Submitted:
01 July 2024
Posted:
01 July 2024
You are already at the latest version
Abstract
Keywords:
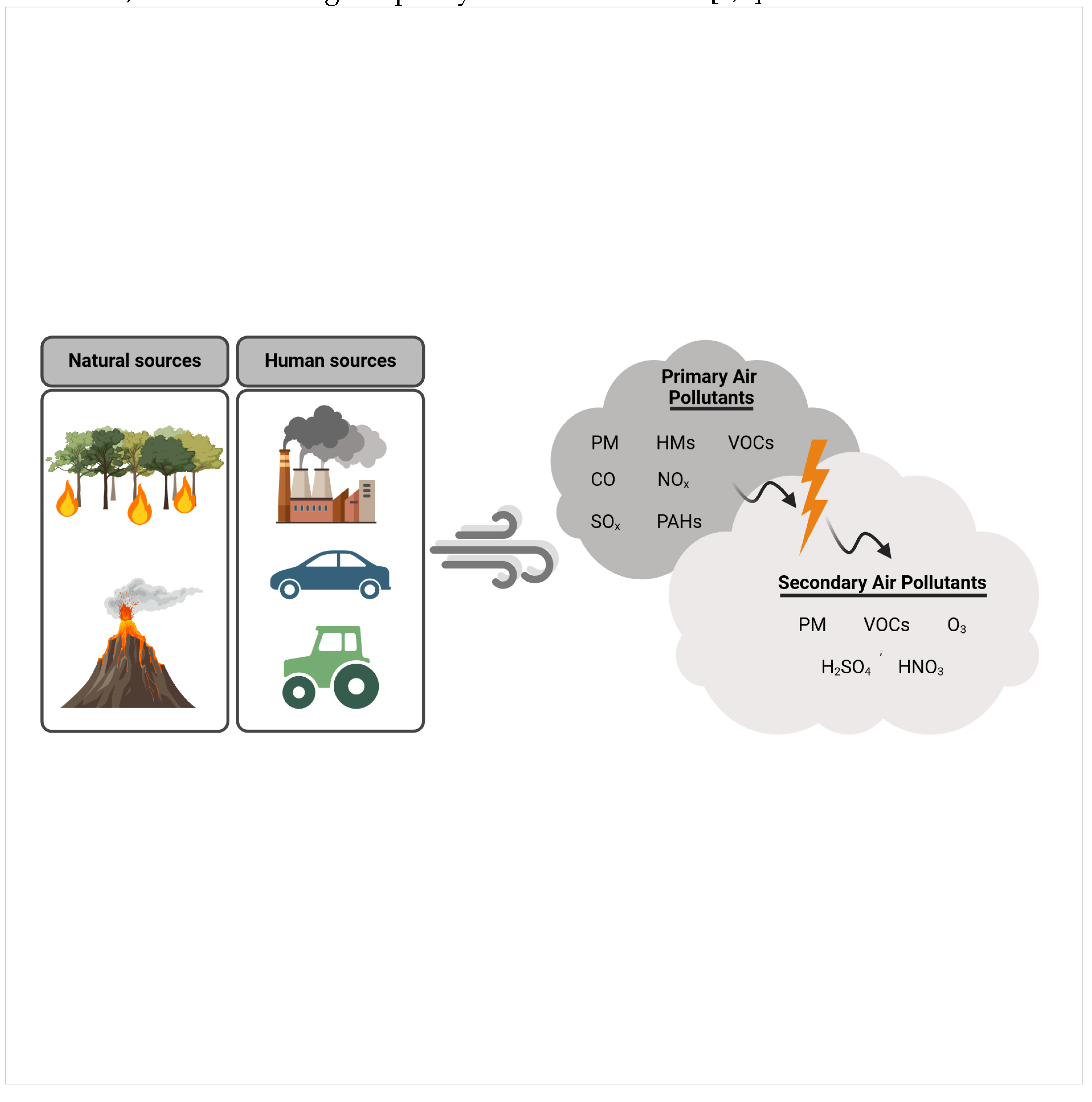
1. Introduction
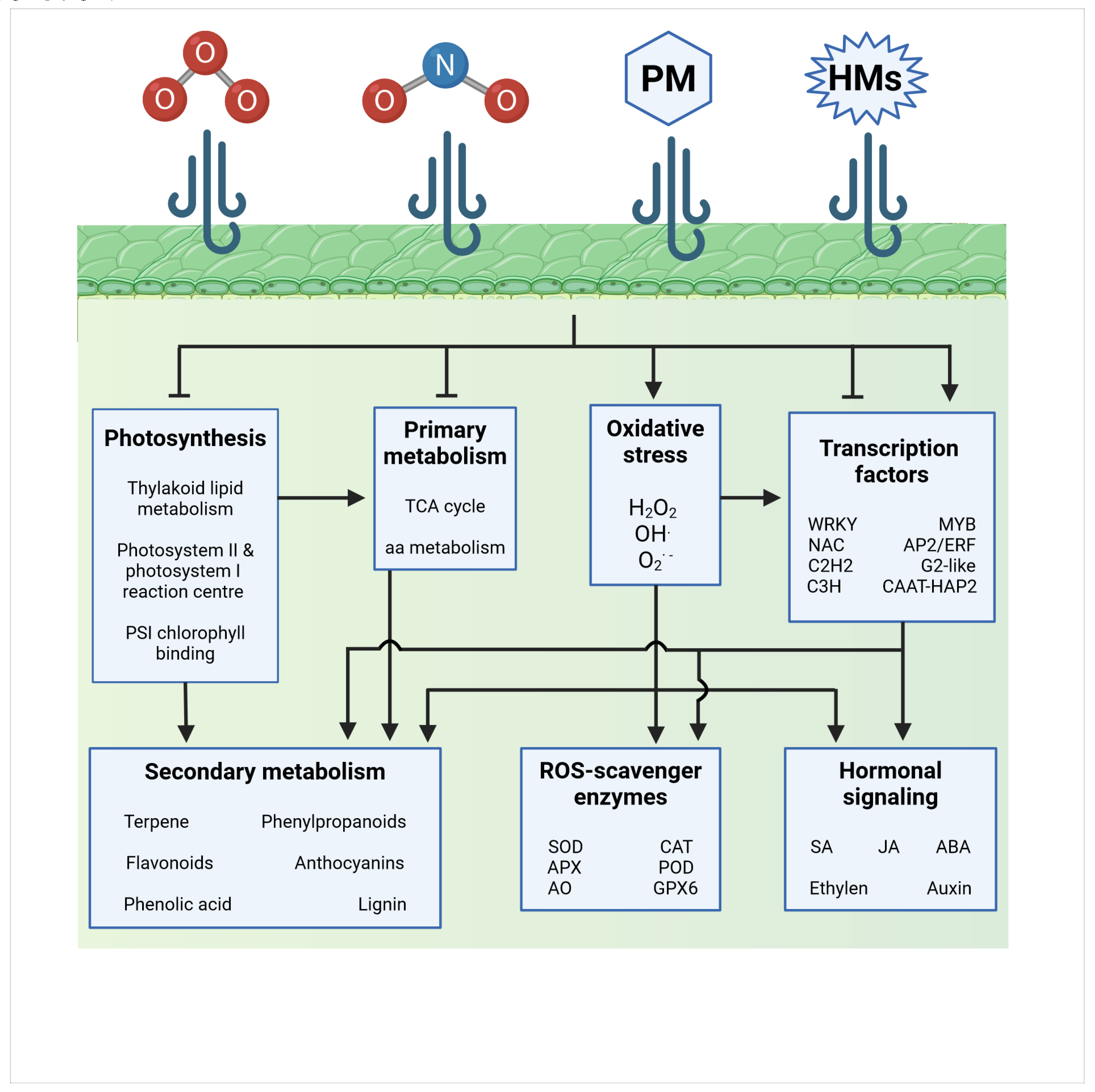
2. Air pollution induces similar but not overlapping molecular responses in plants
| Species | Experimental set-up |
Pollutant | Exposure | Omics Platform | Enriched Pathways | Reference |
|---|---|---|---|---|---|---|
|
Arabidopsis thaliana L. |
Controlled environment growth chambers |
O3 350–423 nL L–1 |
2-6 h | Transcriptomics (RNA-seq) | Photosynthesis Response to SA Response to ROS Response to JA Response to ethylene ABA signalling pathway |
[23] |
|
Medicago truncatula L. |
Controlled environment growth chambers |
O3 70 nmol mol-1 |
6 h d-1 for 6 d | Transcriptomics (Microarray hybridization) |
Phenylalanine biosynthesis Sugar metabolism Photosynthetic electron transport Responses to inorganic substances |
[30] |
| Malus L. | Open-top growth chamber |
O3 300 nL L−1 |
3 h | Transcriptomics (RNA-seq) Metabolomics (ultra-performance liquid chromatography coupled to tandem mass spectrometry UPLC MS/MS) |
Chloroplast thylakoid membrane Chloroplast photosystem I H2O2 dehydratase activity Chalcone synthase activity Flavonoid metabolism Hormone pathways |
[31] |
|
Pisum sativum L. Glycine max L. Phaseolus vulgaris L. |
Controlled environment growth chambers |
O3 ~151.2 nL L−1 |
8 h d-1 for 45 d | Transcriptomics (RNA-seq) | Phenilpropanoid metabolism Ascorbate–glutathione cycling Glycolysis TCA cycle |
[32] |
|
Rosa hybrida L. |
Controlled environment growth chambers |
O3 80 ppb |
10 h | Transcriptomics (RNA-seq) | Phenylpropanoid biosynthesis, Starch and sucrose metabolism Sesquiterpenoid biosynthesis Triterpenoid biosynthesis. |
[33] |
|
Abies religiosa Schltdl. & Cham. |
Urban environment |
O3 87-170 ppb |
3 years | Transcriptomics (RNA-seq) | Carbohydrate metabolism Gene regulation Transcription factors Defense regulation Terpenes |
[36] |
|
Bougainvillea Spectabilis Willd. |
Controlled environment growth chambers |
NO2 8 μL L− 1 |
8 h | Metabolomics (UPLC) | Biosynthesis of amino acids Phenylalanine metabolism Phenylpropanoid biosynthesis Starch and sucrose metabolism Glutathione metabolism TCA cycle |
[37] |
|
Arabidopsis thaliana L. |
Controlled environment growth chambers |
O3 350 ppb NO2 10-30 ppm |
6 h O3 1 h NO2 |
Transcriptomic data Microarray data |
Pathogen resistance Cell death Ethylene signalling |
[38] |
|
Ambrosia artemisiifolia L. |
Controlled environment growth chambers |
O3 NO2 40-80 ppb |
61 d | Transcriptomics (RNA-seq) | Jasmonic acid pathway Response to ethylene stimulus Response to auxin stimulus Abscisic acid signalling pathway |
[39] |
|
Wrightia religiosa (Teijsm.&BINN.)Hook.F. |
Controlled environment growth chambers |
PM 470–500 μg m-3 |
24 h | Proteomics (LC-MS/MS) | Photosynthetic proteins |
[44] |
| Sansevieria trifasciata (Dracaena trifasciata Prain.) | Controlled environment growth chambers |
PM Up to 980 μg m-3 |
24 h | Proteomics (LC-MS/MS) Metabolomics |
Precursor metabolites Photosynthesis Alternative carbon metabolism Brassinosteroid signalling Stress-related proteins Metal and cadmium ion stimuli |
[43, 45] |
|
Laurus nobilis L. |
Urban environment |
PM up to 150 μg m-2 |
3 months | Transcriptomics (RNA-seq) |
Primary metabolism Secondary metabolism Hormone-related pathways Environmental stress response Transcription factors |
[41] |
|
Photinia x fraseri L. |
Urban environment |
PM up to 150 μg m-2 |
3 months | Transcriptomics (RNA-seq) |
Leaf primary metabolism Biotic stress response Abiotic stress response Cell cycle and cell division Transcription factors |
[40] |
2.1. Ozone
2.2. Nitrogen Dioxide
2.3. Particulate matter and heavy metals
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization WHO Global Air Quality Guidelines: Particulate Matter (PM2. 5 and PM10), Ozone, Nitrogen Dioxide, Sulfur Dioxide and Carbon Monoxide; World Health Organization: Geneva, 2021; ISBN 9789240034228. [Google Scholar]
- Wang, Q.; Kwan, M.P.; Zhou, K.; Fan, J.; Wang, Y.; Zhan, D. The Impacts of Urbanization on Fine Particulate Matter (PM2.5) Concentrations: Empirical Evidence from 135 Countries Worldwide. Environmental Pollution 2019, 247, 989–998. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization WHO Ambient Air Quality Database (Update 2024). Version 6.1. 2024.
- Addis Alemayehu, Y.; Leta Asfaw, S.; Alemu Terfie, T. Exposure to Urban Particulate Matter and Its Association with Human Health Risks. Environmental Science and Pollution Research 2020, 27491–27506. [Google Scholar] [CrossRef] [PubMed]
- Mühlfeld, C.; Rothen-Rutishauser, B.; Blank, F.; Vanhecke, D.; Ochs, M.; Gehr, P.; hlfeld, M.C. Interactions of Nanoparticles with Pulmonary Structures and Cellular Responses. Am J Physiol Lung Cell Mol Physiol 2008, 294, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhan, C.; Liu, H.; Liu, S.; Quan, J.; Liu, X.; Zhang, J.; Qu, C. Source-Specific Health Risk of PM2.5-Bound Metals in a Typical Industrial City, Central China, 2021–2022. Atmosphere (Basel) 2023, 14. [Google Scholar] [CrossRef]
- Lee, J.K.; Woo, S.Y.; Kwak, M.J.; Park, S.H.; Kim, H.D.; Lim, Y.J.; Park, J.H.; Lee, K.A. Effects of Elevated Temperature and Ozone in Brassica Juncea L.: Growth, Physiology, and Ros Accumulation. Forests 2020, 11. [Google Scholar] [CrossRef]
- Du, Y.; Gao, B.; Zhou, H.; Ju, X.; Hao, H.; Yin, S. Health Risk Assessment of Heavy Metals in Road Dusts in Urban Parks of Beijing, China. Procedia Environ Sci 2013, 18, 299–309. [Google Scholar] [CrossRef]
- Masselot, P.; Sera, F.; Schneider, R.; Kan, H.; Lavigne, É.; Stafoggia, M.; Tobias, A.; Chen, H.; Burnett, R.T.; Schwartz, J.; et al. Differential Mortality Risks Associated With PM2.5 Components A Multi-Country, Multi-City Study. Epidemiology 2022, 33, 167–175. [Google Scholar] [CrossRef]
- Lloyd, A.C.; Cackette, T.A. Diesel Engines: Environmental Impact and Control. J Air Waste Manage Assoc 2001, 51, 809–847. [Google Scholar] [CrossRef]
- Prigioniero, A.; Zuzolo, D.; Niinemets, Ü.; Postiglione, A.; Mercurio, M.; Izzo, F.; Trifuoggi, M.; Toscanesi, M.; Scarano, P.; Tartaglia, M.; et al. Particulate Matter and Polycyclic Aromatic Hydrocarbon Uptake in Relation to Leaf Surface Functional Traits in Mediterranean Evergreens: Potentials for Air Phytoremediation. J Hazard Mater 2022, 435. [Google Scholar] [CrossRef]
- Tan, X.Y.; Liu, L.; Wu, D.Y. Relationship between Leaf Dust Retention Capacity and Leaf Microstructure of Six Common Tree Species for Campus Greening. Int J Phytoremediation 2022, 24, 1213–1221. [Google Scholar] [CrossRef]
- Lee, J.K.; Kwak, M.J.; Park, S.H.; Han Dong Kim, H.D.; Lim, Y.J.; Jeong, S.G.; Choi, Y.S.; Woo, S.Y. Ozone Response of Leaf Physiological and Stomatal Characteristics in Brassica Juncea L. at Supraoptimal Temperatures. Land (Basel) 2021, 10. [Google Scholar] [CrossRef]
- Lee, J.K.; Kim, D.Y.; Park, S.H.; Woo, S.Y.; Nie, H.; Kim, S.H. Particulate Matter (Pm) Adsorption and Leaf Characteristics of Ornamental Sweet Potato (Ipomoea Batatas l.) Cultivars and Two Common Indoor Plants (Hedera Helix l. and Epipremnum Aureum Lindl. & Andre). Horticulturae 2022, 8. [Google Scholar] [CrossRef]
- Weyens, N.; Thijs, S.; Popek, R.; Witters, N.; Przybysz, A.; Espenshade, J.; Gawronska, H.; Vangronsveld, J.; Gawronski, S.W. The Role of Plant–Microbe Interactions and Their Exploitation for Phytoremediation of Air Pollutants. Int J Mol Sci 2015, 16, 25576–25604. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Liu, Y.; Feng, S.; Liu, C.; Zhong, X.; Ren, Y.; Liu, Y.; Huang, Y.; Yang, M. The Relationship between Atmospheric Particulate Matter, Leaf Surface Microstructure, and the Phyllosphere Microbial Diversity of Ulmus L. BMC Plant Biol 2024, 24. [Google Scholar] [CrossRef]
- Huchzermeyer, B.; Menghani, E.; Khardia, P.; Shilu, A. Metabolic Pathway of Natural Antioxidants, Antioxidant Enzymes and ROS Providence. Antioxidants 2022, 11. [Google Scholar] [CrossRef]
- Prigioniero, A.; Zuzolo, D.; Niinemets, Ü.; Guarino, C. Nature-Based Solutions as Tools for Air Phytoremediation: A Review of the Current Knowledge and Gaps. Environmental Pollution 2021, 277. [Google Scholar] [CrossRef] [PubMed]
- Li, S.M.; Zheng, H.X.; Zhang, X.S.; Sui, N. Cytokinins as Central Regulators during Plant Growth and Stress Response. Plant Cell Rep 2021, 40, 271–282. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, C.B.; Ren, R.M.; Jiang, J.H. Salicylic Acid Had the Potential to Enhance Tolerance in Horticultural Crops against Abiotic Stress. Front Plant Sci 2023, 14. [Google Scholar] [CrossRef]
- Chen, H.; Bullock, D.A.; Alonso, J.M.; Stepanova, A.N. To Fight or to Grow: The Balancing Role of Ethylene in Plant Abiotic Stress Responses. Plants 2022, 11. [Google Scholar] [CrossRef]
- Marzi, D.; Brunetti, P.; Saini, S.S.; Yadav, G.; Puglia, G.D.; Dello Ioio, R. Role of Transcriptional Regulation in Auxin-Mediated Response to Abiotic Stresses. Front Genet 2024, 15. [Google Scholar] [CrossRef] [PubMed]
- Morales, L.O.; Shapiguzov, A.; Safronov, O.; Leppälä, J.; Vaahtera, L.; Yarmolinsky, D.; Kollist, H.; Brosché, M. Ozone Responses in Arabidopsis: Beyond Stomatal Conductance. Plant Physiol 2021, 186, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Muneer, S.; Kim, T.H.; Choi, B.C.; Lee, B.S.; Lee, J.H. Effect of CO, NOx and SO2 on ROS Production, Photosynthesis and Ascorbate-Glutathione Pathway to Induce Fragaria×annasa as a Hyperaccumulator. Redox Biol 2014, 2, 91–98. [Google Scholar] [CrossRef]
- Li, L.; Yi, H. Differential Expression of Arabidopsis Defense-Related Genes in Response to Sulfur Dioxide. Chemosphere 2012, 87, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yi, H. Genome-Wide Transcriptome Analysis of Arabidopsis Response to Sulfur Dioxide Fumigation. Mol Genet Genomics 2014, 289, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cheng, X.; Yin, D.; Chen, D.; Luo, C.; Liu, H.; Huang, C. Advances in the Research on Plant WRKY Transcription Factors Responsive to External Stresses. Curr Issues Mol Biol 2023, 45, 2861–2880. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple Functions of Myb Transcription Factors in Abiotic Stress Responses. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF Super-Family Transcription Factors in Plant. Crit Rev Biotechnol 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Iyer, N.J.; Tang, Y.; Mahalingam, R. Physiological, Biochemical and Molecular Responses to a Combination of Drought and Ozone in Medicago Truncatula. Plant Cell Environ 2013, 36, 706–720. [Google Scholar] [CrossRef]
- Wu, S.; Wang, Y.; Zhang, J.; Wang, Y.; Yang, Y.; Chen, X.; Wang, Y. How Does Malus Crabapple Resist Ozone? Transcriptomics and Metabolomics Analyses. Ecotoxicol Environ Saf 2020, 201. [Google Scholar] [CrossRef]
- Yendrek, C.R.; Koester, R.P.; Ainsworth, E.A. A Comparative Analysis of Transcriptomic, Biochemical, and Physiological Responses to Elevated Ozone Identifies Species-Specific Mechanisms of Resilience in Legume Crops. J Exp Bot 2015, 66, 7101–7112. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, M.; Yang, Y.; Sun, P.; Zhou, S.; Kang, Y.; Xu, Y.; Yuan, X.; Feng, Z.; Jin, W. Physiological and Molecular Responses of Different Rose (Rosa Hybrida L.) Cultivars to Elevated Ozone Levels. Plant Direct 2023, 7. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The Plant Heat Stress Transcription Factors (HSFS): Structure, Regulation, and Function in Response to Abiotic Stresses. Front Plant Sci 2016, 7. [Google Scholar] [CrossRef]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC Transcription Factors in Plant Abiotic Stress Responses. Biochim Biophys Acta Gene Regul Mech 2012, 1819, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Galindo, V.; Jaramillo-Correa, J.P.; Shishkova, S.; Sandoval-Zapotitla, E.; Flores-Ortiz, C.M.; Piñero, D.; Spurgin, L.G.; Martin, C.A.; Torres-Jardón, R.; Zamora-Callejas, C.; et al. Histologic, Metabolomic, and Transcriptomic Differences in Fir Trees from a Peri-Urban Forest under Chronic Ozone Exposure. Ecol Evol 2024, 14. [Google Scholar] [CrossRef]
- Sheng, Q.; Zhou, C.; Liang, Y.; Zhang, H.; Song, M.; Zhu, Z. Elevated NO2 Induces Leaf Defensive Mechanisms in Bougainvillea Spectabilis Seedlings. Ecotoxicol Environ Saf 2022, 248. [Google Scholar] [CrossRef] [PubMed]
- Leppälä, J.; Gaupels, F.; Xu, E.; Morales, L.O.; Durner, J.; Brosché, M. Ozone and Nitrogen Dioxide Regulate Similar Gene Expression Responses in Arabidopsis but Natural Variation in the Extent of Cell Death Is Likely Controlled by Different Genetic Loci. Front Plant Sci 2022, 13. [Google Scholar] [CrossRef]
- Zhao, F.; Durner, J.; Winkler, J.B.; Traidl-Hoffmann, C.; Strom, T.M.; Ernst, D.; Frank, U. Pollen of Common Ragweed (Ambrosia Artemisiifolia L.): Illumina-Based de Novo Sequencing and Differential Transcript Expression upon Elevated NO2/O3. Environmental Pollution 2017, 224, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Vergata, C.; Contaldi, F.; Baccelli, I.; Santini, A.; Pecori, F.; Buti, M.; Mengoni, A.; Vaccaro, F.; Moura, B.B.; Ferrini, F.; et al. How Does Particulate Matter Affect Plant Transcriptome and Microbiome? Environ Exp Bot 2023, 209. [Google Scholar] [CrossRef]
- Vergata, C.; Contaldi, F.; Baccelli, I.; Buti, M.; Vangelisti, A.; Giordani, T.; Moura, B.; Ferrini, F.; Martinelli, F. The Transcriptional Mechanism Responding to Air Particulate Matter in Laurus Nobilis (L.). Environ Exp Bot 2023, 210. [Google Scholar] [CrossRef]
- Zhao, J.; Yi, H. Genome-Wide Transcriptome Analysis of Arabidopsis Response to Sulfur Dioxide Fumigation. Mol Genet Genomics 2014, 289, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Permana, B.H.; Krobthong, S.; Yingchutrakul, Y.; Saithong, T.; Thiravetyan, P.; Treesubsuntorn, C. Evidence of Brassinosteroid Signalling and Alternate Carbon Metabolism Pathway in the Particulate Matter and Volatile Organic Compound Stress Response of Sansevieria Trifasciata. Environ Exp Bot 2023, 205. [Google Scholar] [CrossRef]
- Treesubsuntorn, C.; Setiawan, G.D.; Permana, B.H.; Citra, Y.; Krobthong, S.; Yingchutrakul, Y.; Siswanto, D.; Thiravetyan, P. Particulate Matter and Volatile Organic Compound Phytoremediation by Perennial Plants: Affecting Factors and Plant Stress Response. Science of the Total Environment 2021, 794. [Google Scholar] [CrossRef]
- Permana, B.H.; Krobthong, S.; Yingchutrakul, Y.; Thiravetyan, P.; Treesubsuntorn, C. Sansevieria Trifasciata’s Specific Metabolite Improves Tolerance and Efficiency for Particulate Matter and Volatile Organic Compound Removal. Environmental Pollution 2024, 355. [Google Scholar] [CrossRef] [PubMed]
- Kerchev, P.; van der Meer, T.; Sujeeth, N.; Verlee, A.; Stevens, C. V.; Van Breusegem, F.; Gechev, T. Molecular Priming as an Approach to Induce Tolerance against Abiotic and Oxidative Stresses in Crop Plants. Biotechnol Adv 2020, 40. [Google Scholar] [CrossRef] [PubMed]
- Dinh, Q.P.; Novirsa, R.; Jeong, H.; Cahya Nugraha, W.; Addai-Arhin, S.; Viet, P.H.; Tominaga, N.; Ishibashi, Y.; Arizono, K. Mercury, Cadmium, and Lead in Cigarettes from International Markets: Concentrations, Distributions and Absorption Ability of Filters; 2021.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





