 10, 678. [Google Scholar] [CrossRef]
- Hsu, M.H.; Yang, Y.L.; Wu, M.L.; Wang, L.J. Host Plants of the Immature Stages of the Invasive Longan Lanternfly, Pyrops candelaria (L.) (Hemiptera, Fulgoridae) in Taiwan. Insects 2021, 12, 1022. [Google Scholar] [CrossRef]
- Wu, Y.H.; Kamiyama, M.T.; Chung, C.C.; Tzeng, H.Y.; Hsieh, C.H.; Yang, C.S. Population Monitoring, Egg Parasitoids, and Genetic Structure of the Invasive Litchi Stink Bug, Tessaratoma papillosa in Taiwan. Insects 2020, 11, 690. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Huang, J.; Huang, B. Preliminary study on the biological characteristics of Fulgora candelaria (L.). J. East China Entomol. 2000, 61–65. [Google Scholar]
- Urban, J.M.; Leach, H. Biology and Management of the Spotted Lanternfly, Lycorma delicatula (Hemiptera: Fulgoridae), in the United States. Annu Rev Entomol 2023, 68, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Toni, L.S.; Garcia, A.M.; Jeffrey, D.A.; Jiang, X.; Stauffer, B.L.; Miyamoto, S.D.; Sucharov, C.C. Optimization of phenol-chloroform RNA extraction. MethodsX 2018, 5, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Affeldt, K.J.; Carrig, J.; Amare, M.; Keller, N.P. Global Survey of Canonical Aspergillus flavus G Protein-Coupled Receptors. Mbio 2014, 5. [Google Scholar] [CrossRef]
- Ryabov, E.V. Invertebrate RNA virus diversity from a taxonomic point of view. J Invertebr Pathol 2017, 147, 37–50. [Google Scholar] [CrossRef]
- Ribière, M.; Olivier, V.; Blanchard, P. Chronic bee paralysis: A disease and a virus like no other? J Invertebr Pathol 2010, 103 (Suppl. 1), S120–S131. [Google Scholar] [CrossRef]
- Jia, W.; Wang, F.; Li, J.; Chang, X.; Yang, Y.; Yao, H.; Bao, Y.; Song, Q.; Ye, G. A Novel Iflavirus Was Discovered in Green Rice Leafhopper Nephotettix cincticeps and Its Proliferation Was Inhibited by Infection of Rice Dwarf Virus. Front Microbiol 2020, 11, 621141. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.M.; Vaissiere, B.E.; Cane, J.H.; Steffan-Dewenter, I.; Cunningham, S.A.; Kremen, C.; Tscharntke, T. Importance of pollinators in changing landscapes for world crops. Proc Biol Sci 2007, 274, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Biesmeijer, J.C.; Roberts, S.P.; Reemer, M.; Ohlemuller, R.; Edwards, M.; Peeters, T.; Schaffers, A.P.; Potts, S.G.; Kleukers, R.; Thomas, C.D.; et al. Parallel declines in pollinators and insect-pollinated plants in Britain and the Netherlands. Science 2006, 313, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Goulson, D.; Lye, G.C.; Darvill, B. Decline and conservation of bumble bees. Annu Rev Entomol 2008, 53, 191–208. [Google Scholar] [CrossRef] [PubMed]
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol Evol 2010, 25, 345–353. [Google Scholar] [CrossRef] [PubMed]
- McMahon, D.P.; Furst, M.A.; Caspar, J.; Theodorou, P.; Brown, M.J.F.; Paxton, R.J. A sting in the spit: Widespread cross-infection of multiple RNA viruses across wild and managed bees. J Anim Ecol 2015, 84, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Kemp, E.M.; Woodward, D.T.; Cory, J.S. Detection of single and mixed covert baculovirus infections in eastern spruce budworm, Choristoneura fumiferana populations. J Invertebr Pathol 2011, 107, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Millan-Leiva, A.; Jakubowska, A.K.; Ferre, J.; Herrero, S. Genome sequence of SeIV-1, a novel virus from the Iflaviridae family infective to Spodoptera exigua. J Invertebr Pathol 2012, 109, 127–133. [Google Scholar] [CrossRef]
- Silva, L.A.; Ardisson-Araujo, D.M.; Tinoco, R.S.; Fernandes, O.A.; Melo, F.L.; Ribeiro, B.M. Complete genome sequence and structural characterization of a novel iflavirus isolated from Opsiphanes invirae (Lepidoptera: Nymphalidae). J Invertebr Pathol 2015, 130, 136–140. [Google Scholar] [CrossRef]
- Carballo, A.; Murillo, R.; Jakubowska, A.; Herrero, S.; Williams, T.; Caballero, P. Co-infection with iflaviruses influences the insecticidal properties of Spodoptera exigua multiple nucleopolyhedrovirus occlusion bodies: Implications for the production and biosecurity of baculovirus insecticides. PLoS ONE 2017, 12, e0177301. [Google Scholar] [CrossRef]
- Jakubowska, A.K.; Murillo, R.; Carballo, A.; Williams, T.; van Lent, J.W.; Caballero, P.; Herrero, S. Iflavirus increases its infectivity and physical stability in association with baculovirus. PeerJ 2016, 4, e1687. [Google Scholar] [CrossRef]
- Huang, S.; Han, D.; Wang, J.; Guo, D.; Li, J. Floral Induction of Longan (Dimocarpus longan) by Potassium Chlorate: Application, Mechanism, and Future Perspectives. Front Plant Sci 2021, 12, 670587. [Google Scholar] [CrossRef]
- Park, S.J.; Park, D.H.; Kim, D.H.; Lee, S.; Yoon, B.H.; Jung, W.Y.; Lee, K.T.; Cheong, J.H.; Ryu, J.H. The memory-enhancing effects of Euphoria longan fruit extract in mice. J Ethnopharmacol 2010, 128, 160–165. [Google Scholar] [CrossRef]
- Yang, L.; Fu, S.; Khan, M.A.; Zeng, W.; Fu, J. Molecular cloning and development of RAPD-SCAR markers for Dimocarpus longan variety authentication. Springerplus 2013, 2, 501. [Google Scholar] [CrossRef]
- Zhong, K.; Wang, Q.; He, Y.; He, X. Evaluation of radicals scavenging, immunity-modulatory and antitumor activities of longan polysaccharides with ultrasonic extraction on in S180 tumor mice models. Int J Biol Macromol 2010, 47, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Pattamayutanon, P.; Angeli, S.; Thakeow, P.; Abraham, J.; Disayathanoowat, T.; Chantawannakul, P. Volatile organic compounds of Thai honeys produced from several floral sources by different honey bee species. PLoS ONE 2017, 12, e0172099. [Google Scholar] [CrossRef]
- Chen, Y.; Sun, J.; Lin, H.; Lin, M.; Lin, Y.; Wang, H.; Hung, Y.-C. Salicylic acid reduces the incidence of Phomopsis longanae Chi infection in harvested longan fruit by affecting the energy status and respiratory metabolism. Postharvest Biol. Technol. 2020, 160, 111035. [Google Scholar] [CrossRef]
- Seo, J.K.; Kim, M.K.; Kwak, H.R.; Kim, J.S.; Choi, H.S. Complete genome sequence of longan witches' broom-associated virus, a novel member of the family Potyviridae. Arch Virol 2017, 162, 2885–2889. [Google Scholar] [CrossRef]
- Behura, S.K. Insect phylogenomics. Insect Mol Biol 2015, 24, 403–411. [Google Scholar] [CrossRef]
- Klein, A.M.; Vaissière, B.E.; Cane, J.H.; Steffan-Dewenter, I.; Cunningham, S.A.; Kremen, C.; Tscharntke, T. Importance of pollinators in changing landscapes for world crops. Proc Biol Sci 2007, 274, 303–313. [Google Scholar] [CrossRef]
- Kremen, C. The value of pollinator species diversity. Science 2018, 359, 741–742. [Google Scholar] [CrossRef]
- Eggleton, P. The State of the World's Insects. Annu. Rev. Environ. Resour. 2020, 45, 61–82. [Google Scholar] [CrossRef]
- Crespo-Pérez, V.; Kazakou, E.; Roubik, D.W.; Cárdenas, R.E. The importance of insects on land and in water: A tropical view. Curr Opin Insect Sci 2020, 40, 31–38. [Google Scholar] [CrossRef]
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol Evol 2004, 19, 535–544. [Google Scholar] [CrossRef]
- Rashidi, M.; Lin, C.Y.; Britt, K.; Batuman, O.; Al Rwahnih, M.; Achor, D.; Levy, A. Diaphorina citri flavi-like virus localization, transmission, and association with Candidatus Liberibacter asiaticus in its psyllid host. Virology 2022, 567, 47–56. [Google Scholar] [CrossRef]
- Patton, M.F.; Hansen, A.K.; Casteel, C.L. Potato leafroll virus reduces Buchnera aphidocola titer and alters vector transcriptome responses. Sci. Rep. 2021, 11, 23931. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Lu, H.; Wang, W.; Zhu, J.; Zhao, W.; Cui, F. Membrane association of importin α facilitates viral entry into salivary gland cells of vector insects. Proc. Natl. Acad. Sci. United States Am. 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Kemenesi, G.; Dallos, B.; Görföl, T.; Boldogh, S.; Estók, P.; Kurucz, K.; Kutas, A.; Földes, F.; Oldal, M.; Németh, V.; et al. Molecular survey of RNA viruses in Hungarian bats: Discovering novel astroviruses, coronaviruses, and caliciviruses. Vector Borne Zoonotic Dis 2014, 14, 846–855. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




