1. Introduction
Candida albicans is a pathogenic fungus that causes oral mucosal, vaginal, and systemic candidiasis in millions worldwide. According to the Centers for Disease Control (CDC), 5-7% of infants <1 month of age develop oral candidiasis, while the prevalence among AIDS patients is estimated to range from 9-31% [
1]. Furthermore, the clinical incidence of oral candidiasis is nearly 90% in cancer patients, with
Candida albicans isolated from 58% of patients [
2]. The risk of systemic/invasive candidiasis is also increased in immunocompromised patients and may become life-threatening. Hence, diagnosis and treatment of oral/mucosal candidiasis is critical. However, there are numerous reports of
Candida species becoming resistant to the currently available antifungal agents. As a result, there is a critical need for drug targets to develop novel antifungal drugs.
C. albicans DFG5 and
DCW1 encode for glycosidase/mannanase/mannosyltransferase enzymes (
gh-76 family) that are targeted to the cell wall space by the N-terminal signal for secretion and a C-terminal GPI anchor. Past studies in
Saccharomyces cerevisiae have shown that
dfg5/dcw1 knockout mutation is lethal, indicating that these cell wall proteins fulfill essential cellular functions [
3,
4]. Our study in
Neurospora crassa demonstrated that these proteins serve in cell wall protein incorporation in the wall and thus affect cell wall biogenesis [
5]. These studies clearly indicate that
DFG5 and
DCW1 have highly conserved functions and play an important role in fungal cell physiology. In
C. albicans,
dfg5 and
dcw1 single mutants are viable; however, the
dfg5/dcw1 knockout mutant is lethal indicating a functional redundancy [
6]. In addition, Dfg5 has been shown to be in the cell membrane and the expression of
HWP1, a hypha specific gene, is affected in the
dfg5 knockout mutant [
6]. Such alterations in specific gene expression occur when signal transduction pathways are affected. It was confirmed in
Saccharomyces cerevisiae confirmed that Hog1 and Slt2 cell signaling pathways are affected in the
dfg5Δ mutant [
7]. Further evidence in the mycoparasite
Trichoderma atroviride, also suggests that Dfg5 plays critical role in hyphal morphogenesis and osmoregulation via MAPK signaling [
8]
Our past studies in
C. albicans have utilized the
pMET3 modulated
dfg5/dcw1 conditional mutant [
6]. We showed that
DFG5 and
DCW1 function in covalent incorporation of cell wall proteins and thus play critical roles in cell wall biogenesis [
9]. Our data also indicate that
dfg5/
dcw1 mutants are affected in hyphal morphogenesis and biofilm formation. Additionally, we also showed that basal Hog1 MAPK levels are reduced in the
dfg5/dcw1 mutants and that
dfg5/dcw1 mutants have a cell separation phenotype similar to
hog1 knock-out mutant [
10]. This study focused on delineating the roles of Dfg5 and Dcw1 in chitin synthesis and disease pathogenesis.
2. Materials & Methods
2.1. Strains and Growth Conditions
The genetic background of the strains used in this study can be found in
Table 1. The strains were cultured in Yeast Nitrogen Base (YNB) medium with ammonium sulfate and 2% glucose. Synthetic complete supplement mixture (MP Biomedicals) was added as amino acid supplement to YNB. 5mM Methionine and 2 mM cysteine were added to the medium for ES195 strain for conditional repression (85%) of the chimeric
MET3::DFG5 gene to generate a Dfg5p-deficient condition.
2.2. Light & Fluorescence Microscopy Analysis
Overnight cultures were diluted to OD
600 of 1.5 (approximately 5x10
7 CFU/ml) in a total volume of 1 mL of YNB either with or without 1.668 µg/ml Chitinase (Sigma # C8241-25UN) as described previously [
11]. The ES195 strain was grown with and without Methionine and Cysteine for control cultures and chitinase-treated cultures. Cultures were allowed to incubate at room temperature with shaking at 225 rpm for 3 hours. Cells were pelleted by centrifugation at 900 x g for 2 min. The media was removed and the cells were re-suspended in 1x PBS containing 100 µg/ml Calcofluor White [
12]. 3 µl of each sample were immediately placed on microscope slides with a coverslip and imaged with a Nikon Eclipse TE2000-U at 400x total magnification using Spot Advanced 4.0.4 software. Fluorescence microscopy of CFW was done using a UV filter. False color was added to the fluorescent images with ImageJ software. Calcofluor white
(CFW) fluorescence intensity of 100 cells/strain and the background directly next to each of those cells was measured using ImageJ software as described previously [
13]. Corrected Total Cell Fluorescence (CTCF) calculations were performed to quantify chitin accumulation in each strain as follows: CTCF=Integrated density−[(Area of selected cell) ×(Mean fluorescence of background readings)].
2.3. Scanning Electron Microscopy (SEM) Analysis
SEM analysis was performed as described previously [
14]. The cultures were prepared for light microscopy and then transferred to 6-well polystyrene plates where the cells were allowed to settle for 90 minutes at 37°C on Fetal Bovine Serum (FBS, Seradigm) coated glass squares. The cells were fixed and dried. The samples were coated with evaporated carbon at high vacuum (Denton 502 Evaporator). SEM images were acquired with a Hitachi SU70 FESEM at 2.0 KeV using the lower detector and no tilt.
2.4. Quantitative Real Time PCR (qRT-PCR) Analysis
qPCR analysis was performed as described previously [
15]. Primers were prepared for
CHS genes (
CHS1,
CHS2,
CHS3 and
CHS8), positive hyphal transcriptional regulators (
CST20,
HST7,
CPH1 &
CPH2), negative hyphal regulators (
TUP1,
NRG1,
RBF1,
RFG1 &
MIG1) and housekeeping gene
EFB1 (
Table 2). The qPCR reactions were performed using the Applied Biosystems 7500 Real Time PCR machine with standard cycling protocol from the SYBR Green FastMix product manual: denaturation at 95°C for 1 minute, annealing 40 cycles of 95°C for 5 seconds, extension 60C for 34 seconds. Data was collected at the end of the extension step. To analyze the data, the values for the
CHS genes were normalized to the
EFB1 housekeeping gene for the 2
-ΔΔCt calculations using Microsoft Excel.
2.5. Mouse Model Protocol for Oral Candidiasis
Oral candidiasis infections were performed in mice as previously described using a protocol approved by University at Buffalo IACUC (Protocol #201700003) [
16]. Five BALB/c mice (11-week-old, male and female mice) (Jackson Labs) were infected for each strain used (5 groups of mice total). Groups infected with SC5314 (group 1) and DAY185 (group 2) were used as controls. These were compared with groups infected with mutant strains ES1 (group 3) and the conditional mutant ES195. For the ES195 strain, one group of mice were infected with untreated cells (group 4) and one group of mice were infected with cells that had been pre-treated with 5mM Methionine and 2mM Cysteine (Bulksupplements.com) for one hour prior to infection (group 5) to achieve 85% repression of the remaining copy of
DFG5 in this strain. To maintain this repression throughout the experiment, group 5 mice received Methionine and Cysteine in their drinking water. Immunosuppression was performed by giving 225 mg/kg cortisone 21-acetate (Sigma Aldrich) subcutaneously on the day prior to infection and on days 1 and 3 post-infection. The infected mice were monitored for changes in behavior and health. Pictures of the tongue were taken at days 1, 3 and 5 post-infection. On day 5, mice were euthanized by cervical dislocation performed under anesthesia (Ketamine/Xylazine as described above). The tongues and surrounding hypoglossal tissue were removed and cut in half lengthwise. One half was used for histopathological analysis by H&E (Hematoxylin and Eosin) staining as well as PAS (Period Acid Schiff) staining after fixation with 10% Neutral Buffered Formalin (IMEB). The other half was weighed and homogenized completely for quantification of infection by colony forming unit (CFU) assessment.
3. Results
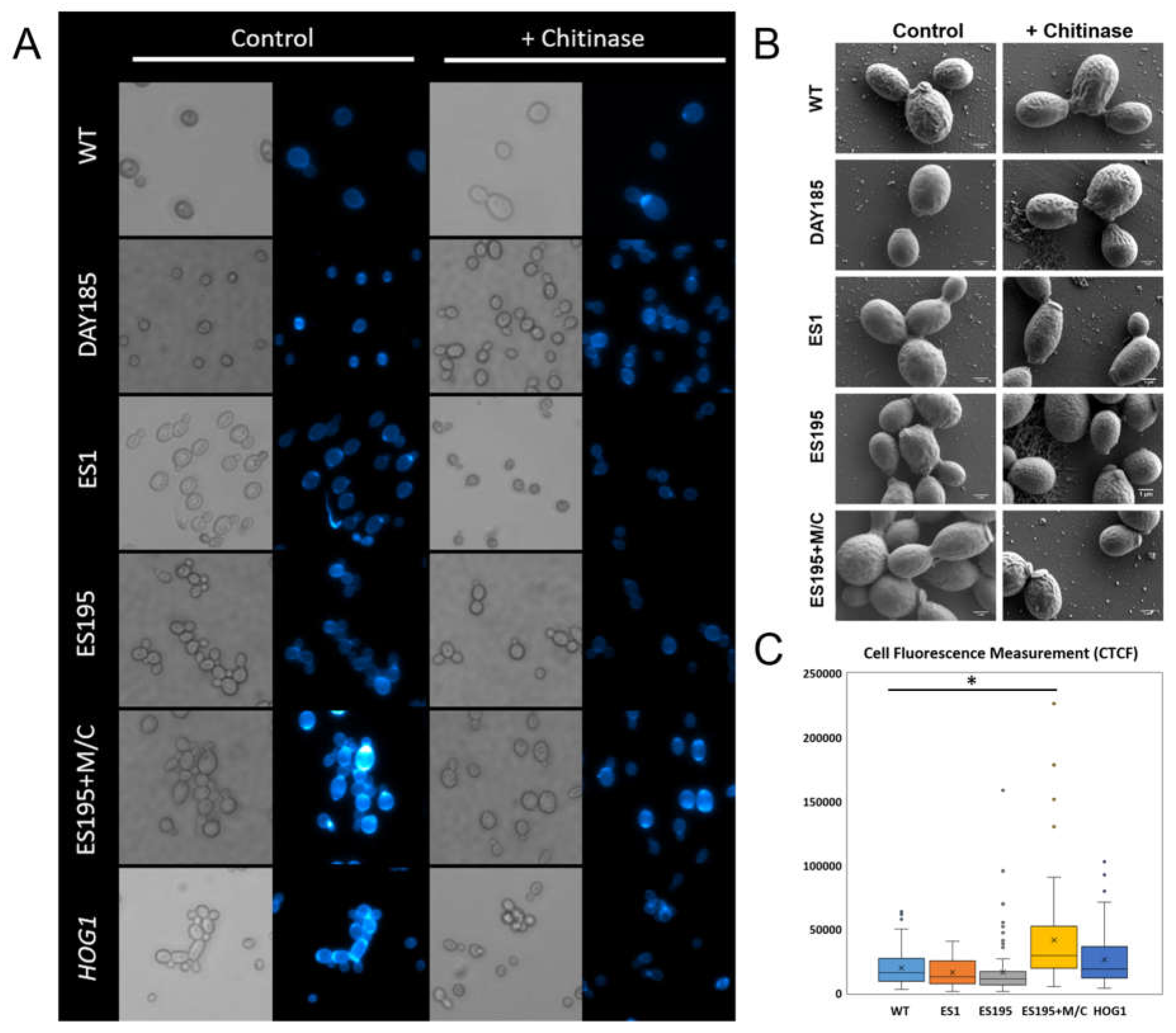
3.1. DFG5 and DCW1 Mutations Result in a Cell Separation Defect Identical to hog1 Mutant
Light microscopy analysis of strains ES195 and ES195+M/C conditional mutant indicated that they show a cell separation defect, as compared to WT and DAY185 strains (
Figure 1). This was also confirmed for
hog1 knock-out mutant (
Figure 1). Fluorescence imaging using CFW that binds chitin revealed higher intensity of fluorescence at the cell septae, indicating increased chitin accumulation (
Figure 1). It is interesting that this cell separation defect or even increased intensity of CFW fluorescence was only minimal for ES1 mutant, in which both copies of
DFG5 are mutated and only one functional copy of
DCW1 is present. This may indicate that the one remaining copy of
DCW1 is sufficient to compensate for the loss of both copies of
DFG5.
3.2. Chitinase Treatment Results in Reversal of Cell Separation Phenotype for dfg5/dcw1 Mutants
A characteristic of the cell separation defect in the
hog1 knock-out mutant is its reversal by treatment with commercially available chitinase. Mutant and control strains were incubated with chitinase for 3 hours which resulted in improvement of cell separation for the ES195, ES195+M/C conditional mutant and
hog1 knock-out mutant strain, indicating that the identical phenotype among the mutant strains was due to abnormal increase in chitin accumulation (
Figure 1). This is further confirmed by SEM analysis which showed that the cell separation defect was due to a lack of separation of the mother-bud neck following cell division (
Figure 1).
3.3. DFG5/DCW1 Mutations Result in Increased Chitin Levels
CFW fluorescence for ES195+M/C conditional mutant and
hog1 knock-out mutant appeared to be increased as compared to WT, DAY185 and ES1 and ES195 strains (
Figure 1). This is suggestive of increased chitin levels in the ES195+M/C conditional mutant and
hog1 knock-out mutant, although the levels were much higher for the former. Correspondingly normal chitin levels were observed for the ES1 mutant as measured by CFW fluorescence.
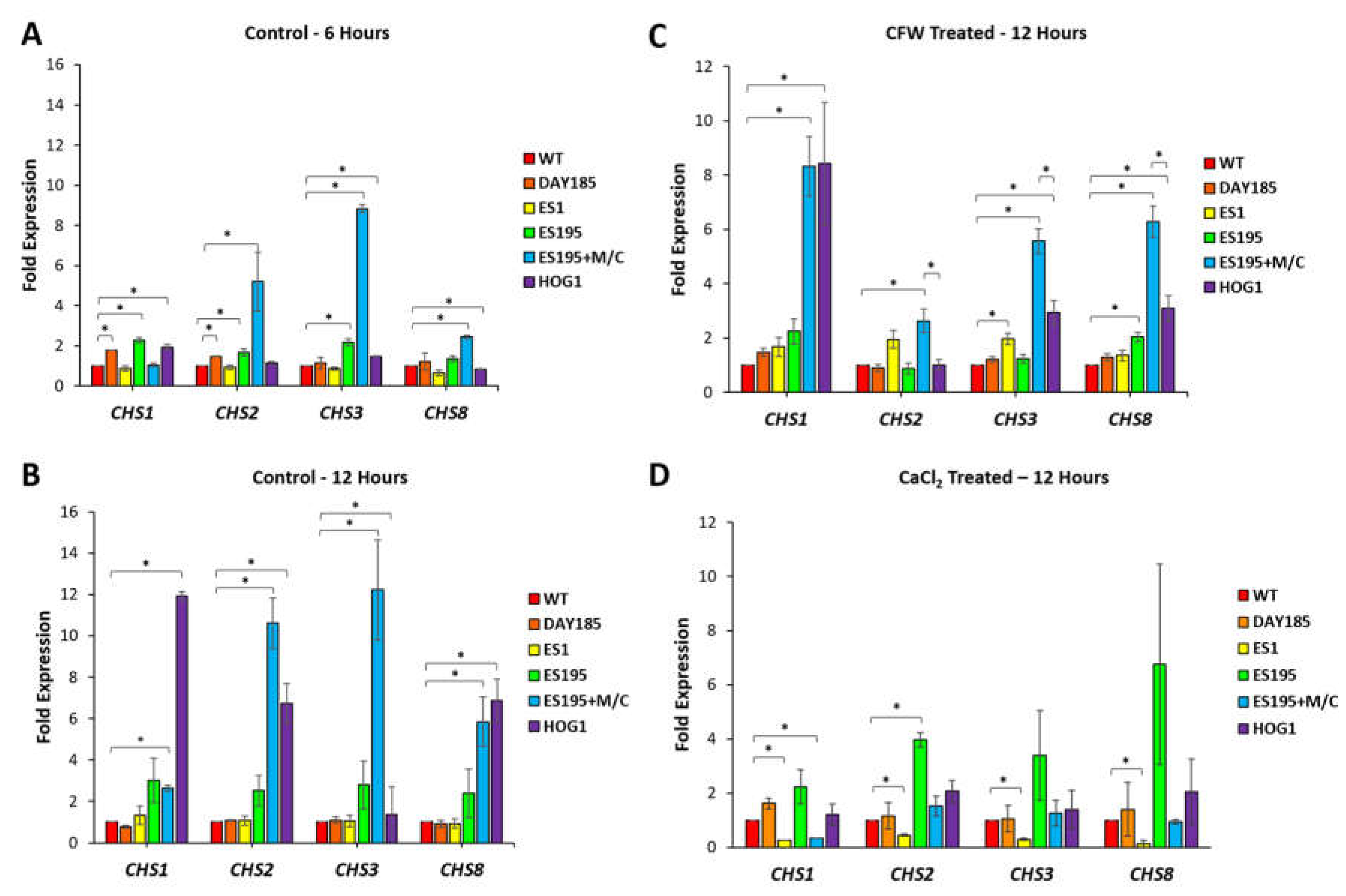
3.4. Dfg5 and Dcw1 Affect Gene Expression of Chitin Synthases CHS1, CHS2, CHS3 and CHS8
Transcriptional analysis of
CHS genes was performed for control and mutant strains under basal and chitin stress (CFW) conditions using qRT-PCR analysis (
Figure 2). Analysis of the control strains - WT and DAY185, indicated that the level of expression of all four chitin synthases
CHS1,
CHS2,
CHS3 and
CHS8 was almost identical indicating that under normal conditions these chitin synthases may be produced in similar quantities. However,
dfg5/dcw1 heterologous mutations resulted in variable regulation of chitin synthase gene expression. Under basal conditions, ES1 mutant was unable to upregulate the expression of all four chitin synthases at either 6 h or 12 h time points. This may indicate that Dfg5 is required for upregulation for all four chitin synthases. This could be important under cell wall stress conditions considering that the ES1 mutant is already under cell wall stress due to inability to cross-link proteins in the cell wall. The ES195 mutant only had modest increase (2-3 fold) in the expression of all four chitin synthase genes at 6 h time point and further increased (3-5 fold) at 12 h time point. This result indicates that one copy of
DFG5 is sufficient to compensate for the lack of
DCW1 and thus can upregulate the chitin synthases modestly. Only a modest increase in expression may occur in the ES195 mutant due to a possible modest activation of the PKC pathway occurring in response to cell wall stress [
17]. On the other hand, the ES195+M/C conditional mutant showed an increase in gene expression for
CHS2 (6-7 fold),
CHS3 (10-11 fold) and
CHS8 (3-4 fold) for the 6 h time point. However, at 12 h time point, this gene expression further increased for
CHS2 (10-12 fold),
CHS3 (12-14 fold) and
CHS8 (6-8 fold). On the other hand,
CHS1 gene expression remained low. This information indicates that very low levels of Dfg5 (15%) trigger a compensatory upregulation of
CHS2,
CHS3 and
CHS8 while ignoring
CHS1.
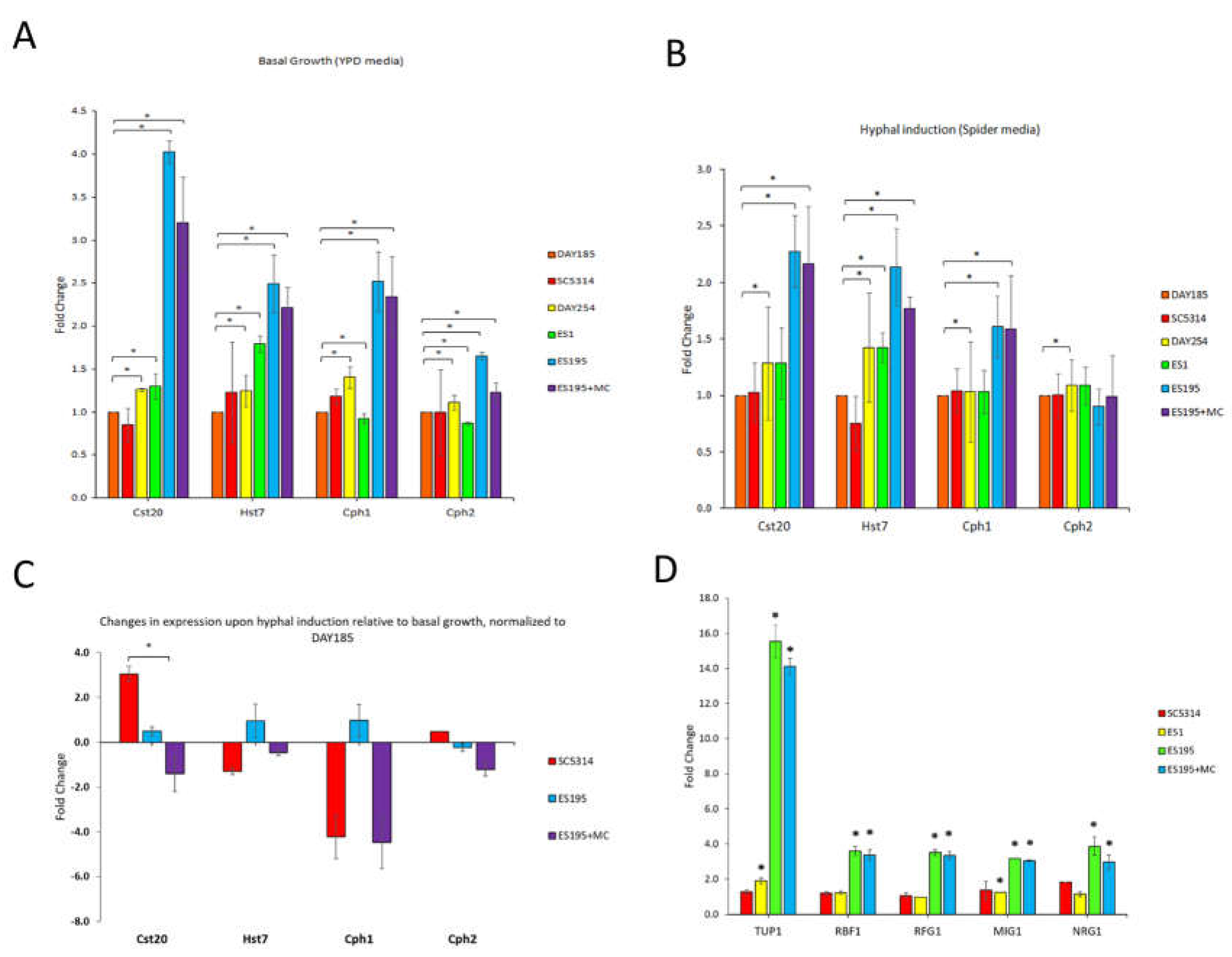
3.5. DFG5/DCW1 Conditional Knockout Mutations Results in Decreased Levels of Cst20, a Positive Transcriptional Regulator of Hyphal Morphogenesis
The mechanism by which
dfg5/
dcw1 heterozygous mutations affect transcriptional regulators of hyphal morphogenesis is depicted in
Figure 3. In
Figure 3A, the gene expression analysis of positive transcriptional regulators under basal conditions (30°C) is shown. The most significant increase in gene expression was observed for the
CST20 transcriptional regulator for the ES195 and ES195 + M/C strains.
Figure 3B demonstrates gene expression analysis under hyphal-inducing conditions (spider media). In this case, there is likewise increased gene expression with respect to the
CST20 transcriptional regulator for the ES195 and ES195 + M/C mutants, but that difference is not nearly as pronounced as that shown under basal conditions in
Figure 3A. It also depicts gene expression increases for
HST7 and
CPH1 for the ES195 and ES195 + M/C strains, but relatively lower expression for
CPH2. When relative gene expression analysis was performed between the hyphal inducing and basal conditions (
Figure 3C), a significant decrease in gene expression was found for
CST20 in ES195 + M/C conditional mutant strains as compared to WT. This result indicates that Dfg5 and Dcw1 may function in hyphal morphogenesis by increasing
CST20 levels.
3.6. DFG5 /DCW1 Conditional Knockout Mutations Result in Increased Levels of TUP1, RBF1, MIG1, RFG1 and NRG1, Negative Transcriptional Regulators of Hyphal Morphogenesis
Figure 3D depicts the impact of several negative transcriptional regulators (
TUP1,
RBF1,
RFG1,
MIG1,
NRG1) on gene expression in the various Candida strains. Tup1 is a major hyphal regulator and serves as a transcriptional suppressor. Of note here are the significantly increased levels of expression of Tup1 in both the ES195 and ES195 + M/C strains, thus leading to constitutive hyphal expression.
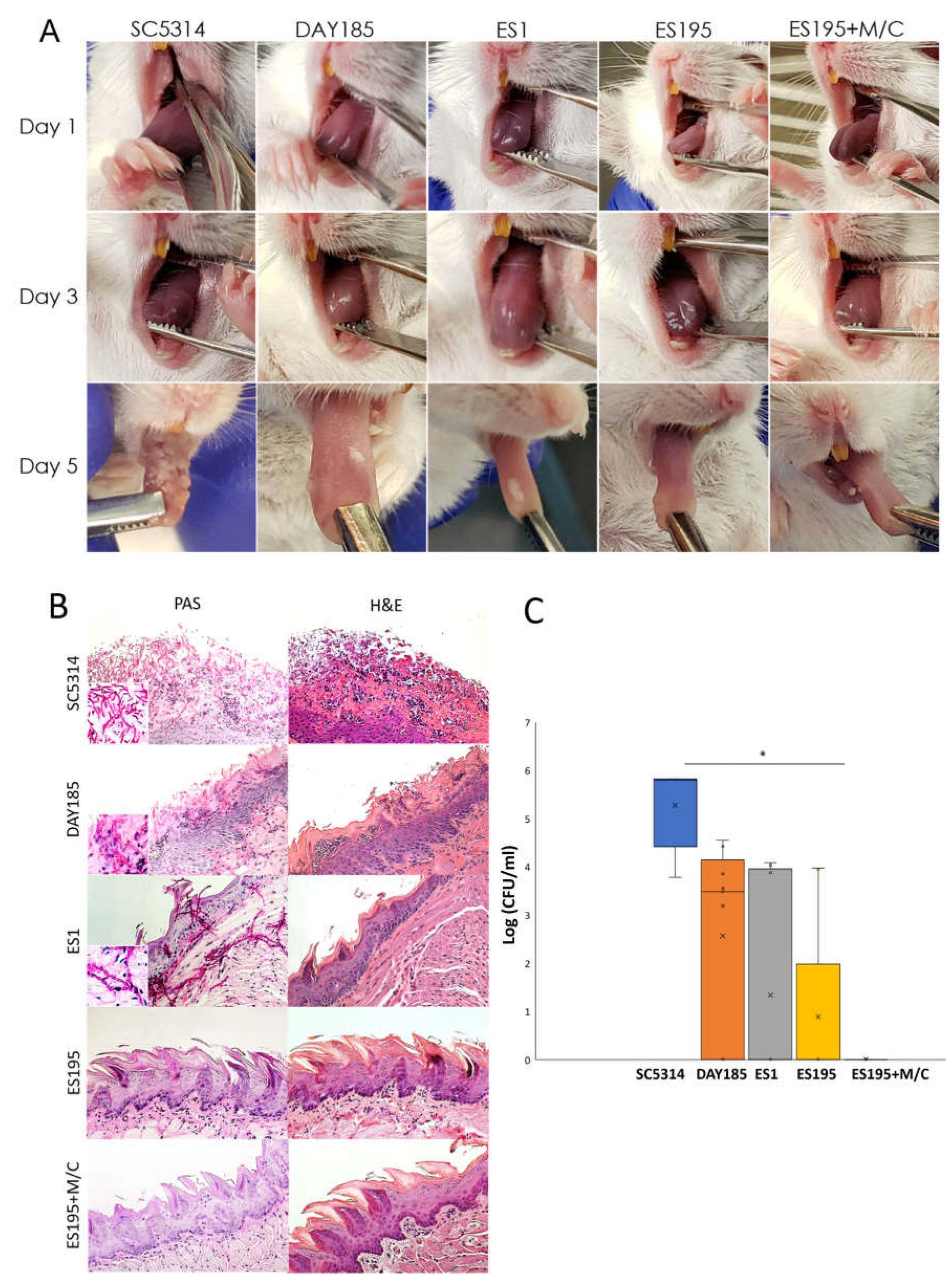
3.7. DFG5 /DCW1 Mutations Cause a Defect in In Vivo Virulence and Pathogenesis of C. albicans in a Mouse Model of Oral Candidiasis
Figure 4 depicts the
in vivo pathogenesis of wild type and heterozygous mutant strains in a mouse model of oral candidiasis. Mice infected with WT strain SC5314 and parental control DAY185 showed oral candidiasis on the tongue. Among the heterozygous mutants, ES1 was able to cause infection. However, the ES195 strain and ES195 + M/C strain did not show any visible plaque formation.
Figure 4B demonstrates the histological analysis of the mouse tongue in the various infection groups. Wild type strain SC5314 and DAY185 strains both depict that there is destruction of epithelium, and clear presence of hyphal structures penetrating into the tissues. In the ES1 mutant, pseudo-hyphal type of structures were noted, but real hyphae were not observed. Minor destruction of epithelium was also observed. In the ES195 and ES195 + M/C mutant groups, there was no colonization observed and the epitheliums were large intact, thus indicating that no infection had occurred. Further quantification of Candida CFUs within the tongue tissue (
Figure 4C) indicated that ES195 strain had reduced CFUs, whereas the ES195 + M/C strain did not have any (
Figure 4C). This demonstrates that cells may be present and survive on the tongue on ES1 and ES195 strains, however due to lack of hyphal structure they are not able to penetrate into tissues and ultimately cause disease. However, for the ES195 + M/C conditional mutant the cells could not survive due to a defect in hyphal morphogenesis.
4. Discussion
The composition and structural organization of the
Candida albicans cell wall is dynamically regulated in response to changing environmental conditions [
18]. The carbohydrates, chitin, and beta-glucan form the structural framework of the fungal cell wall [
18]. Alteration and reconstitution of chitin and beta-glucan within the cell wall occurs in response to disruption of genes in cell wall biosynthetic pathways of
C. albicans [19]. Additionally, upregulation of cell wall chitin levels has been identified as an alternate resistance mechanism against the antifungal drug caspofungin, independent of mutations in the FKS region [
20]. Furthermore, large amounts of chitin in the cell wall correspond to increasing caspofungin resistance in animal models [
13,
20,
21].
C. albicans has four chitin synthases – Chs1, Chs2, Chs3 and Chs8, that play important roles in cell wall formation, septum formation and affect cell wall integrity [
22,
23,
24]. Transcriptional regulation of chitin synthases in
C. albicans is controlled by three signaling pathways in a coordinated manner - Ca
2+ calcineurin pathway, HOG pathway and MKC pathway [
25]. Using
lacZ reporter assay it was found that
hog1∆ mutant had altered expression of chitin synthases
CHS3 and
CHS8 in
C. albicans [
25].
In this study, we compared the dfg5/dcw1 heterologous mutants to the hog1 knock out mutant in relation to chitin synthesis. At the 6 h time point, the levels of expression in the hog1 knock out mutant appeared to be at the WT level. However, at the 12 h time point, hog1 knock out mutant had increased expression of CHS1, CHS2, CHS3 and CHS8 genes. A plausible reason for this could be that by 12 h, which represents the mid-log phase, the glucose present in the media were depleted resulting in reduced beta-glucan synthesis and thus weakening the wall. This in turn could have triggered the alternate pathways i.e., the PKC pathway and/or the calcineurin pathway for chitin synthesis. Additionally, CHS3 expression was not increased for the hog1 knock out mutant even at 12 h time point indicating that Hog1 may be required for its upregulation. Our data is different from the past study by Lenardon (2017) in two ways – the methods used and the time point of gene expression measurement. The Lenardon (2017) study used a promoter-based beta-galactosidase assay to measure gene expression in the presence of CFW as compared to qRT-PCR under basal and CFW conditions in our study. Also, the beta-galactosidase assay was done when the cells reached an OD of 1 which would be past the mid-log phase and may represent a different time point than the 12 h in our study. Furthermore, our study also indicates that gene expression of chitin synthases varies depending upon time of growth.
Our study also determined the functions of Dfg5 and Dcw1 in hyphal morphogenesis and in vivo pathogenesis of C. albicans. Our data indicate that Dfg5 and Dcw1 are required for increased expression of Cst20, a positive transcriptional regulator of hyphal morphogenesis during hyphal induction. However, the most striking data was related to the significantly higher expression of negative transcriptional regulators ((TUP1, RBF1, RFG1, MIG1, NRG1) of hyphal morphogenesis in the DFG5/DCW1 conditional knock out mutants. This data indicates that Dfg5 and Dcw1 are required for repression of negative transcriptional regulators including TUP1 which acts as a co-factor for the others (RBF1, RFG1, MIG1). It is noteworthy that there is no known upstream signaling pathways that have been identified for Tup1. This is the first study that describes novel upstream functions of Dfg5 and Dcw1 in negative transcriptional regulation of hyphal morphogenesis. Further, our animal study experiments indicate that Dfg5 and Dcw1 are required for pathogenesis in a mouse model of oral candidiasis. The heterozygous mutant, ES1, is able to cause disease and forms pseudohyphae as depicted in the histological sections of the tongue. However, the conditional mutants, ES195 and ES195+M/C are unable to form hyphal structures or cause disease.
5. Summary & Conclusions
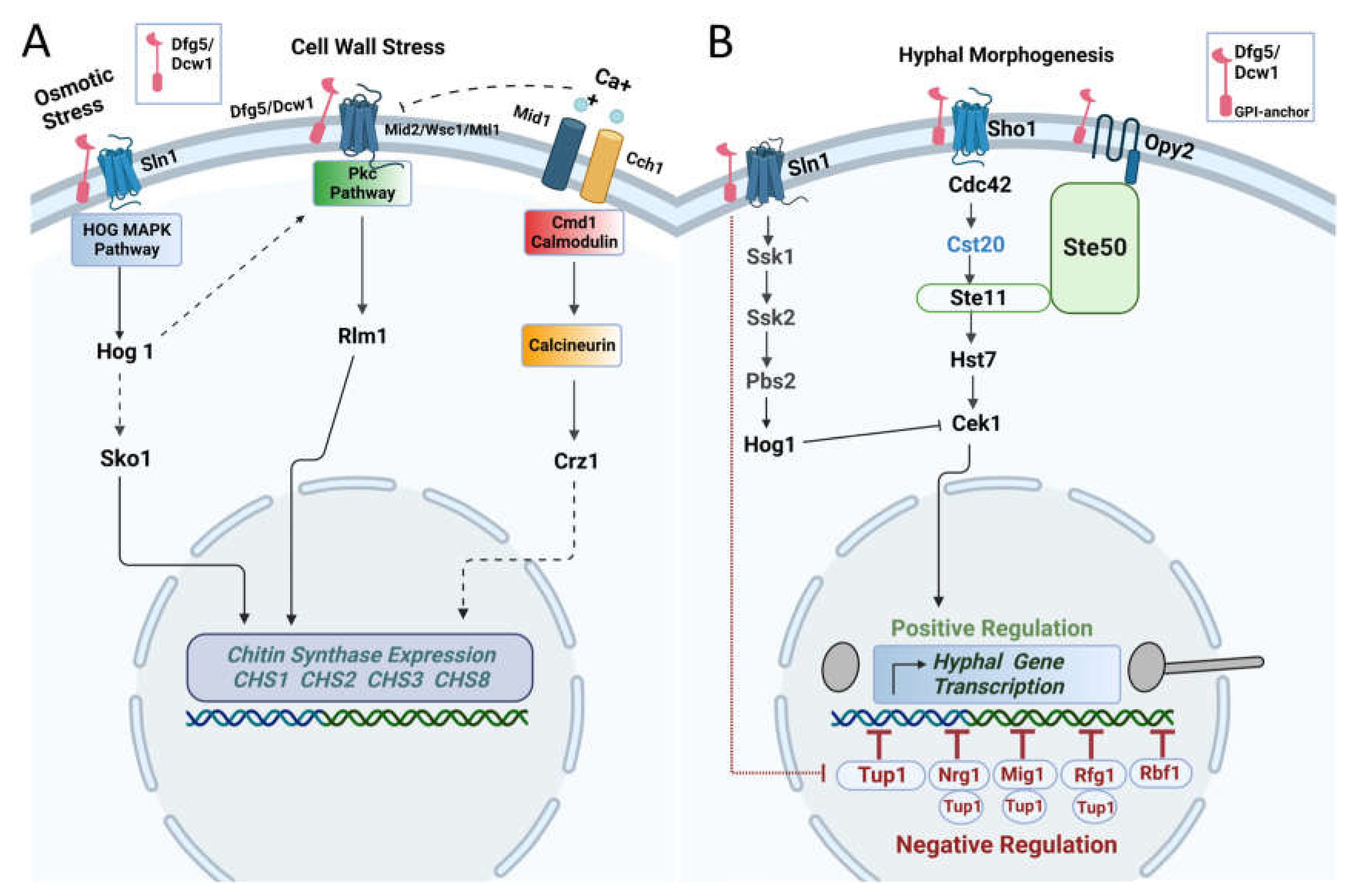
Overall, our data indicate that Dfg5 and Dcw1 cell wall glycosidases regulate cell wall chitin levels by affecting the gene expression of chitin synthases. Furthermore, this phenomenon was similar to the
hog1 knock out mutant but more severe, especially as observed for the
dfg5/dcw1 conditional mutant. Further, the hyphal morphogenesis pathways also appear to be affected by Dfg5 by affecting Cst20, Tup1, Rbf1, Rfg1, Mig1 and Nrg1. Based on our data we hypothesize that Dfg5 and Dcw1 act as cell wall sensors and interact with signaling proteins (Sln1, Sho1 and Opy2) within the cell wall that regulate the aforementioned pathways (
Figure 5).
Author Contributions
Maryam Razmi - Performed experiments and contributed to writing the manuscript; Jaewon Kim – Performed experiments and contributed to writing the manuscript; Jennifer Chinnici - Performed experiments and contributed to writing the manuscript; Sujay Busarajan - Performed experiments and contributed to writing the manuscript; Hema Vuppalapaty - Performed experiments and contributed to writing the manuscript; Deepika Lankipalli - Performed experiments; Rui Li – Performed experiments and contributed to writing and editing the manuscript; Abhiram Maddi – Contributed to designing the study, project management, data acquisition, analysis of results, writing and editing the manuscript.
Acknowledgments
We would like to acknowledge Dr. Peter Bush for assistance with SEM analysis and Mr. Jason Chwirut for help in preparation of the figures for this manuscript. This study was supported by NIDCR 1R03DE028607 awarded to Dr. Abhiram Maddi.
Conflicts of Interests
The authors do not have any financial or other conflicts of interest.
References
- Patil S et al., 2015. Clinical Appearance of Oral Candida Infection and Therapeutic Strategies. Front Microbiol.17;6:1391. [CrossRef]
- Jayachandran AL et al., 2016. Oral Candidiasis among Cancer Patients Attending a Tertiary Care Hospital in Chennai, South India: An Evaluation of Clinicomycological Association and Antifungal Susceptibility Pattern. Can J Infect Dis Med Microbiol. 8758461. [CrossRef]
- Kitagaki H et al., 2002. Two homologous genes, DCW1 (YKL046c) and DFG5, are essential for cell growth and encode glycosylphosphatidylinositol (GPI)-anchored membrane proteins required for cell wall biogenesis in Saccharomyces cerevisiae. Mol Microbiol. 46(4):1011-22. [CrossRef]
- Kitagaki H et al., 2004. A temperature-sensitive dcw1 mutant of Saccharomyces cerevisiae is cell cycle arrested with small buds which have aberrant cell walls. Eukaryot Cell. 3(5):1297-306. [CrossRef]
- Maddi A et al., 2012. The Neurospora crassa dfg5 and dcw1 genes encode α-1,6-mannanases that function in the incorporation of glycoproteins into the cell wall. PLoS One. 7(6):e38872. [CrossRef]
- Spreghini E et al., 2003. Roles of Candida albicans Dfg5p and Dcw1p cell surface proteins in growth and hypha formation. Eukaryot Cell. 2(4):746-55. [CrossRef]
- Nasution O et al., 2015. Loss of Dfg5 glycosylphosphatidylinositol-anchored membrane protein confers enhanced heat tolerance in Saccharomyces cerevisiae. Environ Microbiol. 17(8):2721-34. [CrossRef]
- Atanasova L et al. The GPI-Anchored GH76 Protein Dfg5 Affects Hyphal Morphology and Osmoregulation in the Mycoparasite Trichoderma atroviride and Is Interconnected With MAPK Signaling. Front Microbiol. 2021 Feb 10;12:601113. [CrossRef]
- Ao J et al., 2015. The N-Linked Outer Chain Mannans and the Dfg5p and Dcw1p Endo-α-1,6-Mannanases Are Needed for Incorporation of Candida albicans Glycoproteins into the Cell Wall. Eukaryot Cell. 14(8):792-803. [CrossRef]
- Mancuso R et al., 2018. Functions of Candida albicans cell wall glycosidases Dfg5p and Dcw1p in biofilm formation and HOG MAPK pathway. PeerJ. 2018 6:e5685. [CrossRef]
- Heilmann, Clemens J., et al. 2013. "Surface stress induces a conserved cell wall stress response in the pathogenic fungus Candida albicans." Eukaryotic cell 12.2: 254-264. [CrossRef]
- Liesche J et al., 2015. Cell wall staining with Trypan blue enables quantitative analysis of morphological changes in yeast cells. Front. Microbiol. 6:107. [CrossRef]
- Walker LA et al., 2008. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog 4:e1000040. [CrossRef]
- Chinnici J et al., 2019. Candida albicans cell wall integrity transcription factors regulate polymicrobial biofilm formation with Streptococcus gordonii. PeerJ.11;7:e7870. [CrossRef]
- Finkel JS et al., 2012. Portrait of Candida albicans Adherence Regulators. Cowen LE, ed. PLoS Pathogens. 8(2):e1002525. [CrossRef]
- Solis NV & Filler SG, 2012. Mouse model of oropharyngeal candidiasis. Nat Protoc. 7(4):637-42. [CrossRef]
- Day AM et al., 2017. Stress-induced nuclear accumulation is dispensable for Hog1-dependent gene expression and virulence in a fungal pathogen. Sci Rep. 30;7(1):14340. [CrossRef]
- Chaffin WL, 2008. Candida albicans cell wall proteins. Microbiol Mol Biol Rev. 72(3):495-544. [CrossRef]
- Klis FM et al., 2006. Cell wall construction in Saccharomyces cerevisiae. Yeast. 23(3):185-202. [CrossRef]
- Yang F et al., 2013. Chromosome 5 monosomy of Candida albicans controls susceptibility to various toxic agents, including major antifungals. Antimicrob Agents Chemother. 57:5026 –5036. [CrossRef]
- Lee KK et al., 2012. Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob Agents Chemother 56: 208–217. [CrossRef]
- Munro CA et al., 1998. Regulation of chitin synthesis during dimorphic growth of Candida albicans. Microbiology. 144 ( Pt2):391-401. [CrossRef]
- Munro CA, Gow NA, 2001. Chitin synthesis in human pathogenic fungi. Med Mycol. 39 Suppl 1:41-53.
- Lenardon MD et al., 2007. Individual chitin synthase enzymes synthesize microfibrils of differing structure at specific locations in the Candida albicans cell wall. Mol Microbiol. 66(5):1164-1173. [CrossRef]
- Munro CA et al., 2007. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol Microbiol. 63(5):1399-413. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).