1. Introduction
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was identified as the cause of the coronavirus disease 2019 (COVID-19) in the year 2019 [
1,
2]. Evidence has demonstrated that different variants of SARS-CoV-2 have been circulating globally [
3,
4]. These variants have rapidly undergone mutations, making treatment and vaccination goals difficult to attain and potentially threatening further waves of COVID-19 [
5,
6,
7,
8,
9]. Therefore, there is a need for continuous development of newer and improved diagnostic methods, novel vaccines, and drugs to address COVID-19 [
10].
Since its emergence and spread, many strategies have been employed to improve the surveillance of COVID-19 [
11,
12,
13,
14,
15]. Wastewater surveillance has emerged as a way to monitor the presence of SARS-CoV-2 in communities as the virus is shed in wastewater [
16,
17,
18,
19]. This method can help identify SARS-CoV-2 shed in sewage and other wastewater types and could help to understand communities with a high prevalence of symptomatic or asymptomatic COVID-19 [
20,
21]. Additionally, detecting SARS-CoV-2 ribonucleic acid (RNA) particles in wastewater may indicate emerging COVID-19 clusters in a particular community [
22,
23]. Wastewater surveillance for SARS-CoV-2 involves monitoring wastewater for the presence of the virus to track its spread in a community using polymerase chain reaction and genomic sequencing detection methods [
22,
24,
25,
26]. This approach can provide insights into circulating variants of SARS-CoV-2, act as an early warning system for COVID-19 outbreaks, and help identify areas where the virus is spreading [
24,
27,
28]. Wastewater-based surveillance should be employed as a complement to existing disease surveillance systems such as clinical surveillance [
29,
30,
31]. In clinical surveillance, there is a reliance on symptomatic patients which precludes asymptomatic individuals, thereby underestimating the magnitude of the disease [
32].
During the early stage of the pandemic, a study in the Netherlands was able to isolate SARS-CoV-2 in wastewater close to a week before the first case of COVID-19 providing a proof of concept for the isolation of SARS-CoV-2 in wastewater and as an early indication of clinical onset in a population [
33]. Furthermore, studies in Australia, the Czech Republic, and Italy demonstrated the presence of SARS-CoV-2 in wastewater obtained from water treatment plants providing further credence to using wastewater samples for COVID-19 surveillance [
34,
35,
36]. Another study in the USA found that wastewater surveillance was effective in detecting SARS-CoV-2 and contributed to the diagnosis of more than 85% of COVID-19 cases [
37]. In Pakistan, a study found that 81.5% of SARS-CoV-2 was detected in wastewater [
22]. This evidence shows that the presence of SARS-CoV-2 RNA in wastewater samples can be used as an early warning system for COVID-19 in communities [
38,
39].
In Africa, there are limited SARS-CoV-2 wastewater surveillance activities reported to be important in addressing COVID-19 [
40]. A study conducted in South Africa detected SARS-CoV-2 in wastewater including the Beta, Delta, and Omicron variants [
32]. In Ghana, the presence of SARS-CoV-2 RNA in wastewater was detected in 17% of the samples tested for the presence of viral RNA [
41]. These studies confirmed that SARS-CoV-2 wastewater surveillance could contribute to an improved understanding of SARS-CoV-2 epidemiology and transmission dynamics and contribute to early detection, prevention, and inform intervention strategies to address disease problems [
24,
29,
32].
Zambia is a low- and—middle-income country (LMIC) located in the southern part of the sub-Saharan African region. The first case of COVID-19 in Zambia was reported on March 18, 2020 [
42,
43]. Since the first report, the identification of SARS-CoV-2 using molecular methods has been reported in subsequent studies [
44,
45]. A recent study conducted in Lusaka, Zambia, detected adenoviruses and rotaviruses in wastewater [
46]. However, there are no published studies that have reported on the detection and identification of SARS-CoV-2 in wastewater systems in Zambia. Given the decline in COVID-19 cases and the paucity of information regarding the current circulation of SARS-CoV-2, this study evaluated wastewater surveillance as a potential tool for monitoring SARS-CoV-2 circulation in Zambia.
2. Materials and Methods
2.1. Study Design and Sites
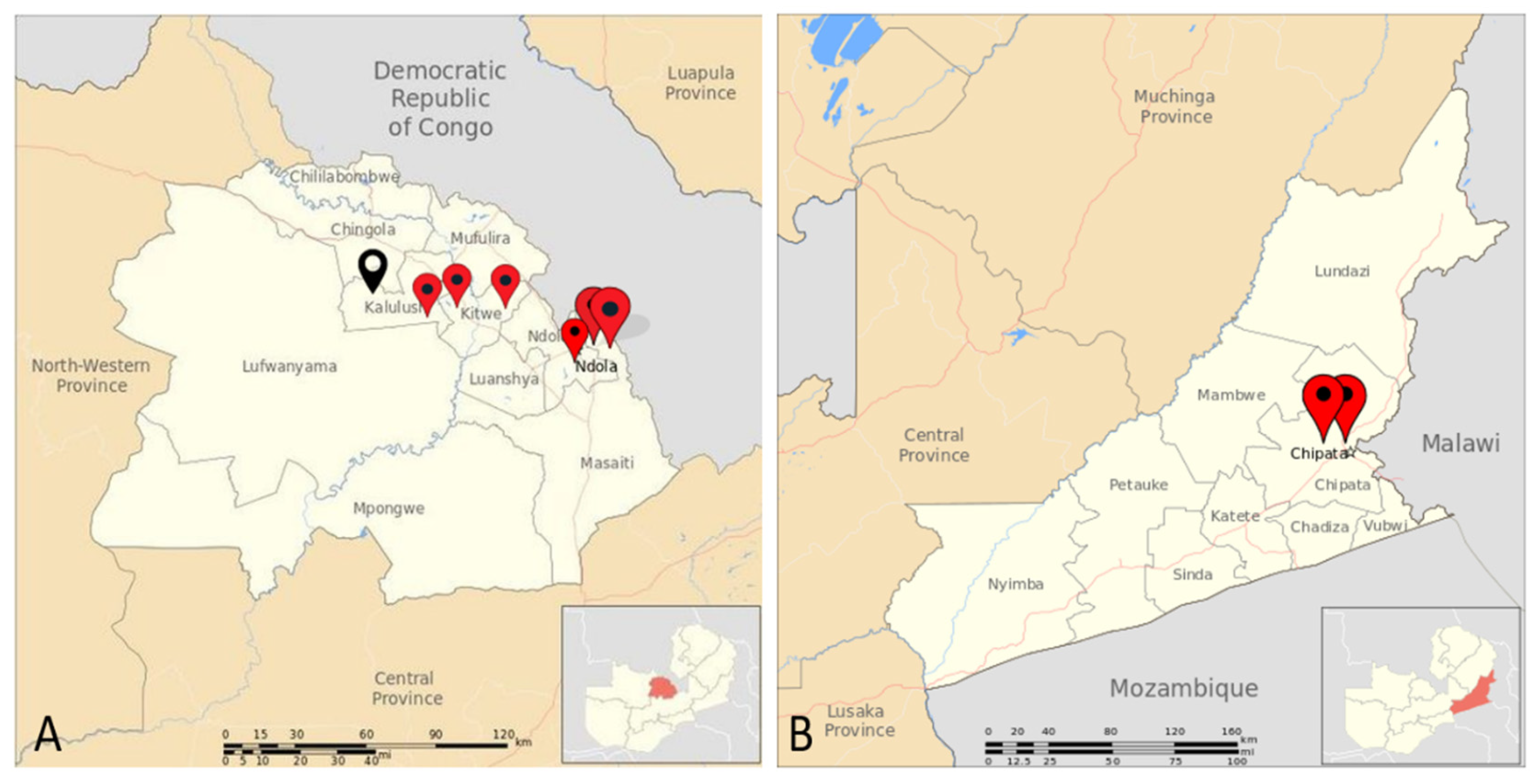
This was a prospective longitudinal study that collected samples between October 2023 and December 2023 from the Copperbelt and Eastern provinces in Zambia. The study sites included Ndola district (Old Kanini, New Kanini and Lubuto), Kitwe (Nkana East, Mindolo, Chambishi, and Kalulushi) (
Figure 1A) and Chipata (Chipata Motel and Chipata ponds) (
Figure 1B). A total of nine weekly collections were done for Ndola and Kitwe while seven collections were conducted for Chipata. The sampling was conducted at influent points of sewer ponds and treatment plants that service selected parts of the districts.
2.2. Sample Collection
A composite-grab sample of one litre was collected from each site; with the aid of a 2-litre bucket as described previously [
23]. A bag-mediated filtration system was also used to collect samples from each township. Sample collection was done following strict safety procedures safeguarding the sample and study staff. Collected samples were immediately stored at 4 to 8 °C using cool boxes with controlled and monitored temperatures and mobile vehicle refrigerators during transportation. Samples were processed immediately as soon as they were received at the Churches Health Association of Zambia (CHAZ) laboratory in Lusaka.
2.3. Viral Concentration
The wastewater samples were subjected to three (3) viral concentration methods before nucleic acid extraction. These included the skimmed milk flocculation, bag-mediated filtration system and polythene glycol-based (PEG) concentration method.
2.3.1. Skimmed Milk Flocculation
A composite wastewater sample (500 mL) was used for the direct skimmed milk flocculation method. Briefly, 0.5g of skimmed milk powder was dissolved in 50 mL of sterile water to obtain 1% (w/v) skimmed milk solution [
47]. The
pH of the solution was carefully adjusted to 3.5 using 1M HCl. We added 5 mL of a 1% skimmed milk solution to 500 mL of the wastewater composite sample to attain a final concentration of 0.01% (w/v). Samples were stirred for 8 hours, and flocs were allowed to settle at room temperature for another 8 hours. The supernatant was carefully removed using serological pipettes without agitating the flocs. A final volume of 50 mL containing the flocs was transferred to 50 mL falcon tubes and centrifuged at 8,000 × g for 30 min. The supernatant was carefully removed, and the pellets were carefully dislodged. The pellets were resuspended in 2mL of 0.2 M phosphate buffer saline (PBS),
pH 7.5. The final concentrated sample was aliquoted in small working volumes (~200 μL) and stored at -80 °C until viral nucleic acid extraction.
2.3.2. Bag-Mediated Filtration System (BMFS)
Samples were collected into the BMFS bag and processed as described previously [
48]. Briefly, wastewater (6,000 mL) from the composite sample was collected in sampling bags with a pre-screen mesh (249-μm pore size) over the opening. The composite sample was filtered into the collection bag mounted on a custom-made tripod stand (BoundaryTEC, Minneapolis, MN, USA). A ViroCap
TM filter (Scientific Methods, Inc., Granger, IN, USA) was attached to the bag’s outlet port and the sample was allowed to flow through the filter by gravity. The average volume filtered was about 5,400 mL, over an average of 90 min to 3 hours (the time for filtration was dependent on the amount and size of faecal debris in the wastewater). The filter was transported under a cold chain to the CHAZ laboratory for viral elution and secondary concentration with skimmed milk. To elute the virus from the viral cup filter, the beef extract eluate was injected into the filter inlet and incubated for 30 min before recovering the eluate through the filter outlet using a peristaltic pump.
2.3.2.1. Secondary Concentration Procedure
A total of three (3) mL of 1% w/v skimmed milk was added to the eluate. The pH of the mixture was adjusted to the range of 3.0-4.0 using sodium hydroxide (NaOH) and hydrochloric acid (HCl). To coagulate the proteins in flocs, the eluate was incubated at room temperature and shaken evenly for 2 hours. The sample was aliquoted into 50 mL conical tubes and centrifuged at 3,500 x g, for 30 min at a temperature of 4°C, and thereafter returned to the biosafety cabinet (BSC) where the supernatant was decanted. The pellets were completely resuspended in 10 mL of sterile PBS, pH 7.4 by vigorous vortexing. The concentrated sample was aliquoted in 1 mL working volumes and stored at -80 °C until use.
2.3.3. Polyethylene Glycol-Based (PEG) Concentration
The PEG precipitation was performed as previously described [
49]. Briefly, 14 g of PEG 8000 and 1.17g of NaCl were added to 100 mL of the composite sample. The PEG 8000 and NaCl were completely dissolved by vigorous shaking. The sample was stirred at 200 rpm, for 4 hours at 4°C followed by centrifugation at 6,500 x g, for 30 min at 4°C. The supernatant was discarded and the pellet was re-suspended in 6 mL of sterile PBS (
pH 7.4). The final concentrated sample was aliquoted in small working volumes (~200 μL) and stored at -80°C until use.
2.4. Nucleic Acid Extraction
Viral RNA was extracted from the 155 wastewater concentrated samples using the MagMAX viral isolation kit (Applied Biosystems, Foster City, CA) on an automated Kingfisher Flex 96 Deep-well magnetic particle processor (Thermo-Fisher Scientific, USA) according to the manufacturer’s recommendation. Briefly, in a class II biosafety cabinet, 200 µL of wastewater concentrated sample was added to 265 µL of lysis/binding solution. 20 µL of bead mix was then added to the sample-lysis buffer mix. Following the removal of the supernatant, samples were washed twice with wash buffer and RNA was eluted in 50 µL of elution buffer and stored at -80 °C.
2.5. SARS-CoV-2 Detection by RT-PCR
To confirm the presence of SARS-CoV-2 nucleic acid in wastewater RNA samples, the TaqPath COVID-19 CE-IVD RT-qPCR master mix (Thermo-Fisher Scientific, Waltham, USA) was used. The 25µL reaction mix contained 15µL of TaqPath qPCR master mix and 10µL of extracted RNA. The TaqPath COVID-19 CE-IVD RT-qPCR kit mix has negative, positive, and internal controls to monitor the reliability of the results from RNA extraction to PCR amplification. As per protocol, the internal control with a Ct ≤37 was considered positive for SARS-CoV-2. Additionally, N, ORF1ab, and S genes with Ct≤37 were considered positive for SARS-CoV-2. In this study, the Ct cutoff of ≤37 was considered positive while samples with no amplification (Ct = 0) for any of the three targets (N, ORF1ab) excluding the S gene (due to mispriming or primer hybridization failure due to mutations) were considered as not valid. The reporter dye and detector dye were set on the ABI 7500 Fast Real-Time PCR system (ThermoFisher, CA, USA) as follows; FAM: ORF1ab, VIC: N gene, ABY: S gene, JUN: MS2. A positive and no template control was added to each run. The samples were incubated for 2 minutes at 25 °C, 10 minutes at 53 °C, and 2 minutes at 95 °C followed by amplification of 40 cycles consisting of 3 seconds at 95 °C and 30 seconds at 60 °C. Fluorescence detection was set during the annealing/extension step (30 seconds at 60 °C). All samples that tested positive for SARS-CoV-2 positive samples were considered for whole genome sequencing.
2.6. Genomic Sequencing
2.6.1. cDNA Synthesis and Amplification of SARS-CoV-2
Random hexamer priming using the first strand cDNA master mix (Illumina) achieved first-strand cDNA synthesis according to the manufacturer’s recommended protocol. In a 96-well PCR plate, 8.5 µL of random hexamers were added to 8.5 µL of extracted RNA and denatured on an ABI 7500 real-time PCR for 3 minutes at 65 °C and then incubated at 4 °C. Ten microliters of First Strand Mix and 1 µL of Reverse Transcriptase were then added to the denatured sample. cDNA synthesis was achieved with the following conditions: 5 minutes at 25 °C, 10 minutes at 50 °C, and 5 minutes at 80 °C. SARS-CoV-2 genome amplification was conducted using ARCTIC network V4 primer pools (
https://github.com/artic-network/primer-schemes; provided by Illumina). ARCTIC V4 primer pool amplification employed two reactions per sample i.e., COVIDseq Primer Pool 1 (CPP1) and COVIDseq Primer Pool 2 (CPP2). The reaction components for each reaction consisted of 12.5µL Illumina PCR Master Mix, 3.5 µL of either CPP1 or CPP, 5 µL of first strand cDNA synthesis and 3.9 µL of nuclease-free water. Thermoprofile were as follows; 1 cycle of 98°C for 3 minutes, followed by 35 cycles of 98°C for 15 s and 63°C for 5 min. For each run, a single positive control (TaqPath COVID-19 Control; ThermoFisher, CA, USA) and negative no template control (nuclease-free water) were included to serve as indicators of extraneous nucleic acid contamination. PCR amplicons for each sample were then combined by transferring 10 µL from each well of the CPP1 and CPP2 into a new well.
2.6.2. Library Preparation and Illumina Sequencing
Library preparation was performed using the Illumina COVIDseq kit (Illumina Inc.) on the automated liquid robotic handler instrument. (Hamilton, NV, USA). Pooled PCR products were processed for tagmentation and adapter ligation using the Illumina COVIDseq Kit with IDT Illumina-PCR indexes. Pooling and library clean-up were performed as per the protocol provided by the manufacturer (Illumina Inc.). Pooled libraries were quantified using the Qubit 4.0 fluorometer (Invitrogen Inc.) using the Qubit dsDNA High Sensitivity kit. The pooled library was normalized to a 4 nM concentration. The normalized library (4 nM) was denatured and neutralized using 0.2N sodium hydroxide and 200 mM Tris-HCL (pH7), respectively. The library was further diluted to a final loading concentration of 0.5 nM and sequenced (301 paired-end) on the Illumina NextSeq 2000 (Illumina, SanDiego, CA, USA) platform.
2.6.3. Genome Assembly and Annotation
To assemble SARS-CoV-2 whole genomes, the Illumina DRAGEN DNA pipeline was used to analyze the sequence reads prepared. The DRAGEN pipeline uses a Kmer reference database to match kmers from the sequencing read to kmers from the SARS-CoV-2 reference genome (Wuhan Hu-1, accession no. NC_045512). The DRAGEN BCL converter was used to generate the FASTQ files which were analyzed using a SARS-CoV-2 specific wastewater sequence data analysis tool called Freyja, available on a cloud-native platform, Terra Bio (Version 1.3.0 [Freyja] Retrieved May 14 2024 from
https://app.terra.bio/#workspaces/aphl-personal/aphl-zambia_wwbs_workstation3/ Freyja).
2.7. Data Analysis
Demographic data were entered into Microsoft Excel version 2013, cleaned then exported to Stata version 14 (Stata Corp, College Station TX) for statistical analysis. The prevalence of SARS-CoV-2 in wastewater was presented as proportions and their 95% confidence intervals. Data were summarized and visualized using the R package dplyr v1.0.7 (Rstudio, Boston, MA, USA).
2.8. Phylogenetic Analysis
For phylogenetic analysis, AudacityInstant, accessed through GISAID was initially used to retrieve SARS-CoV-2 genome sequences from GISAID that closely resembled the sequences generated in this study. These sequences, along with the sequences obtained in this study, underwent multiple sequence alignment using Clustal Omega [
50]. Subsequently, the aligned sequences were trimmed to a length of 3495 bp using a custom Python script. A maximum likelihood phylogenetic tree was then constructed using the Kimura-3-parameter model with unequal base frequencies, estimation of base frequencies, and estimation of the proportion of invariable sites (K3Pu+F+I), as determined by the model test integrated into IQ-TREE (
http://www.iqtree.org/). The phylogenetic tree construction in IQ-TREE included 1000 bootstrap replicates to ascertain genetic tree reliability. The resulting maximum likelihood tree was rooted using TempEst v1.5.3 (
http://tree.bio.ed.ac.uk/software/tempest/), which estimated the best-fitting root of the phylogeny by minimizing the variance of root-to-tip distances using the heuristic residual mean squared function. Finally, the ML tree file underwent editing using the Interactive Tree of Life (iTOL) v5 (
https://itol.embl.de/), an online tool designed for the display and annotation of phylogenetic trees.
2.9. Collection of Clinical Data for COVID-19
Country data for COVID-19 clinical cases was reviewed from Worldometer (Zambia COVID—Coronavirus Statistics—Worldometer, worldometers.info. Accessed on 17th May 2024) as well as from the districts covering the study sites for the period October 2023 to December 2023. Sequencing data for clinical samples during the study period was obtained from GISAID (GISAID Initiative epicov.org. Accessed on 17th May 2024) where 17 sequences were downloaded.
3. Results
3.1. Detection of SARS-CoV-2
A total of nine collections were conducted for the Copperbelt Province and seven for Eastern Province from October 2023 up to December 2023. A total of 155 samples were processed in the laboratory for SARS-CoV-2 nucleic acid detection. Sample collection per site is summarized in
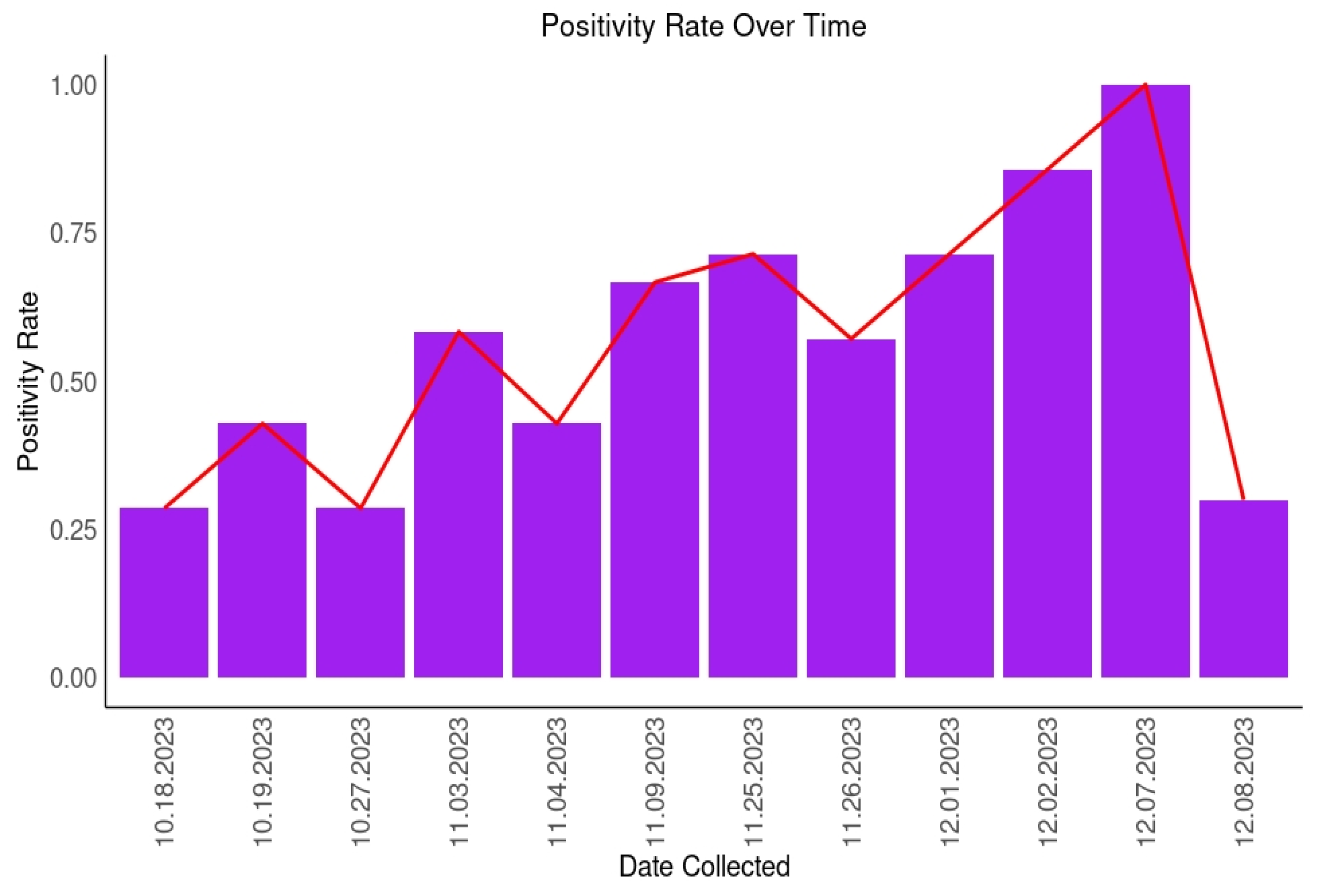
Table 1. Sixty-two (62) samples tested positive for SARS-CoV-2 representing an overall positivity rate of 40% of the study period. Of the positives, the RT PCR Ct value average was 33, while the lowest Ct value was 30.1.
This study found that there was a general trend towards increasing in positivity of wastewater samples for SARS-CoV-2 over the 9-week study time between October and December 2023 (
Figure 2).
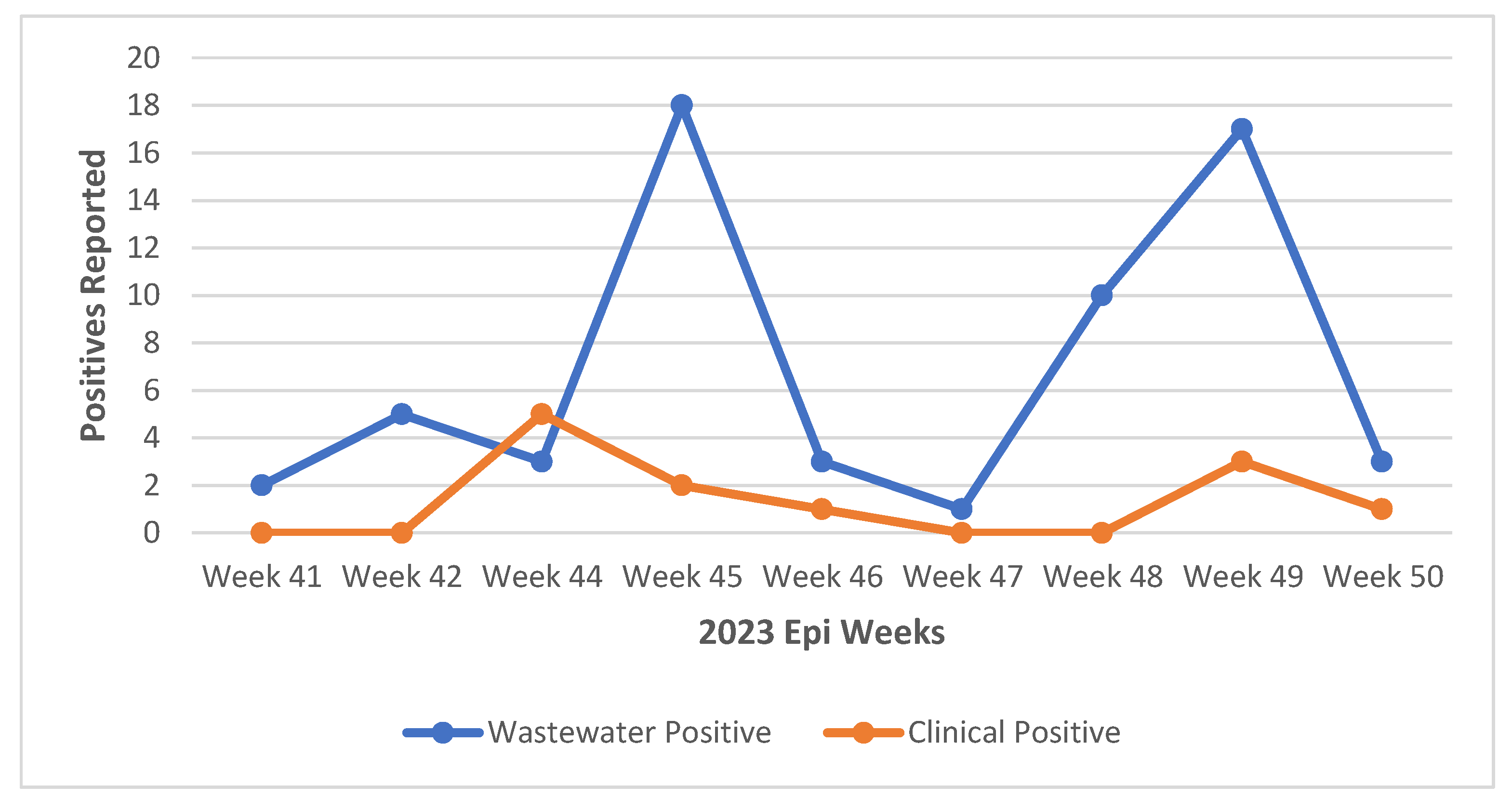
During the study period, aggregated data from clinical testing for SARS-CoV-2 showed no positive cases in the initial two intervals of wastewater sample collection. Positive clinical cases were recorded only in epidemiological weeks 44, 45, and 46. Clinical cases of SARS-CoV-2 were reported in both Copperbelt and Eastern provinces during epidemiological weeks 49 and 50. When we compared the wastewater sampling results to clinical cases reported in the respective catchment populations, we noted a potential consistency in an increase in positivity around epidemiological week 44 and week 49, similarly to that observed in the wastewater samples (
Figure 3).
3.2. SARS-COV-2 Sequencing of Wastewater Samples
From all the 62 SARS-CoV-2 positive samples, sequence data was successfully recovered from 30 samples. As expected from wastewater samples, which contain highly fragmented virus genomic material, sequencing data presented with multiple short fragments of various sequence lengths. However, data from 13 samples with an average genome coverage of 53% was sufficient for confident lineage assignment and two for phylogenetic analysis. The sequences obtained covered a region which corresponds to the Spike protein.
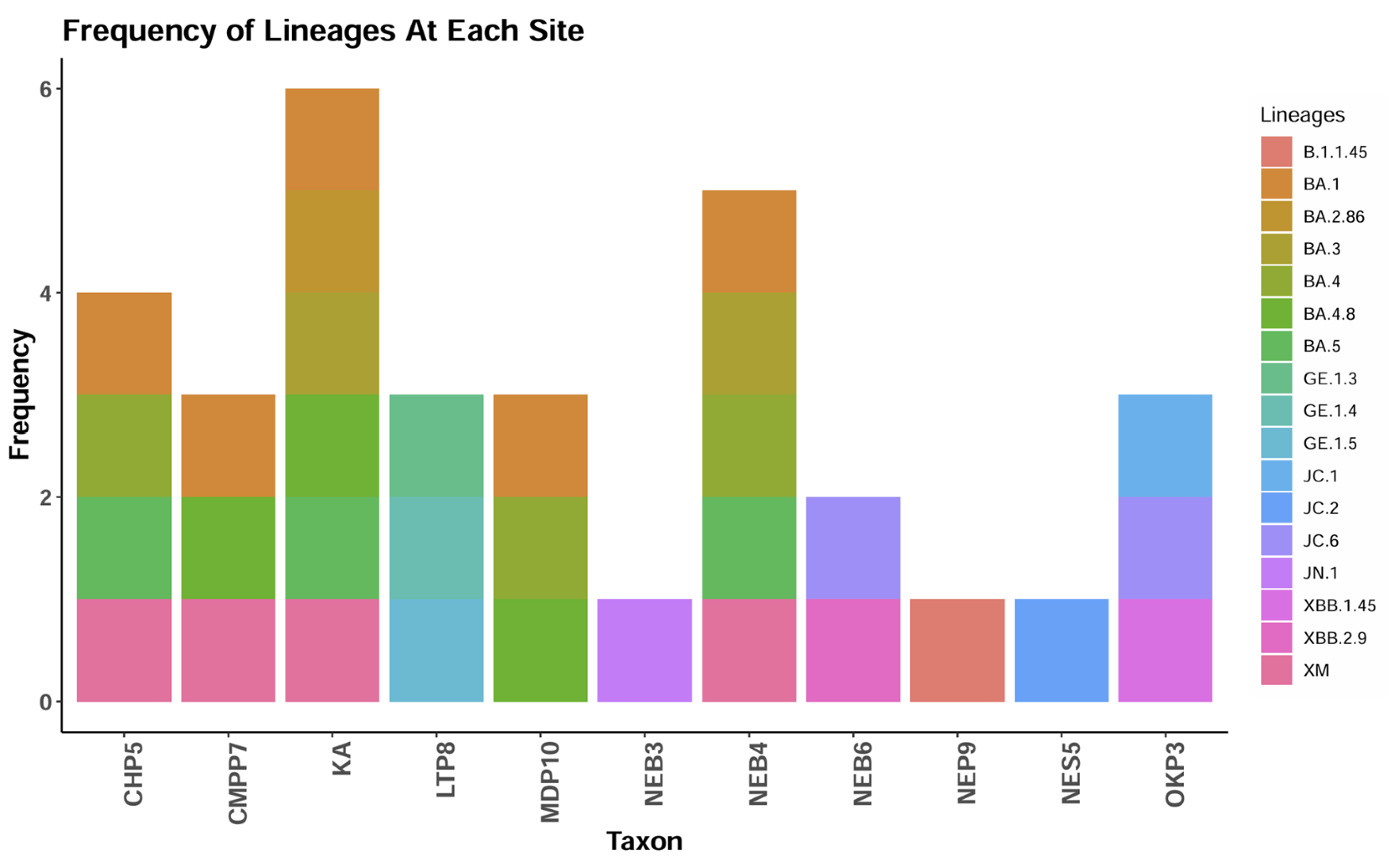
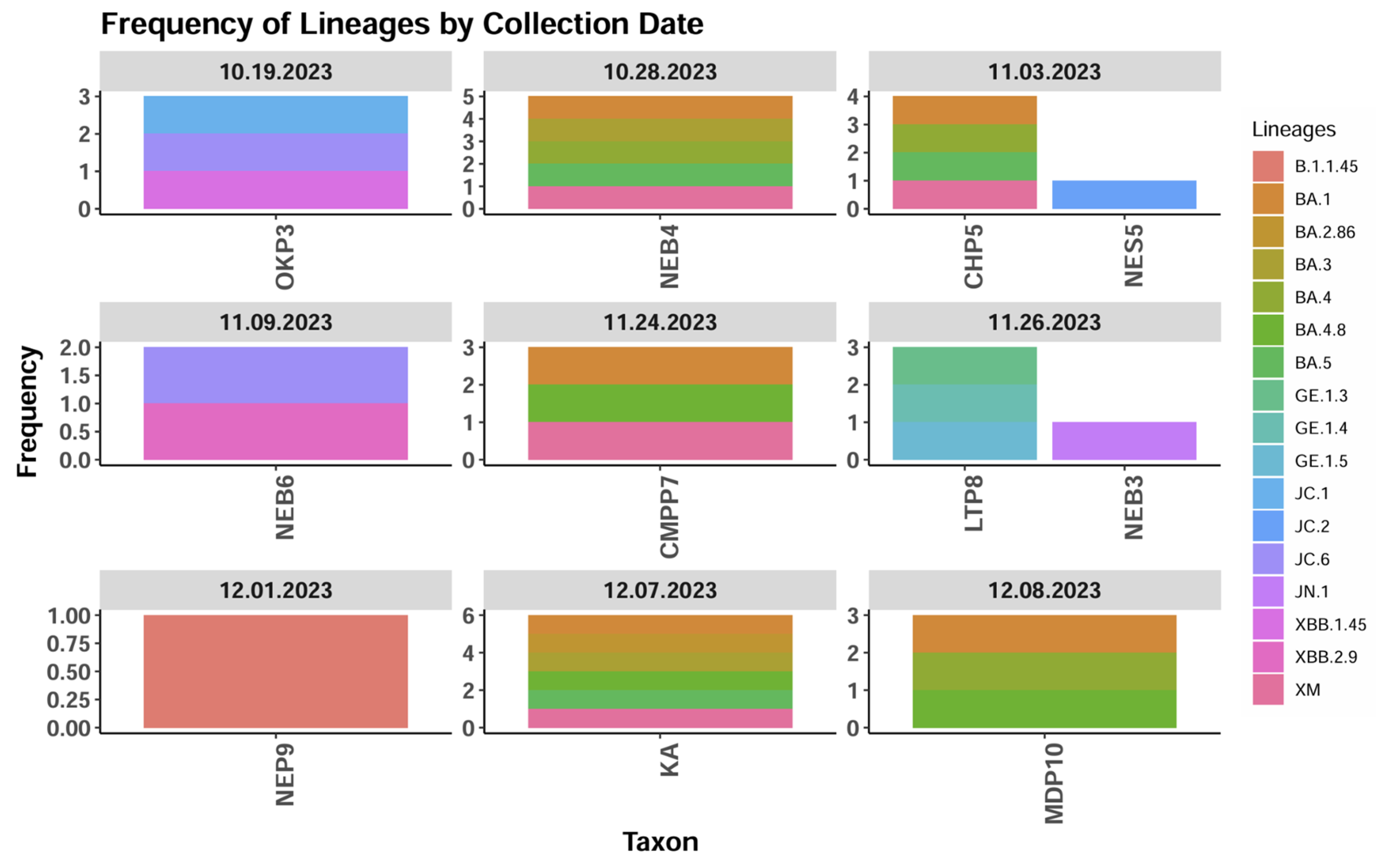
All the SARS-CoV-2 variants detected from this study were Omicron subvariants. This study did not detect a specific pattern or trend in variants from the various collection sites. JN.1 was isolated from the Nkana East treatment plant, XBB.1.45 was isolated from the Old Kanini treatment plant and XBB.2.9 was isolated from the Nkana East treatment plant. BA.2.86 was isolated from the Chipata Motel Ponds and the Kalulushi treatment plant (
Figure 4).
The time distribution trends also did not demonstrate any specific pattern or trend. XBB.1.45 was detected in October 2023 from the Old Kanini treatment plant. JN.1 and XBB.2.9 were detected from the Nkana East treatment plant in November 2023. In December 2023, BA.1 and BA.2.86 were detected from wastewater samples collected from Mindolo Ponds and Kalulushi treatment plants, respectively (
Figure 5).
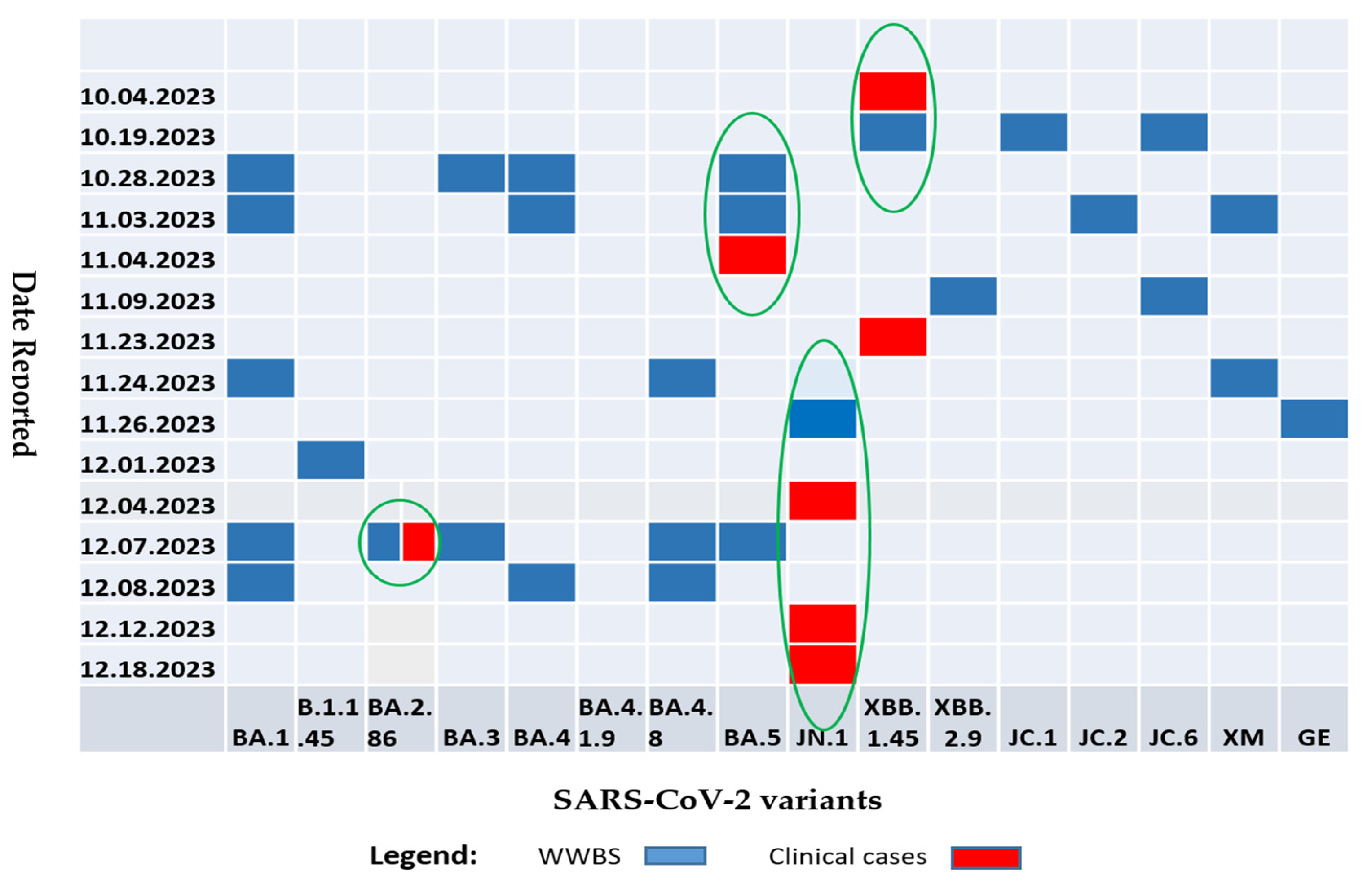
The sequences for the reported clinical cases of SARS-CoV-2 variants collected between October 2023 and December 2023 were downloaded from GISAID. The XBB.1.45 subvariant was detected in wastewater as well as in clinical samples in October 2023. The BA.2.86 subvariant was detected in both wastewater and clinical samples in December 2023. Equally, the BA.5 subvariant was detected in wastewater and clinical samples in November 2023. The JN.1 was isolated in wastewater two weeks before it was reported in clinical cases in the Copperbelt Province. The study also detected other subvariants of public health importance such as BA.1, BA.3, and suspected recombinant variant XM in wastewater which were absent in clinical cases (
Figure 6).
3.3. Mutation and Phylogenetic Analysis
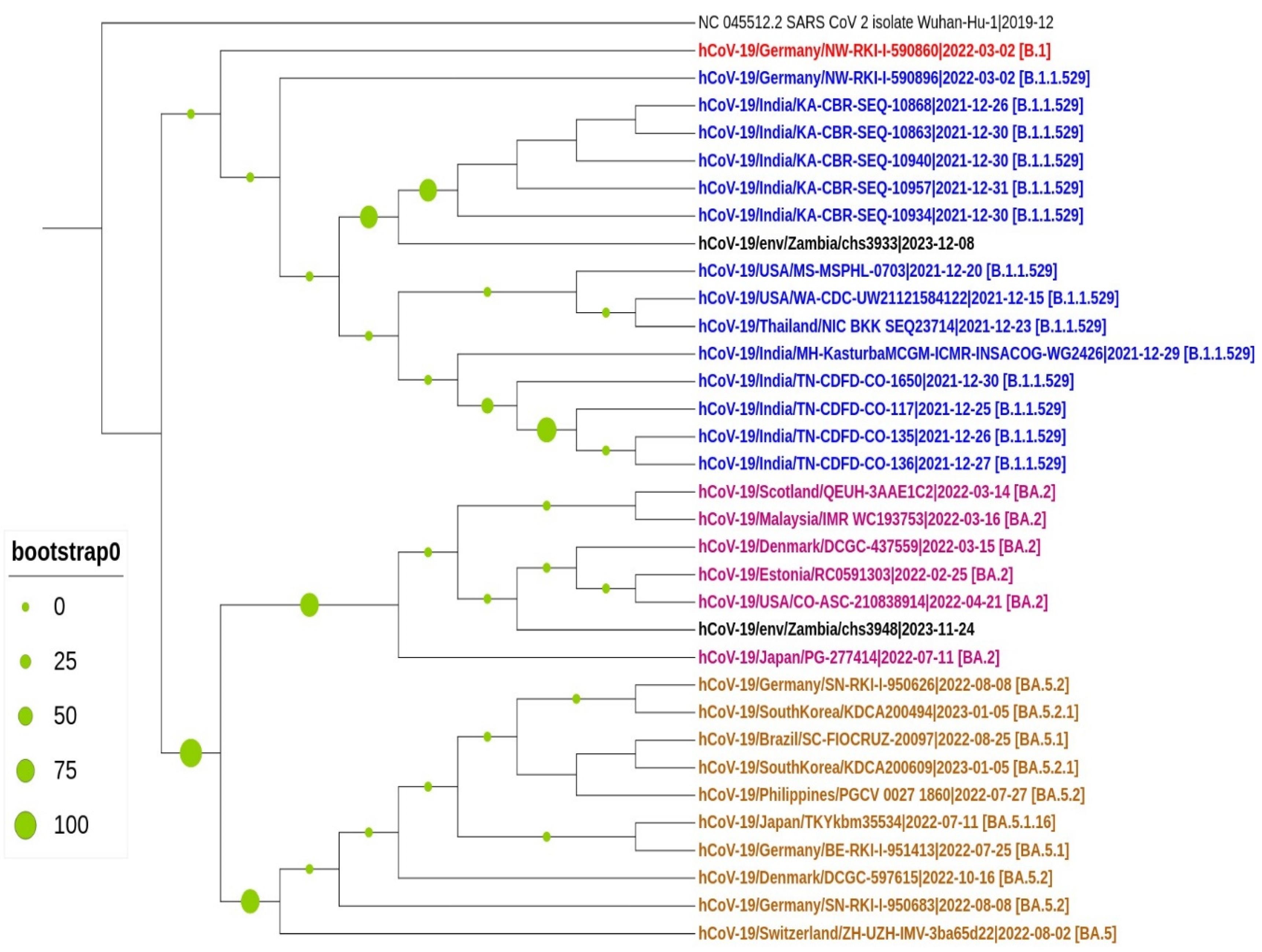
The 3495 bp spike gene segment, spanning the region 21651 to 25360 of the SARS-CoV-2 genome, was successfully isolated from a wastewater sample (GISAID accession ID EPI_ISL_19159151) collected at the Mindolo wastewater site. This sequence contained 1.98% ambiguous nucleotides (N) and thirteen mutations: N679K, Y145del, P681H, D614G, H655Y, A570V, N856K, Y144del, E554K, Q954H, P621S, G142D, and V143del. Phylogenetic analysis revealed that the Mindolo/2023 wastewater sequence was distinguishable but clustered within the B.1.1.529 clade (
Figure 7). The B.1.1.529 emerged in November 2021 [
51]. Meanwhile, the sequence obtained from the wastewater sample in Chipata (GISAID accession ID EPI_ISL_19157057) clustered closely with the BA.5.2.1 lineage, which emerged in early 2022 (
Figure 7).
4. Discussion
This study was conducted to investigate the presence of SARS-CoV-2 in wastewater in Zambia. To the best of our knowledge, this is the first wastewater surveillance of SARS-CoV-2 being reported in Zambia. Our study found that the overall positivity rate of SARS-CoV-2 was 40% and increased with time from October 2023 to December 2023.
The townships with the highest positivity rates of SARS-CoV-2 in wastewater samples were Nkana East (62%), Mindolo (61%), and New Kanini (46%). This study demonstrated that SARS-CoV-2 was being shed into the wastewater consistently over time. Higher positivity rates of SARS-CoV-2 in wastewater have been reported in previous studies including 50% in Italy [
36], 67% in Japan [
52] and 81.5% in Pakistan [
22]. In comparison to the current study, lower detection of SARS-CoV-2 in wastewater was reported in the USA where the positivity rate was 33.5% [
23]. Other lower positivity rates of SARS-CoV-2 detected in wastewater samples have been reported in other studies including 17% in Ghana [
41], 12.28% in Spain [
53] and 11.6% in the Czech Republic [
35]. Our study findings and those reported in similar studies demonstrate that wastewater surveillance can be used for early detection of SARS-CoV-2 in communities.
In this study, the detection of SARS-CoV-2 in wastewater is in line with reports concerning clinical cases. For instance, this study detected SARS-CoV-2 RNA in wastewater which subsequently increased from October 2023 to December 2023. Similarly, when wastewater analysis detected higher levels of SARS-CoV-2, a similar notice was observed in clinical cases reported in the community. As the COVID-19 pandemic rescinded and communities experienced less severe clinical manifestations, their health-seeking behaviour to test for COVID-19 as well as clinicians’ demand to request for COVID-19 testing reduced [
54]. Notably, our study detected SARS-CoV-2 from the first collection, suggesting a sustained viral shedding pattern from the communities despite the low to no clinical cases of COVID-19 being reported at that time. This is similar to the findings from Denmark where SARS-CoV-2 variants were detected from wastewater samples despite the discontinuation of testing for COVID-19 at the community level [
55]. During our study, an appreciable increase in the detection of SARS-CoV-2 in wastewater was noted and increased with time. Some studies have demonstrated that upon identification of SARS-CoV-2 in wastewater samples, the detection increased with time similar to what has been reported in clinical cases, and suggests that wastewater surveillance can be used as early warning to predict positive clinical cases for SARS-CoV-2 in communities before clinical presentations and detection [
36,
52,
56,
57]. A Japanese study found a lag period of 3 and 7 days indicating the importance of wastewater surveillance in early detection and spread of SARS-CoV-2 [
52]. This indeed highlights the potential of wastewater surveillance as an early indicator of COVID-19 outbreaks and emphasizes the importance of incorporating this method into existing disease surveillance systems [
27,
38,
56,
58].
Our study detected Omicron as the main variant in wastewater across the sampled communities. The Omicron variant was first discovered in South Africa [
59]. Its spread and clinical features affecting many countries in Southern Africa have been reported [
60]. Consequently, many variants of Omicron have surfaced [
61]. Previous studies that were conducted in Zambia revealed the presence of various variants of SARS-CoV-2 among clinical cases including Alpha, Beta, Delta, and many Omicron variants among clinical cases [
44,
45]. In Zambia, a previous study demonstrated that most variants were few with only Omicron being the majority [
44]. A recent study conducted in Zambia only detected Omicron in clinical cases potentially indicating the replacement of other variants [
62]. It does seem that Omicron dominated and replaced other variants and our study findings corroborate with those reported from studies that found the Omicron variant in the wastewater samples [
36,
52]. An Egyptian study found that the Delta variant was replaced by the Omicron variant which became a predominant variant [
63], similar to findings from Puerto Rico [
64], and the USA [
65]. Similarly, a study in Italy found that wastewater samples tested positive for the SARS-CoV-2 Omicron variant on December 7 2021, and increased to 66% in the last week of December 2021, indicating predominance of Omicron and spread across the country [
36]. Yet another study in Mexico also reported that Omicron became a predominant variant and replaced the Delta variant [
66]. Understanding the predominant variants is very critical in guiding mitigation strategies to address COVID-19 [
67,
68].
The present study detected BA.5 subvariant in wastewater samples which were also identified in clinical cases. Again, our findings are consistent with other studies that reported the detection of BA.5 in wastewater in Germany [
69] and Italy [
70,
71]. Other studies, however, have reported contrasting results showing the presence of other subvariants other than BA.5 during the same period [
72]. The differences could indicate a possible temporal shift in the predominance of variants with replacements of the BA.5 by other variants with time. Our study also detected XBB variants in wastewater, with the first detection of XBB.1.4.5 on 19th October 2023 which was also reported in clinical cases. The detection of these variants in wastewater indicates that there was indeed circulation of these variants. This is similar to findings from other countries where they detected XBB variants circulating in wastewater [
73]. Another study in Italy detected XBB subvariants in November 2022 [
72].
In Zambia, the BA.2.86 subvariants were detected in wastewater samples on December 7, 2023, in this study. The BA.2.86 was also identified in clinical cases in Zambia. The first global report of the BA.2.86 variant was in August 2023 [
74,
75]. The presence of BA.2.86 in wastewater samples is due to the increased shedding of the virus in faecal material [
76]. Our findings corroborate with other studies that also detected SARS CoV-2 BA.2.86 in wastewater and clinical cases including reports from Sweden [
77], Thailand [
78], China [
79], and Israel [
80].
The highly mutating BA2.86 evolved into JN.1 and other subvariants [
81]. The JN.1 subvariant was first reported in Luxemburg/ Iceland on 25th August 2023 and was initially tracked as part of BA2.86 [
82]. However, its continued report in many countries led to it being the majority among the BA2.86 descendent lineages. The growth advantage of the JN.1 subvariant and its ability for antibody escape increasing its potential for lethal clinical outcomes led to the WHO classifying it as a variant of interest separate from BA2.86 [
23,
83]. The first clinical report of the JN.1 variant in Zambia was on 4th December 2023 in Nakonde, Muchinga province. The present study reported the presence of the JN.1 variant in wastewater from the Copperbelt province earlier on 24th November 2023 which was followed by its detection in clinical cases on 12th December 2023 in the same province. This coincided with reports of spikes in COVID-19 cases in Zambia from December 2023 to January 2024. Similar findings were reported in India and Germany where the JN.1 became the predominant variant towards December 2023 replacing the BA.2.86 and other subvariants [
84,
85].
The presence of possible recombinant strains (XM and GE) in this study allows us to speculate that there could be active SARS-CoV-2 recombination among circulating variants. Therefore, the continued circulation of the BA.1 variant may be dormant in the environment and yet allows us to assert its possible contribution to ongoing recombination, posing a risk of brewing more virulent strains. Furthermore, this ‘dormant’ variant could be a source for future mutated versions of clinical significance even without recombination. Hence, it is key to strengthen environmental surveillance to detect the dormant variant in the environment as part of viral mutation vigilance for outbreak preparedness.
Our study found no unique mutations observed in the Mindolo wastewater sequence. However, it exhibited an evolutionary mutation signature: A570V, D614G, G142D, H655Y, P681H, V143del, Y144del, and Y145del, that can be classified as rare, indicating that it only occurred once. Interestingly, phylogenetic analysis positioned this sequence from the Copperbelt Province in the B.1.1.529 clade, suggesting that earlier Omicron variants detected in late 2021 could still be circulating and may not have been wholly replaced by newer subvariants. It seems reasonable to assert that the B.1.1.529 may not have been completely replaced by newer omicron subvariants and thus could be circulating and possibly contributing to community transmission of SARS-CoV-2. It is unlikely that the SARS-CoV-2 RNA integrity may have been maintained for close to three years in wastewater. It’s important to note that this sequence may not fully represent the viral composition at the Mindolo wastewater site, as several other lineages were identified within the same sample. As for the sequence obtained from the Chipata Motel Pond, it could not be assigned to a specific lineage because it contained a high proportion of ambiguous nucleotides in its sequence. However, phylogenetic analysis placed it close to the BA.5 series of omicron subvariants.
We acknowledge the limitation of our study, particularly the small sample size, as it only covered two provinces across Zambia and the short duration of the study thereby rendering a weakness to detect time-seasonal trends. However, our findings are of public health importance as they demonstrate the utility of monitoring SARS-CoV-2 in wastewater in LMICs including Zambia.
5. Conclusions
This study demonstrated the presence of SARS-CoV-2 in the wastewater samples from several districts in Zambia. The detection of the JN.1 Omicron variant was a critical public health finding, as it was a globally recognized variant of interest, indicating its circulation in some surveyed communities in Zambia. There is a need to strengthen wastewater surveillance in the monitoring of SARS-CoV-2 in communities in Zambia. Laboratory-based molecular detection of SARS-CoV-2 in the collected wastewater samples in this study indicated a successful ‘proof of concept’ for our wastewater surveillance activities in Zambia. Therefore, strengthening wastewater surveillance of SARS-CoV-2 is critical in identifying circulating SARS-CoV-2 variants and other threats that may emerge in the future.
Author Contributions
Conceptualization, D.M.S.; methodology, D.M.S., J.S. and K.S.; software, J.S. and W.M; validation, D.M.S., J.S. and K.S.; formal analysis, D.M.S., W.M., S.M., V.D., J.S., M.K. (Mapeesho Kamayani) and D.S.; investigation, D.M.S., J.S., D.S., C.C., and N.S.; resources, D.M.S. and K.S.; data curation, D.M.S. and J.S.; writing—original draft preparation, D.M.S., W.M., S.M., V.D., J.S., M.K. (Mapeesho Kamayani) and E.S.; writing—review and editing, D.M.S., S.M., V.D., J.S. and M.K. (Mapeesho Kamayani), D.S., C.C., K.M. (Kapina Muzala), K.M. (Kunda Musonda), J.Y.C., C.M., G.K., M.K. (Maisa Kasanga), M.N., OK, F.C., B.N., L.M., E.S., N.S., J.B.M., K.S. and R.C.; visualization, D.M.S., S.M., V.D., J.S. and M.K. (Mapeesho Kamayani), D.S., C.C., K.M. (Kapina Muzala), K.M. (Kunda Musonda), J.Y.C., C.M., G.K., M.K. (Maisa Kasanga), M.N., F.C., B.N., L.M., E.S., N.S., J.B.M., K.S. and R.C.; supervision, J.B.M., E.S., N.S., K.S. and R.C.; project administration, D.M.S.; funding acquisition, D.M.S. and K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was approved by the National Health Research Authority (NHRA) Ref No: NHRA00013/06/09/2023; “Wastewater Surveillance of SARS-CoV-2 and other Pathogens on the Copperbelt and Eastern Provinces, Zambia”.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in the study is available on request from the corresponding author.
Acknowledgments
We are grateful to the technical staff at the Churches Association of Zambia (CHAZ) laboratory who participated in data collection and laboratory analysis of the samples. The authors would also want to acknowledge the Zambia National Public Health Institute (ZNPHI) and CHAZ Management for the project oversight.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sharma A, Tiwari S, Deb MK, Marty JL. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): a global pandemic and treatment strategies. Int J Antimicrob Agents 2020;56:106054. [CrossRef]
- Liu YC, Kuo RL, Shih SR. COVID-19: The first documented coronavirus pandemic in history. Biomed J 2020;43:328–33. [CrossRef]
- Carabelli AM, Peacock TP, Thorne LG, Harvey WT, Hughes J, de Silva TI, et al. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol 2023;21:162–77. [CrossRef]
- Bostanghadiri N, Ziaeefar P, Mofrad MG, Yousefzadeh P, Hashemi A, Darban-Sarokhalil D. COVID-19: An Overview of SARS-CoV-2 Variants—The Current Vaccines and Drug Development. Biomed Res Int 2023;2023:1879554. [CrossRef]
- Sah R, Rais MA, Mohanty A, Chopra H, Chandran D, Bin Emran T, et al. Omicron (B.1.1.529) variant and its subvariants and lineages may lead to another COVID-19 wave in the world? -An overview of current evidence and counteracting strategies. Int J Surg Open 2023;55:100625. [CrossRef]
- Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nat 2022 6087923 2022;608:593–602. [CrossRef]
- Shah M, Woo HG. Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies. Front Immunol 2022;12:830527. [CrossRef]
- Cavanaugh AM, Fortier S, Lewis P, Arora V, Johnson M, George K, et al. COVID-19 Outbreak Associated with a SARS-CoV-2 R.1 Lineage Variant in a Skilled Nursing Facility After Vaccination Program — Kentucky, March 2021. MMWR Recomm Reports 2021;70:639–43. [CrossRef]
- Sarkar P, Banerjee S, Saha SA, Mitra P, Sarkar S. Genome surveillance of SARS-CoV-2 variants and their role in pathogenesis focusing on second wave of COVID-19 in India. Sci Rep 2023;13:4692. [CrossRef]
- Fernandes Q, Inchakalody VP, Merhi M, Mestiri S, Taib N, Moustafa Abo El-Ella D, et al. Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann Med 2022;54:524–40. [CrossRef]
- D’Agostino Y, Rocco T, Ferravante C, Porta A, Tosco A, Cappa VM, et al. Rapid and sensitive detection of SARS-CoV-2 variants in nasopharyngeal swabs and wastewaters. Diagn Microbiol Infect Dis 2022;102:115632. [CrossRef]
- Kevadiya BD, Machhi J, Herskovitz J, Oleynikov MD, Blomberg WR, Bajwa N, et al. Diagnostics for SARS-CoV-2 infections. Nat Mater 2021;20:593–605. [CrossRef]
- Tembo J, Egbe NF, Maluzi K, Mulonga K, Chilufya M, Kapata N, et al. Evaluation of SARS-CoV-2 diagnostics and risk factors associated with SARS-CoV-2 infection in Zambia. Int J Infect Dis 2022;120:150–7. [CrossRef]
- Peeling RW, Heymann DL, Teo YY, Garcia PJ. Diagnostics for COVID-19: moving from pandemic response to control. Lancet 2022;399:757–68. [CrossRef]
- Gangavarapu K, Latif AA, Mullen JL, Alkuzweny M, Hufbauer E, Tsueng G, et al. Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Nat Methods 2023;20:512–22. [CrossRef]
- Amato E, Hyllestad S, Heradstveit P, Langlete P, Moen LV, Rohringer A, et al. Evaluation of the pilot wastewater surveillance for SARS-CoV-2 in Norway, June 2022–March 2023. BMC Public Health 2023;23:1714. [CrossRef]
- Brunner FS, Brown MR, Bassano I, Denise H, Khalifa MS, Wade MJ, et al. City-wide wastewater genomic surveillance through the successive emergence of SARS-CoV-2 Alpha and Delta variants. Water Res 2022;226:119306. [CrossRef]
- Flood MT, Sharp J, Bruggink J, Cormier M, Gomes B, Oldani I, et al. Understanding the efficacy of wastewater surveillance for SARS-CoV-2 in two diverse communities. PLoS One 2023;18:e0289343. [CrossRef]
- Conway MJ, Yang H, Revord LA, Novay MP, Lee RJ, Ward AS, et al. Chronic shedding of a SARS-CoV-2 Alpha variant in wastewater. BMC Genomics 2024;25:59. [CrossRef]
- Smith T, Cassell G, Bhatnagar A. Wastewater Surveillance Can Have a Second Act in COVID-19 Vaccine Distribution. JAMA Heal Forum 2021;2:E201616. [CrossRef]
- Wolfe M, Hughes B, Duong D, Chan-Herur V, Wigginton KR, White BJ, et al. Detection of SARS-CoV-2 Variants Mu, Beta, Gamma, Lambda, Delta, Alpha, and Omicron in Wastewater Settled Solids Using Mutation-Specific Assays Is Associated with Regional Detection of Variants in Clinical Samples. Appl Environ Microbiol 2022;88:e00045-22. [CrossRef]
- Ansari N, Kabir F, Khan W, Khalid F, Malik AA, Warren JL, et al. Environmental surveillance for COVID-19 using SARS-CoV-2 RNA concentration in wastewater—a study in District East, Karachi, Pakistan. Lancet Reg Heal—Southeast Asia 2024;20:100299. [CrossRef]
- Liu P, Ibaraki M, VanTassell J, Geith K, Cavallo M, Kann R, et al. A sensitive, simple, and low-cost method for COVID-19 wastewater surveillance at an institutional level. Sci Total Environ 2022;807:151047. [CrossRef]
- World Health Organization. Wastewater surveillance of SARS-CoV-2. World Heal Organ 2022:1–12.
- Kallem P, Hegab H, Alsafar H, Hasan SW, Banat F. SARS-CoV-2 detection and inactivation in water and wastewater: review on analytical methods, limitations and future research recommendations. Emerg Microbes Infect 2023;12:2222850. [CrossRef]
- Shafer MM, Bobholz MJ, Vuyk WC, Gregory DA, Roguet A, Haddock Soto LA, et al. Tracing the origin of SARS-CoV-2 omicron-like spike sequences detected in an urban sewershed: a targeted, longitudinal surveillance study of a cryptic wastewater lineage. The Lancet Microbe 2024;5:e335–44. [CrossRef]
- Jahn K, Dreifuss D, Topolsky I, Kull A, Ganesanandamoorthy P, Fernandez-Cassi X, et al. Early detection and surveillance of SARS-CoV-2 genomic variants in wastewater using COJAC. Nat Microbiol 2022;7:1151–60. [CrossRef]
- Fontenele RS, Kraberger S, Hadfield J, Driver EM, Bowes D, Holland LRA, et al. High-throughput sequencing of SARS-CoV-2 in wastewater provides insights into circulating variants. Water Res 2021;205:117710. [CrossRef]
- McClary-Gutierrez JS, Mattioli MC, Marcenac P, Silverman AI, Boehm AB, Bibby K, et al. SARS-CoV-2 wastewater surveillance for public health action. Emerg Infect Dis 2021;27:E1–9. [CrossRef]
- Maryam S, Ul Haq I, Yahya G, Ul Haq M, Algammal AM, Saber S, et al. COVID-19 surveillance in wastewater: An epidemiological tool for the monitoring of SARS-CoV-2. Front Cell Infect Microbiol 2023;12:978643. [CrossRef]
- Kirby AE, Walters MS, Jennings WC, Fugitt R, LaCross N, Mattioli M, et al. Using Wastewater Surveillance Data to Support the COVID-19 Response — United States, 2020-2021. MMWR Recomm Reports 2021;70:1242–4. [CrossRef]
- Yousif M, Rachida S, Taukobong S, Ndlovu N, Iwu-Jaja C, Howard W, et al. SARS-CoV-2 genomic surveillance in wastewater as a model for monitoring evolution of endemic viruses. Nat Commun 2023;14:6325. [CrossRef]
- Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ Sci Technol Lett 2020;7:511–6. [CrossRef]
- Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O’Brien JW, et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ 2020;728:138764. [CrossRef]
- Mlejnkova H, Sovova K, Vasickova P, Ocenaskova V, Jasikova L, Juranova E. Preliminary study of Sars-Cov-2 occurrence in wastewater in the Czech Republic. Int J Environ Res Public Health 2020;17:5508. [CrossRef]
- La Rosa G, Iaconelli M, Mancini P, Bonanno Ferraro G, Veneri C, Bonadonna L, et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci Total Environ 2020;736:139652. [CrossRef]
- Karthikeyan S, Nguyen A, McDonald D, Zong Y, Ronquillo N, Ren J, et al. Rapid, Large-Scale Wastewater Surveillance and Automated Reporting System Enable Early Detection of Nearly 85% of COVID-19 Cases on a University Campus. MSystems 2021;6:e0079321. [CrossRef]
- Bar-Or I, Weil M, Indenbaum V, Bucris E, Bar-Ilan D, Elul M, et al. Detection of SARS-CoV-2 variants by genomic analysis of wastewater samples in Israel. Sci Total Environ 2021;789:148002. [CrossRef]
- Albastaki A, Naji M, Lootah R, Almeheiri R, Almulla H, Almarri I, et al. First confirmed detection of SARS-COV-2 in untreated municipal and aircraft wastewater in Dubai, UAE: The use of wastewater based epidemiology as an early warning tool to monitor the prevalence of COVID-19. Sci Total Environ 2021;760:143350. [CrossRef]
- Street R, Malema S, Mahlangeni N, Mathee A. Wastewater surveillance for Covid-19: An African perspective. Sci Total Environ 2020;743:140719. [CrossRef]
- Duker EO, Obodai E, Addo SO, Kwasah L, Mensah ES, Gberbi E, et al. First Molecular Detection of SARS-CoV-2 in Sewage and Wastewater in Ghana. Biomed Res Int 2024;2024:9975781. [CrossRef]
- Simulundu E, Mupeta F, Chanda-Kapata P, Saasa N, Changula K, Muleya W, et al. First COVID-19 case in Zambia — Comparative phylogenomic analyses of SARS-CoV-2 detected in African countries. Int J Infect Dis 2021;102:455–9. [CrossRef]
- Chipimo PJ, Barradas DT, Kayeyi N, Zulu PM, Muzala K, Mazaba ML, et al. First 100 Persons with COVID-19 — Zambia, March 18–April 28, 2020. MMWR Morb Mortal Wkly Rep 2020;69:1547–8. [CrossRef]
- Katowa B, Kalonda A, Mubemba B, Matoba J, Shempela DM, Sikalima J, et al. Genomic Surveillance of SARS-CoV-2 in the Southern Province of Zambia: Detection and Characterization of Alpha, Beta, Delta, and Omicron Variants of Concern. Viruses 2022;14:1865. [CrossRef]
- Mwenda M, Saasa N, Sinyange N, Busby G, Chipimo PJ, Hendry J, et al. Detection of B.1.351 SARS-CoV-2 Variant Strain — Zambia, December 2020. MMWR Surveill Summ 2021;70:280–2. [CrossRef]
- Saasa N, M’kandawire E, Ndebe J, Mwenda M, Chimpukutu F, Mukubesa AN, et al. Detection of Human Adenovirus and Rotavirus in Wastewater in Lusaka, Zambia: Demonstrating the Utility of Environmental Surveillance for the Community. Pathogens 2024;13:486. [CrossRef]
- Prata C, Ribeiro A, Cunha Â, Gomes NCM, Almeida A. Ultracentrifugation as a direct method to concentrate viruses in environmental waters: virus-like particle enumeration as a new approach to determine the efficiency of recovery. J Environ Monit 2012;14:64–70. [CrossRef]
- Fagnant-Sperati CS, Ren Y, Zhou NA, Komen E, Mwangi B, Hassan J, et al. Validation of the bag-mediated filtration system for environmental surveillance of poliovirus in Nairobi, Kenya. J Appl Microbiol 2021;130:971–81. [CrossRef]
- Philo SE, Keim EK, Swanstrom R, Ong AQW, Burnor EA, Kossik AL, et al. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci Total Environ 2021;760:144215. [CrossRef]
- Sievers F, Higgins DG. The Clustal Omega Multiple Alignment Package. Methods Mol. Biol., vol. 2231, Humana, New York, NY; 2021, p. 3–16. [CrossRef]
- Khandia R, Singhal S, Alqahtani T, Kamal MA, El-Shall NA, Nainu F, et al. Emergence of SARS-CoV-2 Omicron (B.1.1.529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ Res 2022;209:112816. [CrossRef]
- Shrestha S, Malla B, Angga MS, Sthapit N, Raya S, Hirai S, et al. Long-term SARS-CoV-2 surveillance in wastewater and estimation of COVID-19 cases: An application of wastewater-based epidemiology. Sci Total Environ 2023;896:165270. [CrossRef]
- Fernández-de-Mera IG, Rodríguez del-Río FJ, de la Fuente J, Pérez-Sancho M, Hervás D, Moreno I, et al. Detection of environmental SARS-CoV-2 RNA in a high prevalence setting in Spain. Transbound Emerg Dis 2021;68:1487–92. [CrossRef]
- World Health Organization. Environmental surveillance for SARS-COV-2 to complement public health surveillance—Interim Guidance. 2022.
- Krogsgaard LW, Benedetti G, Gudde A, Richter SR, Rasmussen LD, Midgley SE, et al. Results from the SARS-CoV-2 wastewater-based surveillance system in Denmark, July 2021 to June 2022. Water Res 2024;252:121223. [CrossRef]
- Wu F, Xiao A, Zhang J, Moniz K, Endo N, Armas F, et al. SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Sci Total Environ 2022;805:150121. [CrossRef]
- Gupta P, Liao S, Ezekiel M, Novak N, Rossi A, LaCross N, et al. Wastewater Genomic Surveillance Captures Early Detection of Omicron in Utah. Microbiol Spectr 2023;11:e0039123. [CrossRef]
- Zhang D, Duran SSF, Lim WYS, Tan CKI, Cheong WCD, Suwardi A, et al. SARS-CoV-2 in wastewater: From detection to evaluation. Mater Today Adv 2022;13:100211. [CrossRef]
- Wolter N, Jassat W, Walaza S, Welch R, Moultrie H, Groome M, et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet 2022;399:437–46. [CrossRef]
- Viana R, Moyo S, Amoako DG, Tegally H, Scheepers C, Althaus CL, et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in Southern Africa. Nature 2022;603:679–86. [CrossRef]
- Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021;592:438–43. [CrossRef]
- Shempela DM, Chambaro HM, Sikalima J, Cham F, Njuguna M, Morrison L, et al. Detection and Characterisation of SARS-CoV-2 in Eastern Province of Zambia; A Retrospective Genomic Surveillance Study. Int J Mol Sci 2024;25:6338. [CrossRef]
- Liddor Naim M, Fu Y, Shagan M, Bar-Or I, Marks R, Sun Q, et al. The Rise and Fall of Omicron BA.1 Variant as Seen in Wastewater Supports Epidemiological Model Predictions. Viruses 2023;15:1862. [CrossRef]
- Santiago GA, Volkman HR, Flores B, González GL, Charriez KN, Huertas LC, et al. SARS-CoV-2 Omicron Replacement of Delta as Predominant Variant, Puerto Rico. Emerg Infect Dis 2023;29:855–7. [CrossRef]
- Robles-Escajeda E, Mohl JE, Contreras L, Betancourt AP, Mancera BM, Kirken RA, et al. Rapid Shift from SARS-CoV-2 Delta to Omicron Sub-Variants within a Dynamic Southern U.S. Borderplex. Viruses 2023;15:658. [CrossRef]
- Aguayo-Acosta A, Oyervides-Muñoz MA, Rodriguez-Aguillón KO, Ovalle-Carcaño A, Romero-Castillo KD, Robles-Zamora A, et al. Omicron and Delta variant prevalence detection and identification during the fourth COVID-19 wave in Mexico using wastewater-based epidemiology. IJID Reg 2024;10:44–51. [CrossRef]
- Zabidi NZ, Liew HL, Farouk IA, Puniyamurti A, Yip AJW, Wijesinghe VN, et al. Evolution of SARS-CoV-2 Variants: Implications on Immune Escape, Vaccination, Therapeutic and Diagnostic Strategies. Viruses 2023;15:944. [CrossRef]
- Tao K, Tzou PL, Nouhin J, Gupta RK, de Oliveira T, Kosakovsky Pond SL, et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet 2021;22:757–73. [CrossRef]
- Wilhelm A, Agrawal S, Schoth J, Meinert-Berning C, Bastian D, Orschler L, et al. Early Detection of SARS-CoV-2 Omicron BA.4 and BA.5 in German Wastewater. Viruses 2022;14. [CrossRef]
- Baldovin T, Amoruso I, Fonzo M, Bertoncello C, Groppi V, Pitter G, et al. Trends in SARS-CoV-2 clinically confirmed cases and viral load in wastewater: A critical alignment for Padua city (NE Italy). Heliyon 2023;9:e20571. [CrossRef]
- Triggiano F, De Giglio O, Apollonio F, Brigida S, Fasano F, Mancini P, et al. Wastewater-based Epidemiology and SARS-CoV-2: Variant Trends in the Apulia Region (Southern Italy) and Effect of Some Environmental Parameters. Food Environ Virol 2023;15:331–41. [CrossRef]
- La Rosa G, Brandtner D, Bonanno Ferraro G, Veneri C, Mancini P, Iaconelli M, et al. Wastewater surveillance of SARS-CoV-2 variants in October–November 2022 in Italy: detection of XBB.1, BA.2.75 and rapid spread of the BQ.1 lineage. Sci Total Environ 2023;873:162339. [CrossRef]
- Qvesel AG, Bennedbæk M, Larsen NB, Gunalan V, Krogsgaard LW, Rasmussen M, et al. SARS-CoV-2 Variants BQ.1 and XBB.1.5 in Wastewater of Aircraft Flying from China to Denmark, 2023. Emerg Infect Dis 2023;29:2559–61. [CrossRef]
- Lambrou AS, South E, Ballou ES, Paden CR, Fuller JA, Bart SM, et al. Early Detection and Surveillance of the SARS-CoV-2 Variant BA.2.86 — Worldwide, July–October 2023. MMWR Recomm Reports 2023;72:1162–7. [CrossRef]
- Rasmussen M, Moller FT, Gunalan V, Baig S, Bennedbak M, Christiansen LE, et al. First cases of SARS-CoV-2 BA.2.86 in Denmark, 2023. Eurosurveillance 2023;28:2300460. [CrossRef]
- Wannigama DL, Amarasiri M, Phattharapornjaroen P, Hurst C, Modchang C, Chadsuthi S, et al. Increased faecal shedding in SARS-CoV-2 variants BA.2.86 and JN.1. Lancet Infect Dis 2024;24:e348–50. [CrossRef]
- Espinosa-Gongora C, Berg C, Rehn M, Varg JE, Dillner L, Latorre-Margalef N, et al. Early detection of the emerging SARS-CoV-2 BA.2.86 lineage through integrated genomic surveillance of wastewater and COVID-19 cases in Sweden, weeks 31 to 38 2023. Eurosurveillance 2023;28. [CrossRef]
- Wannigama DL, Amarasiri M, Phattharapornjaroen P, Hurst C, Modchang C, Chadsuthi S, et al. Tracing the new SARS-CoV-2 variant BA.2.86 in the community through wastewater surveillance in Bangkok, Thailand. Lancet Infect Dis 2023;23:e464–6. [CrossRef]
- Du C, Peng Y, Lyu Z, Yue Z, Fu Y, Yao X, et al. Early Detection of the Emerging SARS-CoV-2 BA.2.86 Lineage Through Wastewater Surveillance Using a Mediator Probe PCR Assay — Shenzhen City, Guangdong Province, China, 2023. China CDC Wkly 2024;6:332–8. [CrossRef]
- Erster O, Bar-Or I, Azar R, Assraf H, Kabat A, Mannasse B, et al. Incursion of SARS-CoV-2 BA.2.86.1 variant into Israel: National-scale wastewater surveillance using a novel quantitative real-time PCR assay. Sci Total Environ 2024;933:173164. [CrossRef]
- Planas D, Bruel T, Staropoli I, Guivel-Benhassine F, Porrot F, Maes P, et al. Resistance of Omicron subvariants BA.2.75.2, BA.4.6, and BQ.1.1 to neutralizing antibodies. Nat Commun 2023;14:1–11. [CrossRef]
- World Health Organization. Initial rapid evaluation of JN.1. vol. 1. 2023.
- Li P, Liu Y, Faraone JN, Hsu CC, Chamblee M, Zheng Y-M, et al. Distinct patterns of SARS-CoV-2 BA.2.87.1 and JN.1 variants in immune evasion, antigenicity, and cell-cell fusion. MBio 2024;15:e00751-24. [CrossRef]
- Karyakarte RP, Das R, Rajmane M V, Dudhate S, Agarasen J, Pillai P, et al. Appearance and Prevalence of JN.1 SARS-CoV-2 Variant in India and Its Clinical Profile in the State of Maharashtra. Cureus 2024;16:e56718. [CrossRef]
- Bartel A, Grau JH, Bitzegeio J, Werber D, Linzner N, Schumacher V, et al. Timely Monitoring of SARS-CoV-2 RNA Fragments in Wastewater Shows the Emergence of JN.1 (BA.2.86.1.1, Clade 23I) in Berlin, Germany. Viruses 2024;16:102. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).