Submitted:
21 June 2024
Posted:
24 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
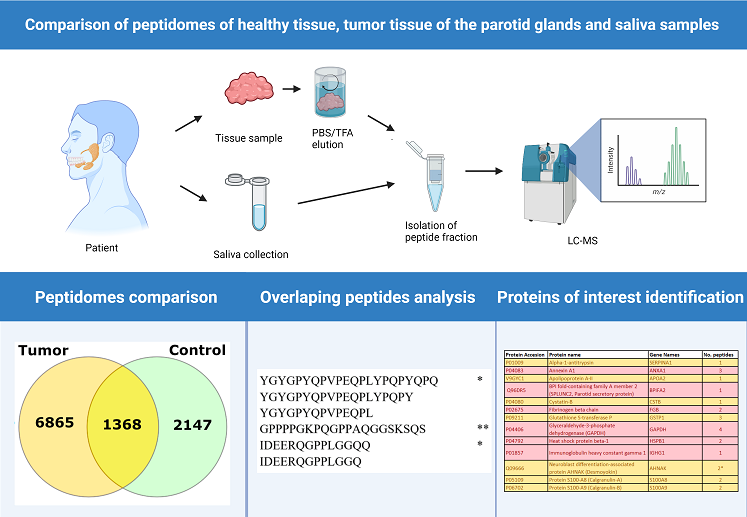
2.1. Comparison of Peptide Extraction Methods
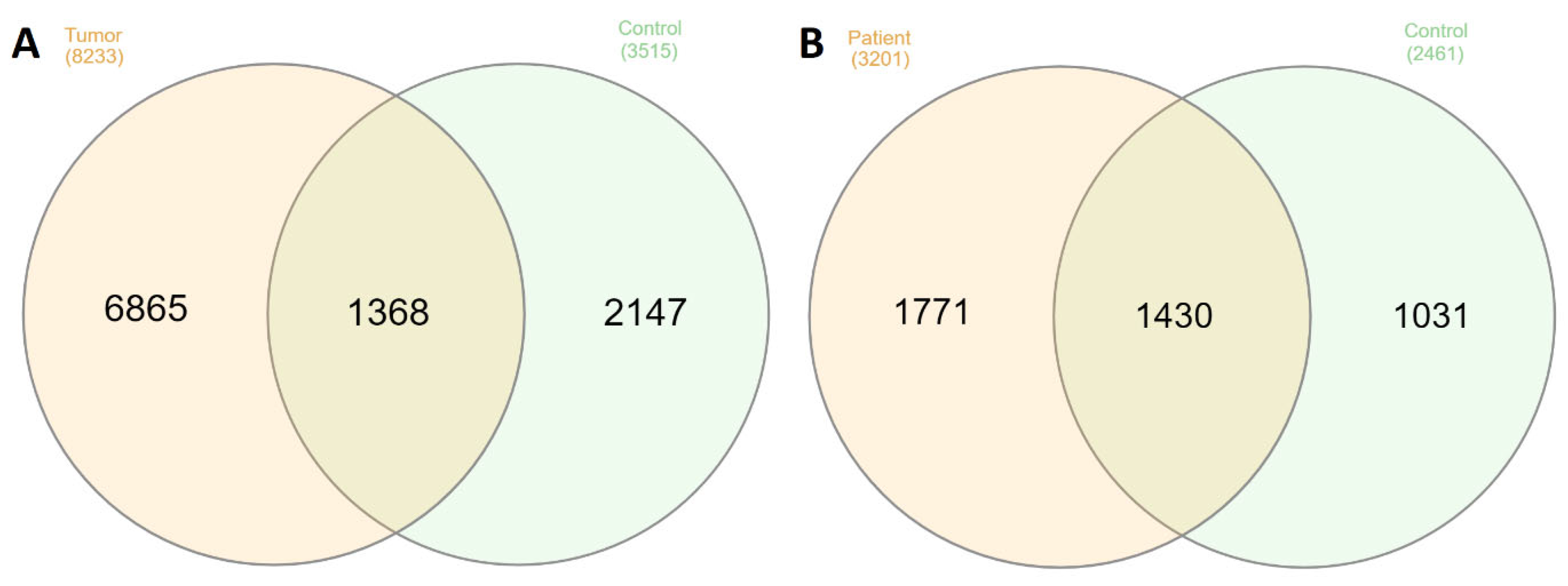
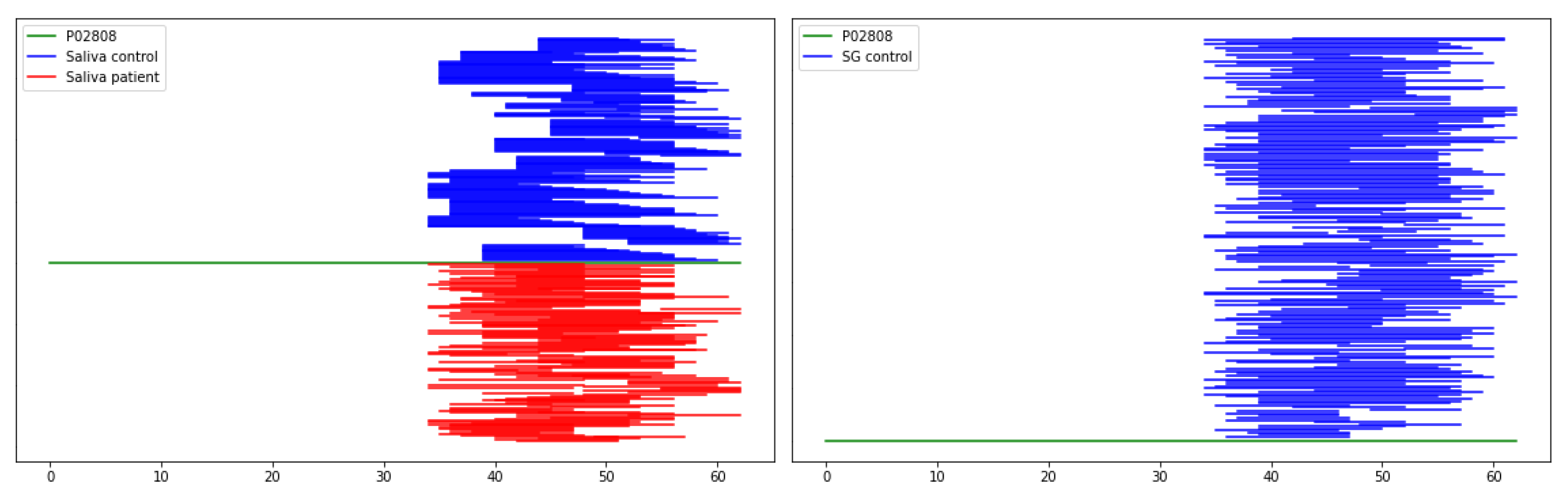
2.2. Comparison Peptidome Composition of Salivary Glands Extracts and Saliva
2.3. Bioinformatic Analysis
2.4. Characterization of Peptide Biomarkers of Salivary Gland Tumors
3. Conclusions
4. Materials and Methods
4.1. Salivary Glands Tissue Collection
4.2. Saliva Samples Collection
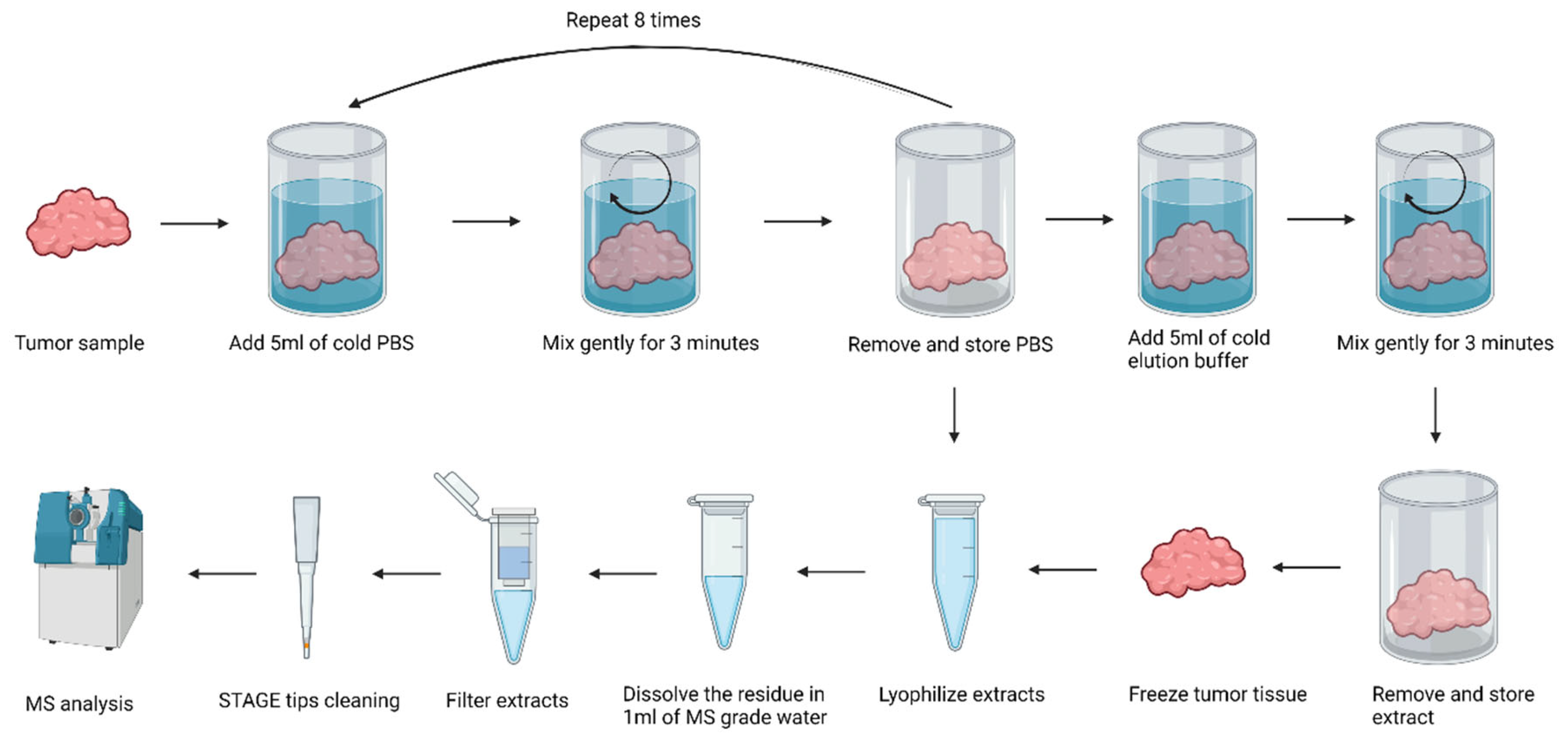
4.3. Isolation of Peptide Fractions from Salivary Gland Tissue
4.4. Peptide Isolation from Saliva
4.5. LC-MS/MS
4.6. Exploratory Data Analysis
4.7. Ethics Committee Approval and Personal Data Handling
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sood, S.; McGurk, M.; Vaz, F. Management of Salivary Gland Tumors: United Kingdom National Multidisciplinary Guidelines. J. Laryngol. Otol. 2016, 130, S142–S149. [Google Scholar] [CrossRef] [PubMed]
- Jegadeesh, N.; Liu, Y.; Prabhu, R.S.; Magliocca, K.R.; Marcus, D.M.; Higgins, K.A.; Vainshtein, J.M.; Trad Wadsworth, J.; Beitler, J.J. Outcomes and prognostic factors in modern era management of major salivary gland cancer. Oral Oncol. 2015, 51, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Mikaszewski, B.; Markiet, K.; Smugała, A.; Stodulski, D.; Szurowska, E.; Stankiewicz, C. Parotid malignancies are not homogenous in terms of their presentation on dynamic MRI - a preliminary report. Acta Otolaryngol. 2017, 137, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Tretiakow, D.; Mikaszewski, B.; Skorek, A. The role of fine-needle aspiration biopsy (FNAB) in the diagnostic management of parotid gland masses with emphasis on potential pitfalls. Eur. Arch. Otorhinolaryngol. 2020, 277, 2939–2940. [Google Scholar] [CrossRef] [PubMed]
- Lammek, K.; Tretiakow, D.; Skorek, A. The first bite syndrome after parotidectomy. Oral Oncol. 2021, 113, 105028. [Google Scholar] [CrossRef] [PubMed]
- Tretiakow, D.; Skorek, A. Regarding to functional outcomes after extracapsular dissection with partial facial nerve dissection for small and large parotid neoplasms. Am. J. Otolaryngol. 2021, 42, 102965. [Google Scholar] [CrossRef]
- Lombardi, D.; McGurk, M.; Vander Poorten, V.; Guzzo, M.; Accorona, R.; Rampinelli, V.; Nicolai, P. Surgical treatment of salivary malignant tumors. Oral Oncol. 2017, 65, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Stodulski, D.; Mikaszewski, B.; Stankiewicz, C. Signs and symptoms of parotid gland carcinoma and their prognostic value. Int. J. Oral Maxillofac. Surg. 2012, 41, 801–806. [Google Scholar] [CrossRef]
- Fonseca, F.P.; Sena Filho, M.; Altemani, A.; Speight, P.M.; Vargas, P.A. Molecular signature of salivary gland tumors: potential use as diagnostic and prognostic marker. J. Oral Pathol. Med. 2016, 45, 101–110. [Google Scholar] [CrossRef]
- Stenman, G. Fusion oncogenes in salivary gland tumors: molecular and clinical consequences. Head Neck Pathol. 2013, 7 (Suppl. S1), S12–9. [Google Scholar] [CrossRef] [PubMed]
- Lima-Souza, R.A. de; Scarini, J.F.; Lavareze, L.; Emerick, C.; Dos Santos, E.S.; Paes Leme, AF; Egal, ESA; Altemani, A. ; Mariano, F.V. Protein markers of primary salivary gland tumors: A systematic review of proteomic profiling studies. Arch. Oral Biol. 2022, 136, 105373. [Google Scholar] [CrossRef]
- Seccia, V.; Navari, E.; Donadio, E.; Boldrini, C.; Ciregia, F.; Ronci, M.; Aceto, A.; Dallan, I.; Lucacchini, A.; Casani, A.P.; et al. Proteomic Investigation of Malignant Major Salivary Gland Tumors. Head Neck Pathol. 2020, 14, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Donadio, E.; Giusti, L.; Seccia, V.; Ciregia, F.; Da Valle, Y.; Dallan, I.; Ventroni, T.; Giannaccini, G.; Sellari-Franceschini, S.; Lucacchini, A. New insight into benign tumors of major salivary glands by proteomic approach. PLoS One 2013, 8, e71874. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, C.M.; Jesus, S.F. de; Souza, M.G. de; Queiroz, L.D.R.P.; Santos, E.M.; Dos Santos, E.P.; Oliveira, L.P.; Cordeiro Santos, C.K.; Santos, S.H.S.; Paula, A.M.B. de; et al. High levels of ANXA2 are characteristic of malignant salivary gland tumors. J. Oral Pathol. Med. 2019, 48, 929–934. [Google Scholar] [CrossRef]
- Mutlu, A.; Ozturk, M.; Akpinar, G.; Kasap, M.; Kanli, A. Proteomics analysis of pleomorphic adenoma of the human parotid gland. Eur. Arch. Otorhinolaryngol. 2017, 274, 3183–3195. [Google Scholar] [CrossRef] [PubMed]
- Lyapina, I.; Ivanov, V.; Fesenko, I. Peptidome: Chaos or Inevitability. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Hellinger, R.; Sigurdsson, A.; Wu, W.; Romanova, E. V.; Li, L.; Sweedler, J. V.; Süssmuth, R. D.; Gruber, C. W. Peptidomics. Nat Rev Methods Primers 2023, 3, 25. [Google Scholar] [CrossRef]
- Latosinska, A.; Frantzi, M.; Siwy, J. Peptides as "better biomarkers"? Value, challenges, and potential solutions to facilitate implementation. Mass Spec Rev. 2023, 1–42. [Google Scholar] [CrossRef]
- Villanueva, J.; Shaffer, D.R.; Philip, J.; Chaparro, C.A.; Erdjument-Bromage, H.; Olshen, A.B.; Fleisher, M.; Lilja, H.; Brogi, E.; Boyd, J.; et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J. Clin. Invest. 2006, 116, 271–284. [Google Scholar] [CrossRef]
- Tanuwidjaya, E.; Schittenhelm, R.B.; Faridi, P. Soluble HLA peptidome: A new resource for cancer biomarkers. Front. Oncol. 2022, 12, 1069635. [Google Scholar] [CrossRef]
- Han, T.; Cong, H.; Yu, B.; Shen, Y. Application of peptide biomarkers in life analysis based on liquid chromatography–mass spectrometry technology. BioFactors. 2022, 48, 725–743. [Google Scholar] [CrossRef] [PubMed]
- Neves, L.X.; Granato, D.C.; Busso-Lopes, A.F.; Carnielli, C.M.; Patroni, F.M.d.S.; Rossi, T. de; Oliveira, A.K.; Ribeiro, A.C.P.; Brandão, T.B.; Rodrigues, A.N.; et al. Peptidomics-Driven Strategy Reveals Peptides and Predicted Proteases Associated With Oral Cancer Prognosis. Mol. Cell. Proteomics 2021, 20, 100004. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, F.P.; Macedo, C.C.S.; Dos Santos Costa, S.F.; Leme, AFP; Rodrigues, R. R.; Pontes, H.A.R.; Altemani, A.; van Heerden, WFP; Martins, M.D.; Almeida, O.P. de; et al. Mass spectrometry-based proteome profile may be useful to differentiate adenoid cystic carcinoma from polymorphous adenocarcinoma of salivary glands. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2019, 128, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Pereira, D.L.; Bingle, L.; Bingle, C.D.; Lopes, M.A.; Vargas, P.A. Expression of BPIF/PLUNC Protein Family and MECT1-MAML2 Fusion in Salivary Mucoepidermoid Carcinomas: a Preliminary Study. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018, 126, e158. [Google Scholar] [CrossRef]
- González-Arriagada, W.A.; Ramos, L.M.A.; Silva, A.A.; Vargas, P.A.; Della Coletta, R.; Bingle, L.; Lopes, M.A. Salivary BPIFA1 (SPLUNC1) and BPIFA2 (SPLUNC2 A) are modified by head and neck cancer radiotherapy. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2015, 119, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Kwon, C.H.; Park, H.J.; Choi, J.H.; Lee, J.R.; Kim, H.K.; Jo, H.-J.; Kim, H.S.; Oh, N.; Song, G. am; Park, D.Y. Snail and serpinA1 promote tumor progression and predict prognosis in colorectal cancer. Oncotarget 2015, 6, 20312–20326. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Hu, L.G. Serpin peptidase inhibitor clade A member 1-overexpression in gastric cancer promotes tumor progression in vitro and is associated with poor prognosis. Oncol. Lett. 2020, 20, 278. [Google Scholar] [CrossRef] [PubMed]
- El-Akawi, Z.J.; Al-Hindawi, F.K.; Bashir, N.A. Alpha-1 antitrypsin (alpha1-AT) plasma levels in lung, prostate and breast cancer patients. Neuro Endocrinol. Lett. 2008, 29, 482–484. [Google Scholar]
- Salem, H.; Ellakwa, D.E.-S.; Fouad, H.; Hamid, M.A. APOA1 AND APOA2 proteins as prognostic markers for early detection of urinary bladder cancer. Gene Reports 2019, 16, 100463. [Google Scholar] [CrossRef]
- Honda, K.; Srivastava, S. Potential usefulness of apolipoprotein A2 isoforms for screening and risk stratification of pancreatic cancer. Biomark. Med. 2016, 10, 1197–1207. [Google Scholar] [CrossRef]
- Feldman, A.S.; Banyard, J.; Wu, C.-L.; McDougal, W.S. ; Zetter, BR Cystatin B as a tissue and urinary biomarker of bladder cancer recurrence and disease progression. Clin. Cancer Res. 2009, 15, 1024–1031. [Google Scholar] [CrossRef]
- Wang, X.; Gui, L.; Zhang, Y.; Zhang, J.; Shi, J.; Xu, G. Cystatin B is a progression marker of human epithelial ovarian tumors mediated by the TGF-β signaling pathway. Int. J. Oncol. 2014, 44, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, G.; Yin, J.; Li, L.; Tan, Y.; Wei, H.; Liu, B.; Deng, L.; Tang, J.; Chen, Y.; et al. GSTP1 and cancer: Expression, methylation, polymorphisms and signaling (Review). Int. J. Oncol. 2020, 56, 867–878. [Google Scholar] [CrossRef]
- Altenberg, B.; Greulich, K.O. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 2004, 84, 1014–1020. [Google Scholar] [CrossRef]
- Lee, H.; Kim, K.; Woo, J.; Park, J.; Kim, H.; Lee, K.E.; Kim, H.; Kim, Y.; Moon, K.C.; Kim, J.Y.; et al. Quantitative Proteomic Analysis Identifies AHNAK (Neuroblast Differentiation-associated Protein AHNAK) as a Novel Candidate Biomarker for Bladder Urothelial Carcinoma Diagnosis by Liquid-based Cytology. Mol. Cell. Proteomics 2018, 17, 1788–1802. [Google Scholar] [CrossRef] [PubMed]
- Sudo, H.; Tsuji, A.B.; Sugyo, A.; Abe, M.; Hino, O.; Saga, T. AHNAK is highly expressed and plays a key role in cell migration and invasion in mesothelioma. Int. J. Oncol. 2014, 44, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, C.; Németh, J.; Angel, P.; Hess, J. S100A8 and S100A9 in inflammation and cancer. Biochem. Pharmacol. 2006, 72, 1622–1631. [Google Scholar] [CrossRef]
- Nayak, S.; Bhatt, M.L.B.; Goel, MM; Gupta, S. ; Mahdi, A.A.; Mishra, A.; Mehrotra, D. Tissue and serum expression of TGM-3 may be prognostic marker in patients of oral squamous cell carcinoma undergoing chemo-radiotherapy. PLoS One 2018, 13, e0199665. [Google Scholar] [CrossRef]
- Uemura, N.; Nakanishi, Y.; Kato, H.; Saito, S.; Nagino, M.; Hirohashi, S.; Kondo, T. Transglutaminase 3 as a prognostic biomarker in esophageal cancer revealed by proteomics. Int. J. Cancer 2009, 124, 2106–2115. [Google Scholar] [CrossRef]
- Zhang, Y.; Feurino, L.W.; Zhai, Q.; Wang, H.; Fisher, W.E.; Chen, C.; Yao, Q.; Li, M. Thymosin Beta 4 is overexpressed in human pancreatic cancer cells and stimulates proinflammatory cytokine secretion and JNK activation. Cancer Biol. Ther. 2008, 7, 419–423. [Google Scholar] [CrossRef]
- Gemoll, T.; Strohkamp, S.; Schillo, K.; Thorns, C.; Habermann, J.K. MALDI-imaging reveals thymosin beta-4 as an independent prognostic marker for colorectal cancer. Oncotarget 2015, 6, 43869–43880. [Google Scholar] [CrossRef] [PubMed]
- Tamesa, M.S.; Kuramitsu, Y.; Fujimoto, M.; Maeda, N.; Nagashima, Y.; Tanaka, T.; Yamamoto, S.; Oka, M.; Nakamura, K. Detection of autoantibodies against cyclophilin A and triosephosphate isomerase in sera from breast cancer patients by proteomic analysis. Electrophoresis 2009, 30, 2168–2181. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Huang, Z.; Tian, Y.; Lin, B.; He, R.; Wang, H.; Ouyang, P.; Chen, H.; Wu, L. Clinical significance and prognostic value of Triosephosphate isomerase expression in gastric cancer. Medicine (Baltimore) 2017, 96, e6865. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W. The PeptideAtlas Project. In: Hubbard, S.; Jones, A. (eds) Proteome Bioinformatics. Methods in Molecular Biology™, 2010, vol 604, Humana Press. [CrossRef]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; Asplund, A.; Sjöstedt, E.; Lundberg, E.; Szigyarto, C. A.; Skogs, M.; Takanen, J. O.; Berling, H.; Tegel, H.; Mulder, J.; Nilsson, P.; Schwenk, J. M.; Lindskog, C.; Danielsson, F.; Mardinoglu, A.; Sivertsson, A.; von Feilitzen, K.; Forsberg, M.; Zwahlen, M.; Olsson, I.; Navani, S.; Huss, M.; Nielsen, J.; Ponten, F.; Uhlén, M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Storkus, W.J. , Zeh H.J., Salter R.D., Lotze M.T. Identification of T-Cell epitopes: Rapid isolation of class i-presented peptides from viable cells by mild acid elution. J. Immunother. 1993, 14, 94–103. [Google Scholar] [CrossRef]
- Purcell, A.W.; Ramarathinam, S.H.; Ternette, N. Mass spectrometry based identification of MHC-bound peptides for immunopeptidomics. Nature Protocols, 2019; 14, 1687–170. [Google Scholar] [CrossRef]
- Brunet, M.A.; Lucier, J.-F.; Levesque, M.; Leblanc, S.; Jacques, J.-F.; Al-Saedi, H.R.H.; Guilloy, N.; Grenier, F.; Avino, M.; Fournier, I.; et al. OpenProt 2021: deeper functional annotation of the coding potential of eukaryotic genomes. Nucleic Acids Res. 2021, 49, D380–D388. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: the Universal Protein Knowledgebase in 2023, Nucleic Acids Research, Volume 51, Issue D1, 6 January 2023, Pages D523–D531. [CrossRef]
- Newfound Coding Potential of Transcripts Unveils Missing Members of Human Protein Communities. Leblanc S, Brunet MA, Jacques JF, Lekehal AM, Duclos A, Tremblay A, Bruggeman-Gascon A, Samandi S, Brunelle M, Cohen AA, Scott MS, Roucou X. Genomics Proteomics Bioinformatics. 2022 Sep 29:S1672-0229(22)00124-3. [CrossRef]
- Wang, ET; Sandberg, R; Luo, S; Khrebtukova, I; Zhang, L; Mayr, C; Kingsmore, SF;, Schroth GP; Burge CB Alternative Isoform Regulation in Human Tissue Transcriptomes Nature. 2008 November 27; 456(7221): 470–476. [CrossRef]
- The Landscape of Isoform Switches in Human Cancers Kristoffer Vitting-Seerup and Albin Sandelin. Mol Cancer Res; 15(9) September 2017.
- Rappsilber, J.; Ishihama, Y.; Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003, 75, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Nobilis, M.; Pour, M.; Kuneš, J.; Kopecký, J.; Květina, J.; Svoboda, Z.; Sládková, K.; Vortel, J. High-performance liquid chromatographic determination of ursodeoxycholic acid after solid phase extraction of blood serum and detection-oriented derivatization. Journal of Pharmaceutical and Biomedical Analysis 2001, 24, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, A.E.; Macur, K.; Czaplewska, P.; Liss, J.; Łukaszuk, K.; Ołdziej, S. Qualitative and Quantitative Analysis of Proteome and Peptidome of Human Follicular Fluid Using Multiple Samples from Single Donor with LC-MS and SWATH Methodology. J. Proteome Res. 2017, 16, 3053–3067. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.A.; Ma, B. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics 2012, 11, M111–010587. [Google Scholar] [CrossRef]
- Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos, D. ,.; Hewapathirana, S.,.; Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M, Walzer M, Wang S, Brazma A, Vizcaíno, J.A. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
- van Rossum, G.; Drake, F.L. Python 3 Reference Manual; CreateSpace: Scotts Valley, CA, 2009; ISBN 1441412697. [Google Scholar]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference. Python in Science Conference, Austin, Texas, 2010, 28 June–3 July 2010; SciPy; pp. 56–61. [Google Scholar]
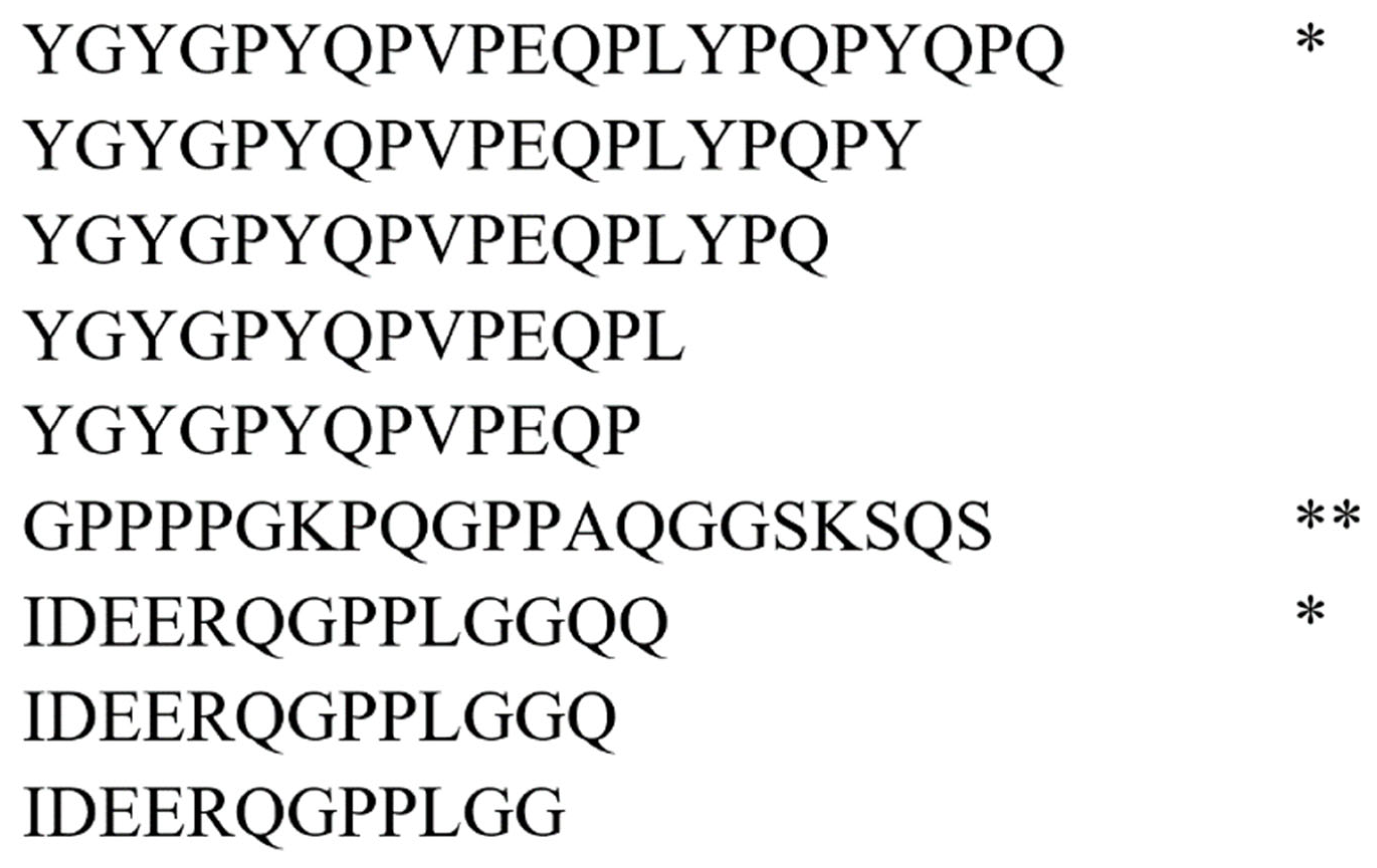
| Salivary glands tissue samples | |||||
| Fresh | Frozen | Total | |||
| PBS | TFA | PBS | TFA | ||
| Tumor | 5 | 4 | 5 | 4 | 18 |
| Control | 5 | 4 | 5 | 4 | 18 |
| Total | 10 | 8 | 10 | 8 | 36 |
| Saliva samples | |||||
| Patient | Control | Total | |||
| 11 | 8 | 19 | |||
| Data sets | Peptides | Unique peptides | Stand-alone peptides |
|---|---|---|---|
| SG tumor | 8233 | 3916 | 1733 |
| SG control | 3515 | 1586 | 589 |
| SG fresh | 6752 | 3205 | 1383 |
| SG frozen | 6913 | 3212 | 1496 |
| SG PBS | 7771 | 3649 | 1667 |
| SG TFA | 3540 | 1770 | 702 |
| SG All | 10380 | 4869 | 2077 |
| Saliva patient | 3201 | 1166 | 325 |
| Saliva control | 2461 | 836 | 258 |
| Saliva all | 4189 | 1490 | 420 |
| Datasets | Overlapping sequences | Common peptides |
|---|---|---|
| Saliva patient - SG tumor | 377 | 86 |
| Saliva patient only - SG tumor only | 109 | 16 |
| Saliva patient - SG control | 960 | 182 |
| Saliva patient only - SG control only | 362 | 42 |
| Saliva control - SG tumor | 425 | 121 |
| Saliva control only - SG tumor only | 154 | 39 |
| Saliva control - SG control | 950 | 209 |
| Saliva control only - SG control only | 351 | 58 |
| Protein Accession | Protein name | Gene Names | No. peptides |
|---|---|---|---|
| P63261, P60709 | Actin, cytoplasmic | ACTB, ACTG | 9 |
| P01009 | Alpha-1-antitrypsin | SERPINA1 | 1 |
| P04083 | Annexin A1 | ANXA1 | 3 |
| V9GYC1 | Apolipoprotein A-II | APOA2 | 1 |
| P04280, P02812, Q04118, P10163 | Basic salivary proline-rich proteins | PRB1/4 | 32 |
| Q96DR5 | BPI fold-containing family A member 2 (SPLUNC2, Parotid secretory protein) | BPIFA2 | 1 |
| P04080 | Cystatin-B | CSTB | 1 |
| P02671 | Fibrinogen alpha chain | FGA | 2 |
| P02675 | Fibrinogen beta chain | FGB | 2 |
| P09211 | Glutathione S-transferase P | GSTP1 | 3 |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | GAPDH | 4 |
| P04792 | Heat shock protein beta-1 | HSPB1 | 2 |
| P69905, P68871 | Haemoglobin | HBA1/HBA2, HBB | 5 |
| P16402, P10412, P16401, P20671 | Histones | H1-3, H1-4, H1-5, H2AC7 | 9 |
| P01857 | Immunoglobulin heavy constant gamma 1 | IGHG1 | 1 |
| P04264, P13647 | Keratin | KRT1 , KRT5 | 4 |
| Q09666 | Neuroblast differentiation-associated protein AHNAK (Desmoyokin) | AHNAK | 2* |
| P05109 | Protein S100-A8 (Calgranulin-A) | S100A8 | 2 |
| P06702 | Protein S100-A9 (Calgranulin-B) | S100A9 | 2 |
| Q08188 | Protein-glutamine gamma-glutamyltransferase E | TGM3 | 1 |
| P02810 | Salivary acidic proline-rich phosphoprotein 1/2 | PRH1; PRH2 | 17 |
| P62328 | Thymosin beta-4 | TMSB4X | 2(1**) |
| P60174 | Triosephosphate isomerase | TPI1 | 2 |
| P08670 | Vimentin | VIM | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





