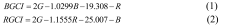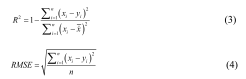1. Introduction
Fraction vegetation coverage (FVC) refers to the ratio of the vertical projection of the canopy area to the ground surface area per unit area. FVC is expressed as a fraction or percentage of the reference area and is influenced by field management practices such as wnitrogen application and irrigation. FVC is also closely related to crop yield [
1]. Monitoring crop vegetation cover is highly important for crop management [
2]. To date, high-resolution remote sensing images have been widely used in many fields to obtain spatial information, providing a more accurate information source for vegetation monitoring [
3]. Remote sensing images facilitate the extraction and monitoring of vegetation information at the field scale.
Estimating crop FVC based on UAV remote sensing technology commonly uses machine learning and threshold-based methods [
4]. Although machine learning methods provide higher accuracy in estimating crop FVC, they often require a large number of samples for training to improve accuracy in large-scale crop coverage estimation. This makes sample selection inefficient and highly susceptible to the influence of human sample selection [
5]. Threshold-based methods are characterized by their simplicity, efficiency, and accuracy; however, the effectiveness of threshold extraction greatly affects the estimation accuracy of crop FVC, making the precision of threshold extraction crucial for the threshold-based estimation of FVC [
6]. The Otsu method is a commonly used technique for determining thresholds and has been widely applied in crop classification, but it can result in undersegmentation in certain situations [
7]. Additionally, the resolution of UAV multispectral remote sensing images is relatively low, making the Otsu method unsuitable. The bimodal histogram method and maximum entropy threshold method are commonly used for estimating crop FVC [
8]. For remote sensing images containing only crops and soil background, there is usually a single peak distribution, whereas for complex images with more than two peak distributions, the bimodal histogram method is not suitable. Noise in the images can affect the maximum entropy value, leading to lower accuracy in FVC estimation [
9]. Therefore, selecting a threshold extraction method that is stable, accurate, and easy to operate is essential for estimating crop FVC.
The Gaussian mixture model (GMM) threshold method can effectively address the issues present in the aforementioned threshold methods [
10]. This method assumes that the target object and background follow a GMM distribution in a colour feature and uses the intersection of the GMM as the classification threshold [
11]. The GMM threshold method can achieve ideal crop FVC estimation accuracy to some extent. However, using colour features for threshold determination often results in low FVC estimation accuracy when crop plants are small or during the near-closure period. Additionally, the GMM threshold method requires appropriate samples to determine the classification threshold, and relying solely on manually selected samples reduces the efficiency of the algorithm [
12]. Combining machine learning with the GMM threshold method can achieve rapid FVC estimation and better accuracy [
13]. The relationship between the number of samples selected by machine learning and the balance between FVC estimation accuracy and efficiency has not yet been explored, and whether the texture features of crop plants and the background conform to a GMM distribution remains unknown. Therefore, further research on the GMM threshold method is needed to propose a stable, efficient, and high-accuracy crop FVC estimation method.
The main research objectives are as follows: (1) To construct a new vegetation index capable of achieving high-precision vegetation cover extraction for potatoes throughout their entire growth period by combining machine learning and threshold extraction methods to determine its classification threshold. (2) To select vegetation indices and texture features that have a high correlation with FVC and achieve high-precision extraction of potato vegetation cover based on the classification threshold. (3) To establish a vegetation cover estimation model for the entire growth period of potatoes.
2. Materials and Methods
2.1. Experimental Design
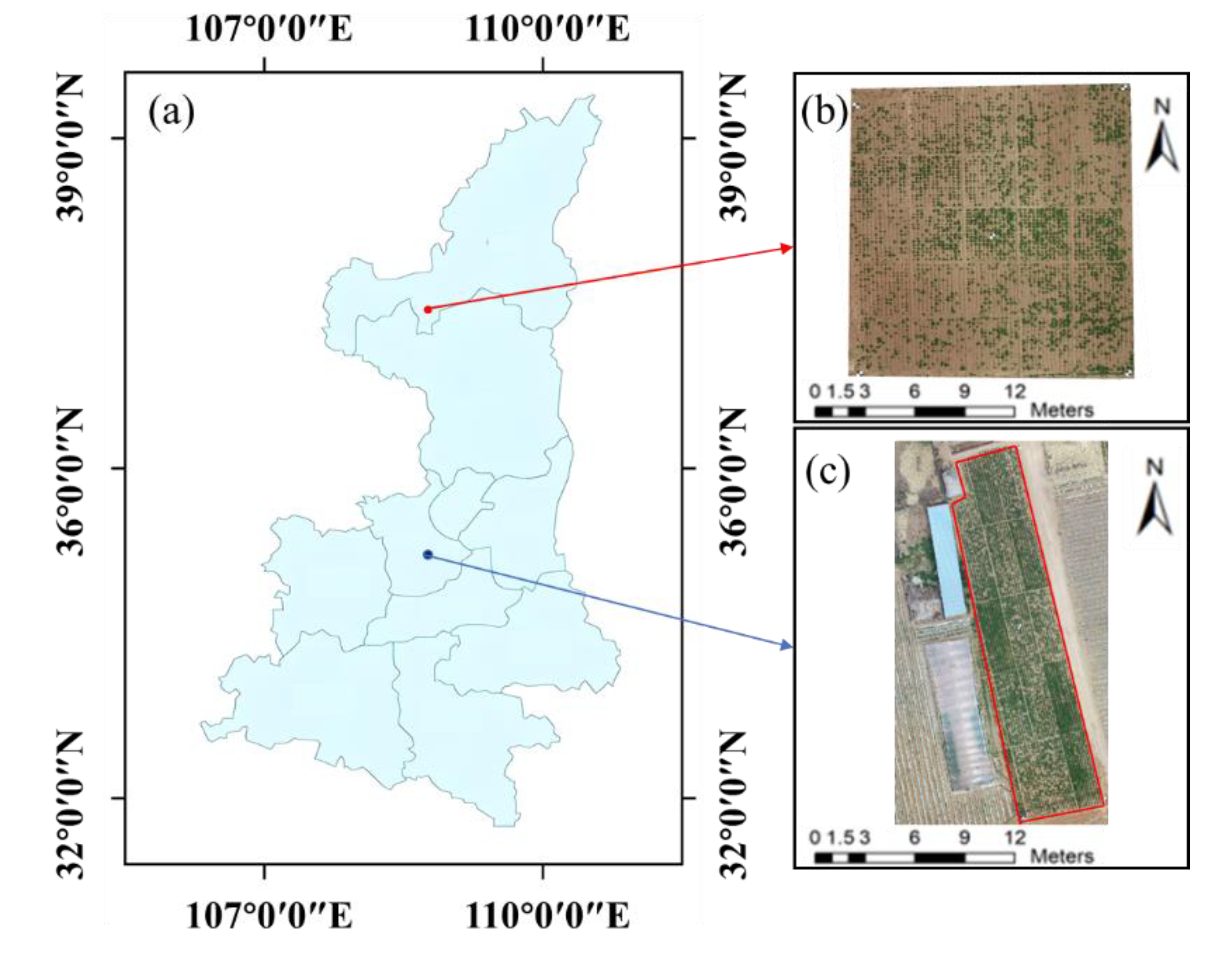
To increase sample diversity and improve the reliability of the FVC estimation model, two experimental areas were designed for field experiments. Experiment Area A is located in the Yangling Demonstration Zone, Shaanxi Province (latitude 34°18'10" N, longitude 108°5'14.33" E). The soil pH, organic matter, and nitrogen concentration in this experimental area were 7.2, 27.31 g/kg, and 1.96 g/kg, respectively. Potatoes of the Jinxu 16 variety were planted in May 2021, with planting spacing, row spacing, and planting depths of 0.6 m, 0.5 m, and 10 cm, respectively, and the potatoes were harvested in September 2021. Five nitrogen levels were set (N1: 0 kg/ha; N2: 75 kg/ha; N3: 150 kg/ha; N4: 225 kg/ha; and N5: 300 kg/ha). Each treatment was repeated five times, totalling 25 plots, each measuring 7.5 metres in length and width. Five ground control points were systematically set around the experimental area for the georeferencing of multiperiod images and the preprocessing of stitched images.
Experiment Area B is located in Ningtiaoliang town, Jingbian County, Yulin city, Shaanxi Province (latitude 37°33'55.60" N, longitude 108°22'7.46" E). The soil pH, organic matter, and nitrogen concentration in this experimental area were 7.7, 16.35 g/kg, and 0.89 g/kg, respectively. Three early-maturing potato varieties (Hisen 6, 226, and V7) were used in this experiment, with five nitrogen levels (N1: 0 kg/ha; N2: 60 kg/ha; N3: 120 kg/ha; N4: 180 kg/ha; and N5: 240 kg/ha). Each treatment was replicated three times, totalling 15 plots, each measuring 112 m2 (14 m × 8 m). The remaining treatments were the same as those in Experiment Area A. Potatoes were planted in May 2022 and harvested in August 2022, with a total growth cycle of approximately 90 days. The information about Experiment Area B is shown in
Figure 1.
2.2. Visible Light Image Acquisition and Preprocessing
Considering the impact of sensors on estimating potato phenotypic information and nitrogen nutrition diagnosis and to further explore the applicability of the model, Experiment Area A utilized a DJI Phantom 4 RTK drone (SZ DJI Technology Co., Ltd., Shenzhen, China) to capture visible light images during the tuber formation and tuber enlargement stages of potatoes. The drone has an aperture range from f/2.8 to f/11, an ISO range of 100-3200 (automatic) and 100-12800 (manual), a maximum photo resolution of 5472×3648, a flight time of 30 minutes, and a positioning accuracy (RMS) of 1.5 cm + 1 ppm vertically and 1 cm + 1 ppm horizontally. DJI GS RTK software was used for drone flight route planning, with the flight altitude set at 30 m and the flight speed set at 3 m/s. The drone captured images vertically downwards with an overlap of 85% in both heading and sideways. All flights were conducted at noon under clear, windless conditions. During the flights, five ground control points were visible in the images, and their GCP values were measured using a differential global positioning system (DGPS) to further correct the image positioning information. The ground resolution of the images is 0.95 cm/pixel.
Experiment B utilized the DJI Phantom 4 Pro drone to capture visible light images of potato canopies during the tuber formation, tuber enlargement, and tuber maturity stages. The drone has a shutter speed range of 1/2000-1/8000 s, with other parameters identical to those of the DJI Phantom 4 RTK drone. Atrtizure software was used for drone flight path planning, with flight parameters identical to those used in Experiment A.
Preprocessing of remote-sensing images is an indispensable process for monitoring crop growth and nutrient diagnosis [
14]. Common preprocessing methods for UAV remote sensing technology include image stitching, image registration, and cropping [
15]. This study utilized Pix4D mapper software developed by the Swiss company Pix4D AG to stitch all visible light images. The main workflow was as follows: (i) importing images, (ii) importing control point coordinates for point cloud generation, (iii) applying a one-click automatic process for point cloud extraction and 3D model generation, and (iv) generating point cloud data, the DSM, and the DOM [
16]. ENVI software Version 5.3 (Exelis Visual Information Solutions, Boulder, CO, USA) was used for image registration of visible light, multispectral orthoimages, and elevation images. The corresponding experimental field areas for the entire growth period of potatoes were cropped using ENVI software.
2.3. Selection and Construction of Remote Sensing Feature Elements
Experiment B included more potato data covering various growth stages and a richer variety of potato species. The potential impact of variety on the accuracy of vegetation cover extraction was considered. This study focused on researching vegetation cover estimation methods using Experiment B and validated them using Experiment A. To reduce the influence of noise on data processing, convolutional low-pass filtering was applied to denoise the visible light images of potatoes captured by drones throughout their growth period. The convolution kernel size was set to 5, with a weighted return value of 0. After denoising, vegetation index calculations and texture feature extraction were performed.
2.3.1. Selection and Construction of Visible Light Band Vegetation Indices
Previous studies have constructed numerous vegetation indices based on visible light images to assess crop vegetation cover. This study selected eight vegetation indices, namely, the GRVI, EXG, RGBVI, MGRVI, NGRVI, NGBDI, GLI, and TRVI[
17,
18,
19,
20]. However, the construction process of these common vegetation indices in the visible light band did not adequately consider the interrelationships among the three bands. Changes in individual bands often had a significant impact on the constructed vegetation indices. Therefore, the construction of new vegetation indices using multiband combinations can significantly improve the effectiveness of cover detection.
The visible light remote sensing images of the tuber enlargement stage of potatoes in Experiment B were visually interpreted. The experimental field encompassed various land cover types, contributing to a complex field environment. To effectively distinguish between these land cover types, 100 regions of interest (ROIs) were selected for each type using ENVI software. Subsequently, the characteristic values of the blue, green, and red bands for each land cover type were calculated and summarized, as shown in
Table 1.
As shown in
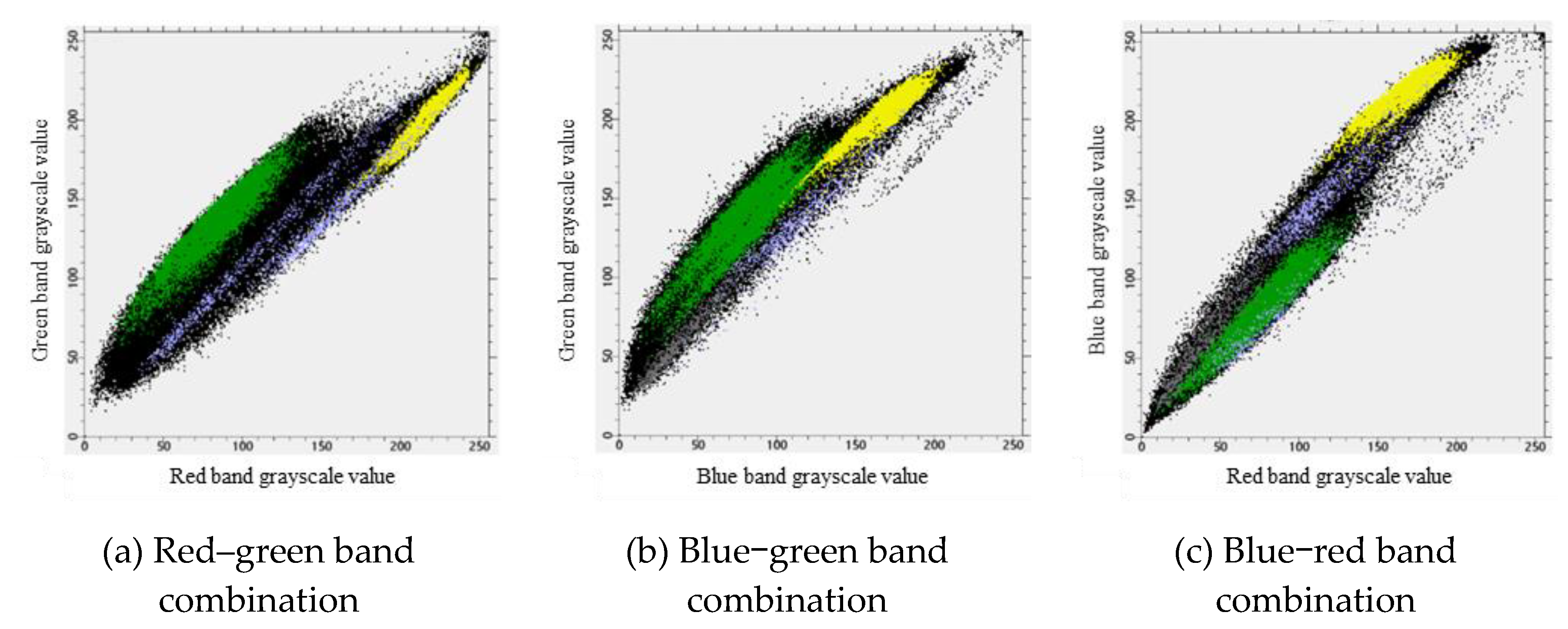
Table 1, there is an overlap between the grayscale values of potato plants in the blue band and shadows and between the grayscale values in the green band and those in the drip irrigation tape band. Additionally, there is a partial overlap in the grayscale values in the blue band with both shadows and drip irrigation tape. It is difficult to distinguish potato plants using a single band. Scatterplots of red‒green–blue for potato plants, soil, shadows, and drip irrigation tape (
Figure 2) were generated.
Figure 2 clearly shows that there are distinct boundaries between potato plants and other land cover categories under the red–green and blue–green combinations. Potato plants are mainly concentrated in the lower-left region of the scatterplots, while other land cover categories are predominantly located in the upper-right region. However, the blue‒red combination was unable to identify potato plants within the shadows.
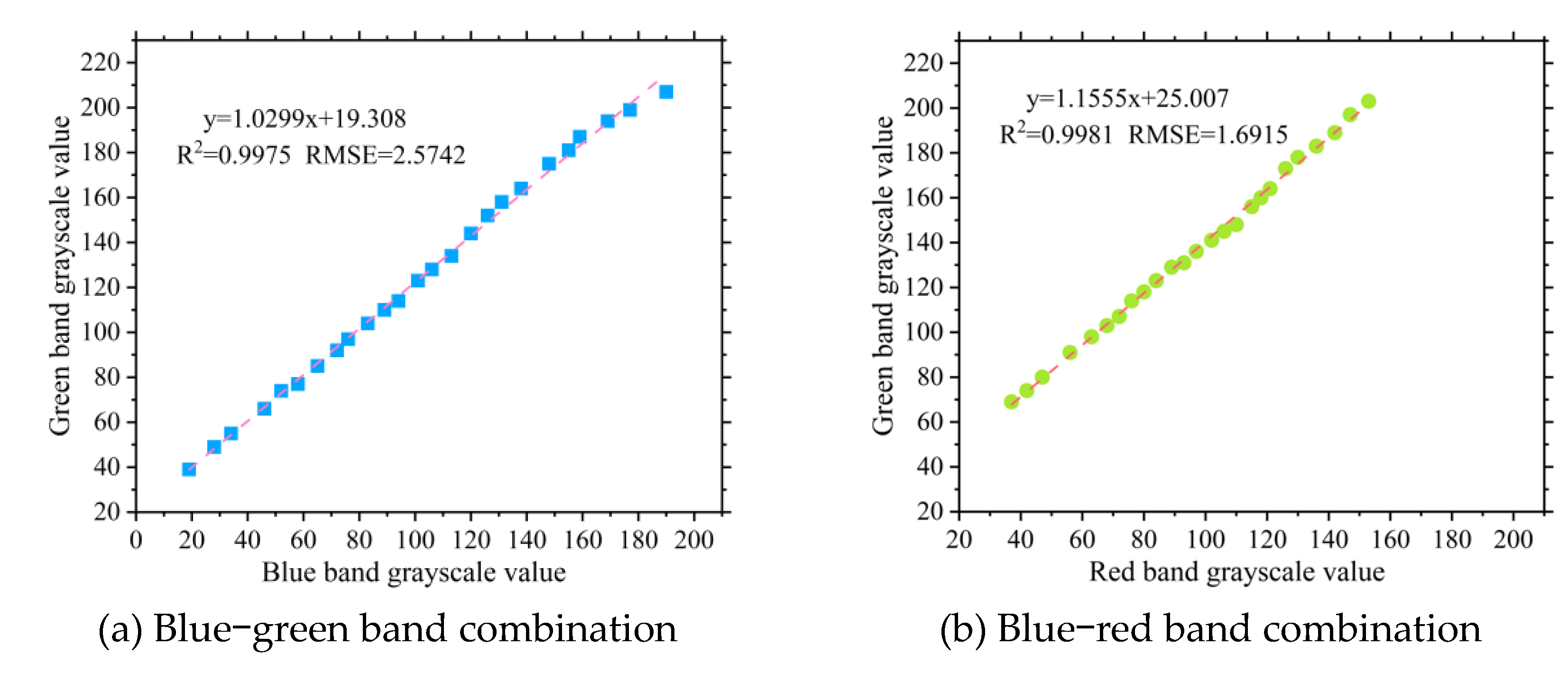
The fitted boundary functions were then used to construct new combination indices: the combination index of blue and green bands (BGCI) and the combination index of red and green bands (RGCI). The corresponding formulas are shown as Equation 1 and Equation 2, respectively.
2.3.2. Texture Feature Extraction
Texture is a visual characteristic that reflects the homogeneous appearance of crops, embodying the structural arrangement attributes of periodic changes on the surface of objects in the image [
21]. Currently, texture features have been widely applied in crop classification and prediction [
22]. The gray level co-occurrence matrix (GLCM) is a common method for calculating image texture features, mainly by extracting texture features through the calculation of conditional density probability functions of the grayscale levels of objects in the image [
9]. In this study, ENVI software was used to calculate eight common texture features, namely, the mean, variance, homogeneity, contrast, dissimilarity, entropy, angular second moment, and correlation, within a 7×7 window.
2.4. Fraction Vegetation Coverage Extraction Method
2.4.1. Determination of Extraction Thresholds
The vegetation index intersection method can effectively achieve vegetation coverage extraction. However, the common applications and vegetation indices of the vegetation index intersection method are currently designed for specific environments. Further investigations are needed to determine the vegetation coverage of potatoes throughout their entire growth period [
23]. In this study, the threshold determination process based on the vegetation index intersection method was divided into two main parts; one part involved clipping the entire experimental potato field and performing supervised classification on the clipped area. The other part combined the supervised classification results with the Gaussian mixture model to further determine the extraction threshold for potato vegetation coverage. This study presents solved thresholds for the constructed RGCI, BGCI, and eight other common vegetation indices solved using the vegetation index intersection method.
The support vector machine (SVM) is currently widely used in remote sensing image classification and effectively addresses issues such as small samples, nonlinearity, and high dimensionality. It also has strong generalization capabilities, with the type of kernel function being the radial basis function. To efficiently process sample data, the SVM was used for supervised classification of the cropped images. The cropping range of the region was set to 7 m × 7 m. The specific process for supervised classification using the SVM was as follows:
(1) The orthophoto images of the potato field were cropped and 40 regions of interest (ROIs) of potato plants and the background on the orthophoto images were selected. The separability of the selected samples was calculated.
(2) Based on the separability of the samples from the three growth stages of potatoes, the reasonableness of the selected samples was assessed.
(3) Additionally, 30 ROIs of potato plants and the background were selected to verify the SVM classification results using a confusion matrix.
2.4.2. Validation of Vegetation Coverage Extraction Accuracy
Using the determined thresholds, the vegetation coverage of potatoes was extracted for each growth stage. In this study, 300 regions of interest (ROIs) of potato plants and the background were selected outside the cropped areas of the potato experimental field using visual interpretation. A confusion matrix was used to validate the vegetation coverage extracted by the vegetation index intersection method for the three potato growth stages. The validation was evaluated using the kappa coefficient and overall accuracy.
2.5. Establishment of the Fraction Vegetation Coverage Estimation Model
2.5.1. Vegetation Index and Texture Feature Selection
Various potential covariates can be used for vegetation coverage estimation, but applying all available covariates can reduce the data processing efficiency. Therefore, it is necessary to filter out important features before establishing the estimation model.
Based on the best potato vegetation coverage results from Experiment B, three sampling areas were selected from each of the 15 experimental plots to measure vegetation coverage during the three growth stages. The visible light image consists of three bands, red, green, and blue, each containing eight texture features. To facilitate presentation, the 24 texture features are shown in
Table 2. The mean values of the corresponding 10 vegetation indices and 24 texture features were calculated for each sampling area.
To improve the computational efficiency of the model, this study utilized random forest and Pearson correlation coefficient methods to select 10 vegetation indices and 24 texture features and obtained important remote sensing factors. Random forest (RF) is an algorithm that measures feature importance by randomly replacing each feature [
24]. When a feature is highly important, the prediction error rate of the RF model will increase. The change in the error rate of the out-of-bag data before and after feature replacement is applied to evaluate each feature, and their importance scores are obtained. This study used the Python 3.6 library (Scikit Learn package) to implement this algorithm and selected repeated tenfold cross-validation to optimize the accuracy of the RF model. Subsets with minimal impact on the random forest model were used as subsets of predictors.
The Pearson correlation coefficient was used to detect the linear correlation between continuous variables, with values ranging from -1 to 1; positive and negative values indicate positive and negative correlations, respectively. The larger the absolute value is, the greater the linear correlation.
2.5.2. Establishment and Validation of the Fraction Vegetation Coverage Estimation Model
This study used RF and Pearson correlation coefficients to assess the importance of vegetation indices and texture features. Based on the top 6 features selected by these algorithms, a model for estimating the FVC was constructed. For each potato growth stage, a total of 45 vegetation coverage extraction results were obtained. Thirty randomly selected data points were used to construct the vegetation coverage estimation model, while the remaining 15 data points were used for model validation. Linear fitting was used to establish a vegetation coverage estimation model for potato plants and validate its performance.
Common model evaluation metrics include the coefficient of determination (R²) and root mean square error (RMSE). A higher R² value and a smaller RMSE value indicate higher model accuracy. The formulas for calculating R² and RMSE are shown in Equations 3 and 4, respectively.
In the equations, xi represents the measured values, yi represents the predicted values, x represents the mean of the measured values, and n represents the sample size.
3. Results
3.1. Extraction of FVC Thresholds Based on the Vegetation Index Intersection Method
To determine the effectiveness of the SVM in classifying plants and the background in images, the classification performance of the SVM was validated using a confusion matrix, as shown in
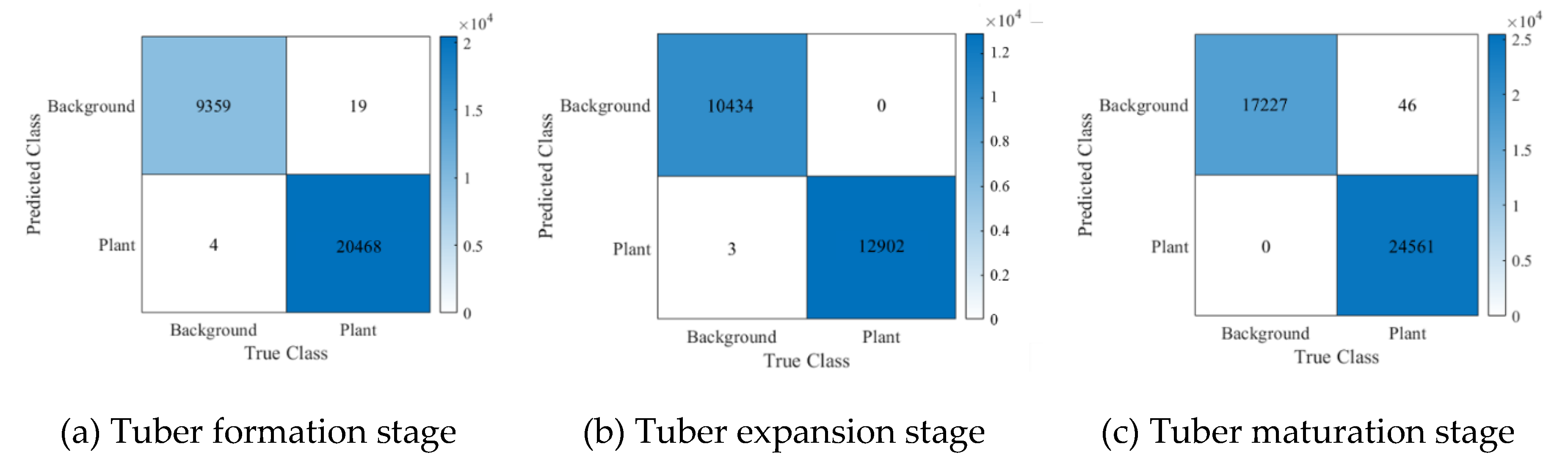
Figure 4. The overall classification accuracy for the experimental field during the tuber formation stage was 99.922%, with a kappa coefficient of 0.9982. During the tuber enlargement stage, the overall classification accuracy was 99.9871%, with a kappa coefficient of 0.9997. For the maturity stage, the overall classification accuracy was 99.8899%, with a kappa coefficient of 0.9977. The validation results from the confusion matrix indicate that the SVM achieved high classification accuracy in the small-potato regions across all three stages. The overall classification accuracy for the three growth stages of potatoes was greater than 99%, and the kappa coefficient was greater than 0.99. Therefore, this method can be used to determine the FVC extraction threshold for potatoes.
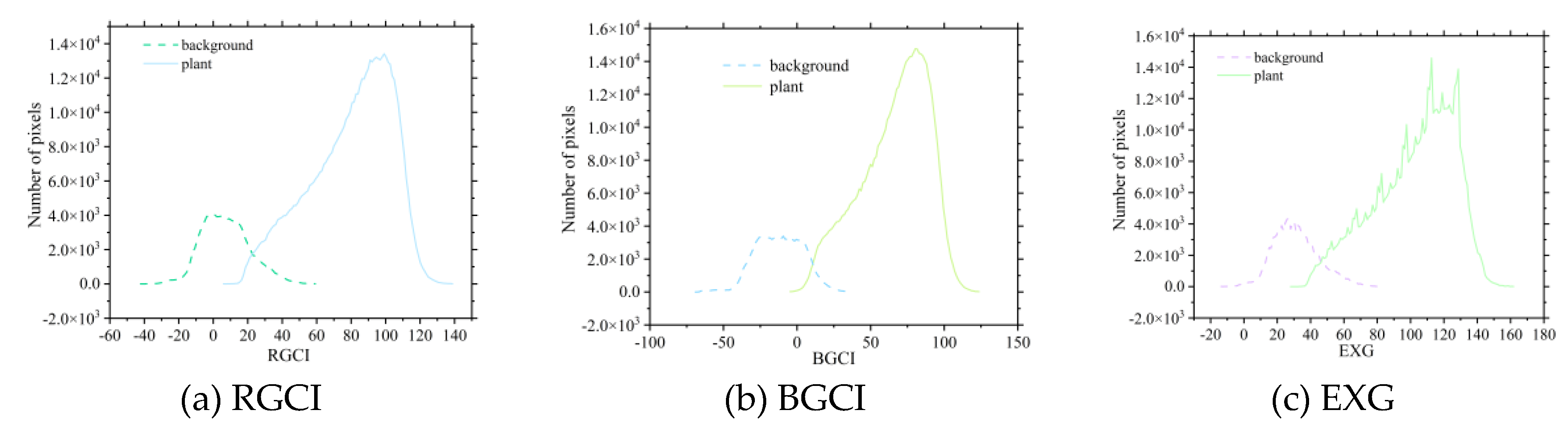
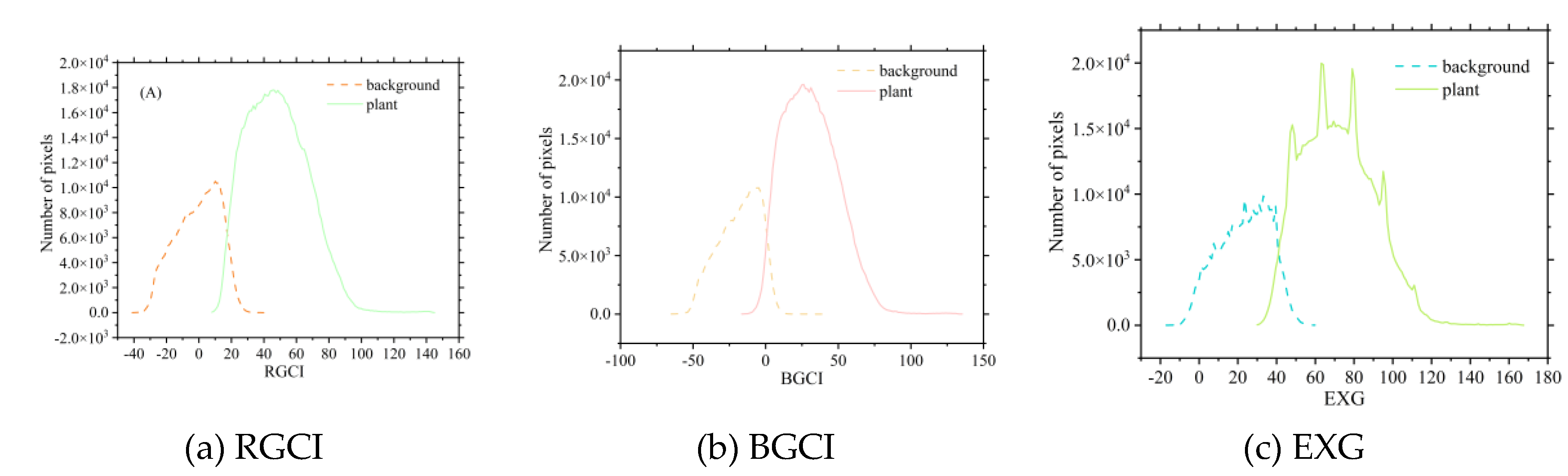
The extraction thresholds for various vegetation indices were calculated based on the vegetation index intersection method. Since the newly constructed vegetation indices, RGCI and BGCI, are similar in principle to the EXG vegetation index, this study focused on comparing the threshold extraction results of these three indices. As shown in
Figure 5,
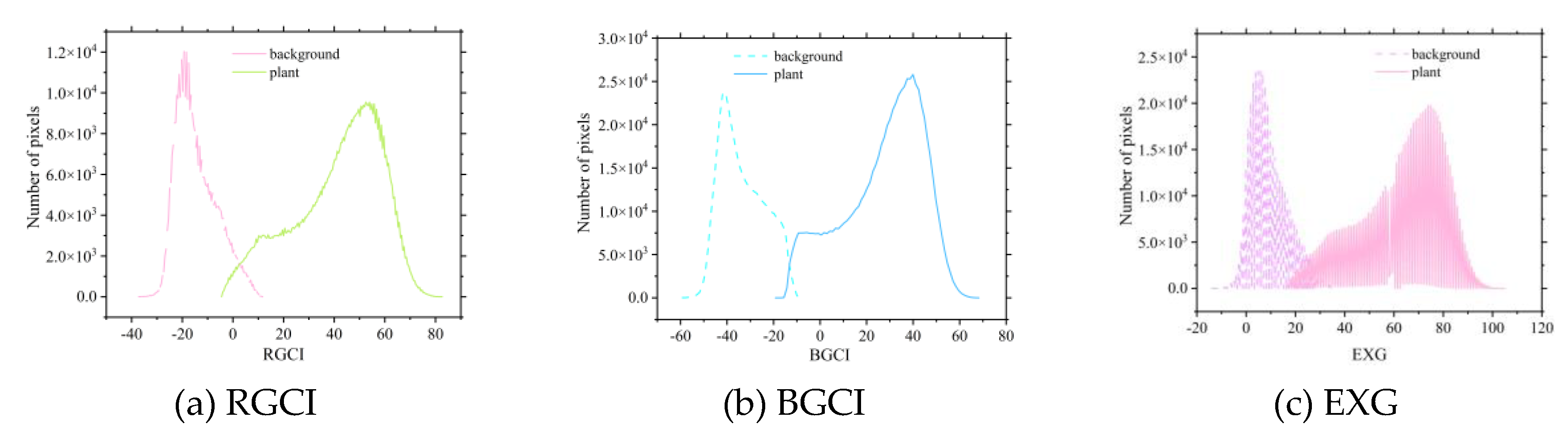
Figure 6, and
Figure 7, the EXG histograms exhibit irregularities during the three growth stages of the potatoes. In particular, in the tuber formation stage, multiple intersections in the EXG histogram prevent effective determination of the extraction threshold for potato vegetation coverage. The newly constructed RGCI and BGCI vegetation indices effectively address the irregularity issues of the EXG index and can better determine the classification thresholds for the background and potato plants. The coverage extraction thresholds of the other selected vegetation indices can also be determined via the vegetation index intersection method. The extraction threshold results for each vegetation index corresponding to the three growth stages of potatoes are shown in
Table 3.
3.2. Extraction Results of Potato FVC
The determined thresholds were applied to extract the vegetation coverage of the entire experimental field for the corresponding three growth stages of potatoes. Pixels with values greater than the threshold were classified as potato plant pixels, while those with values less than the threshold were classified as background pixels. Further calculations were performed using Formula 5 to calculate the vegetation coverage of the potatoes. The extraction results of potato vegetation coverage for the three stages are shown in
Figure 8.
In the formula, , , and represent potato vegetation coverage, the number of potato plant pixels, and the number of soil pixels, respectively.
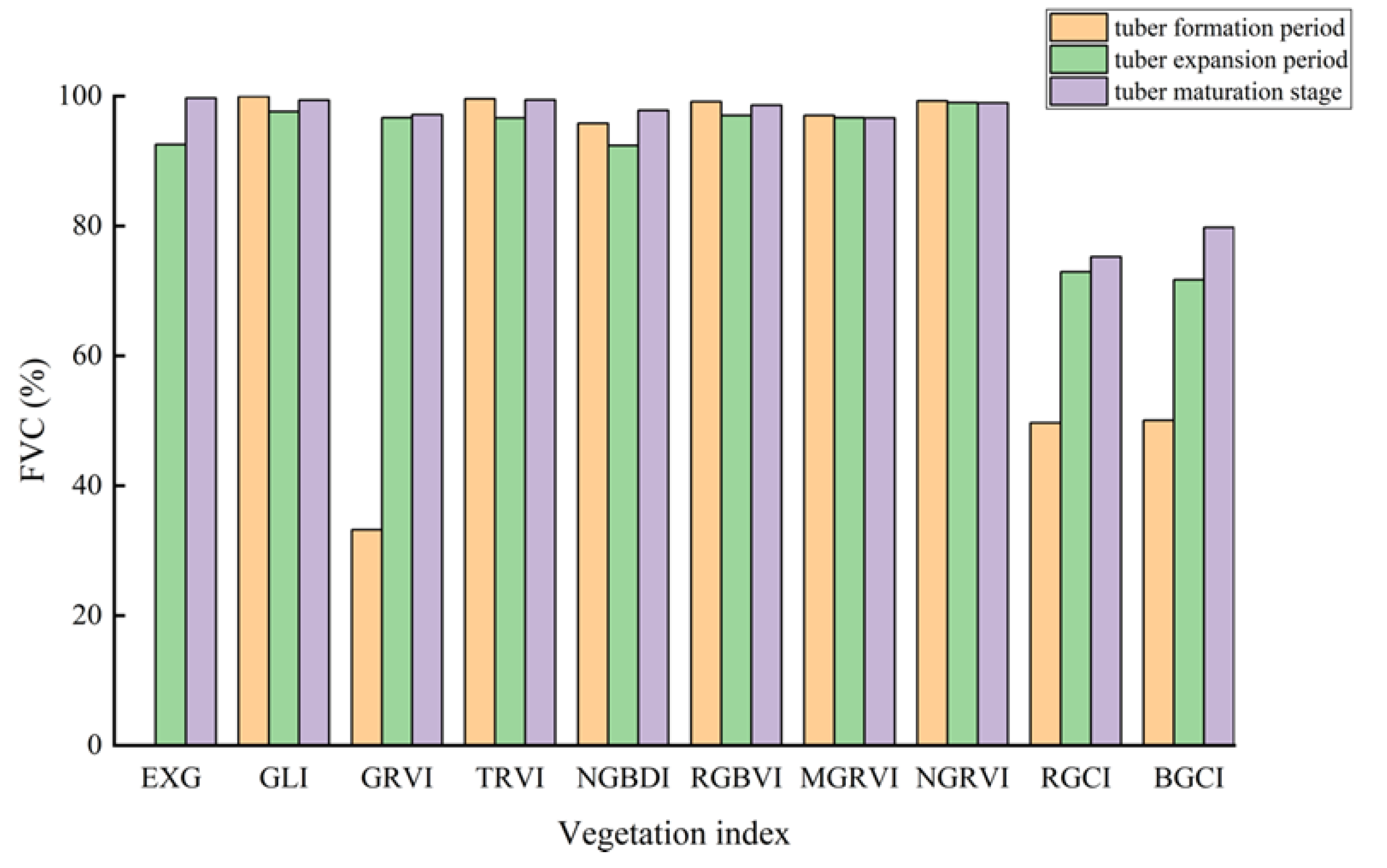
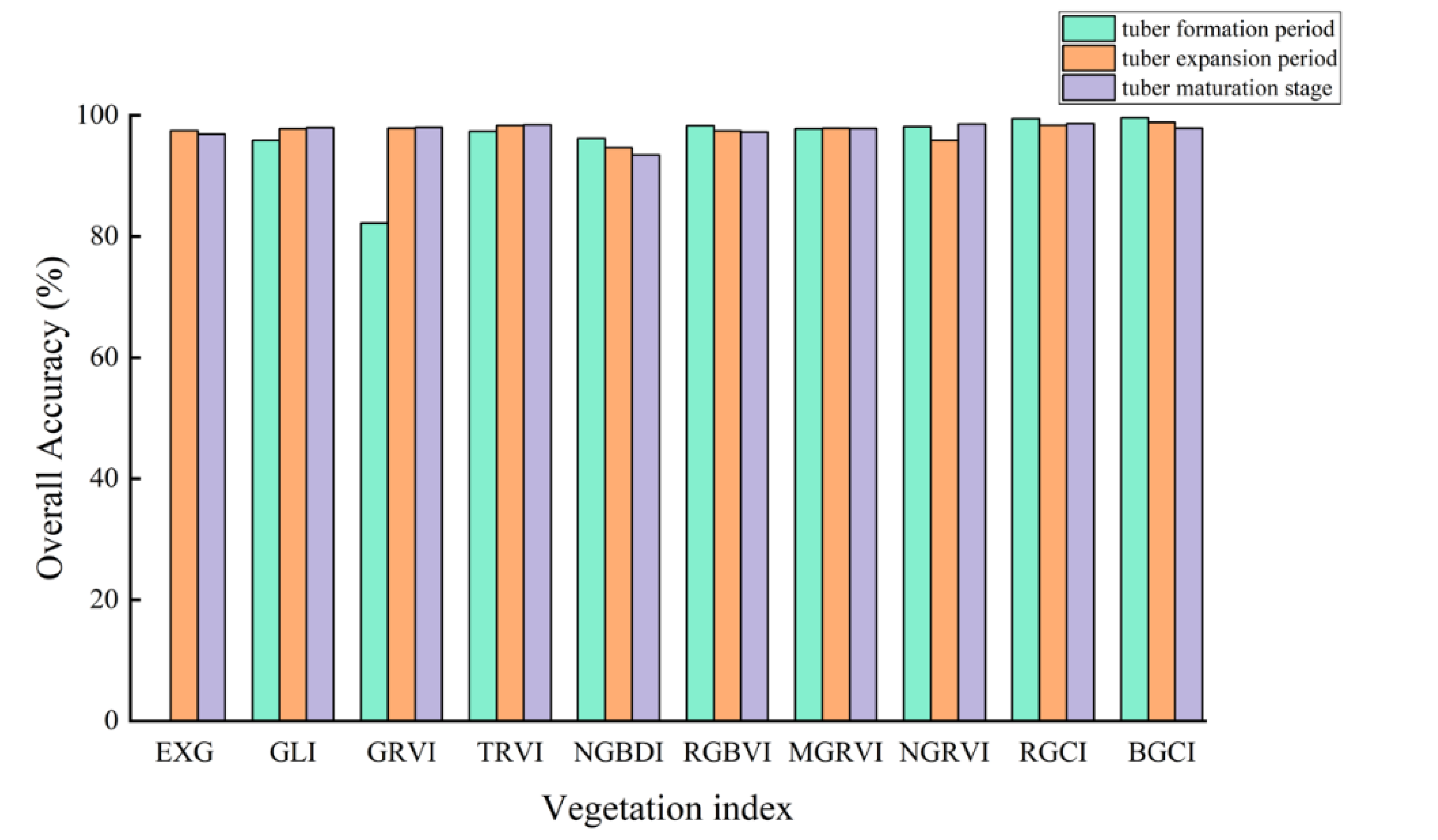
Figure 9 shows the accuracy verification results of potato vegetation coverage extraction based on the vegetation index intersection method. During the three growth stages of the potatoes, except for the GRVI, which had lower accuracy in extracting vegetation coverage during the tuber formation stage with an overall accuracy below 90% and a kappa coefficient below 0.5, the remaining vegetation indices achieved higher accuracy in estimating vegetation coverage.
During the tuber formation stage, the BGCI achieved the highest accuracy in terms of vegetation coverage extraction among all the vegetation indices, with an overall accuracy of 99.6079% and a kappa coefficient of 0.9898. Similarly, during the tuber enlargement stage, the BGCI achieved the highest accuracy in terms of vegetation coverage extraction, with an overall accuracy and kappa coefficient of 98.8405% and 0.9753, respectively. In the potato maturity stage, the RGCI achieved the highest accuracy in estimating vegetation coverage, with an overall accuracy and kappa coefficient of 98.6336% and 0.9712, respectively.
Through comparison, the RGCI and BGCI demonstrated better accuracy in extracting potato vegetation coverage based on the vegetation index intersection method than did the other vegetation indices. Comparing the vegetation coverage extraction results of the first two stages, the overall accuracy of the BGCI was higher than that of the RGCI by 0.1741% and 0.4866%, respectively. However, in the potato maturity stage, the overall classification accuracy of the BGCI was lower than that of the RGCI by 0.7719%.
3.3. Potato Vegetation Coverage Estimation Model
To ensure the effective estimation accuracy of the potato FVC estimation model throughout the entire growth period, this study selected the BGCI combined with the vegetation index intersection method to extract vegetation coverage during the tuber formation and tuber enlargement stages of the potatoes. The RGCI combined with the vegetation index intersection method was chosen to extract vegetation coverage during the maturity stage. The extraction results are shown in
Figure 10.
During the feature selection process for individual potato growth stages, the limited amount of data from each stage may lead to nonrepresentative feature selection results. Therefore, this study conducted feature selection for the entire growth period of the potato plants. As shown in
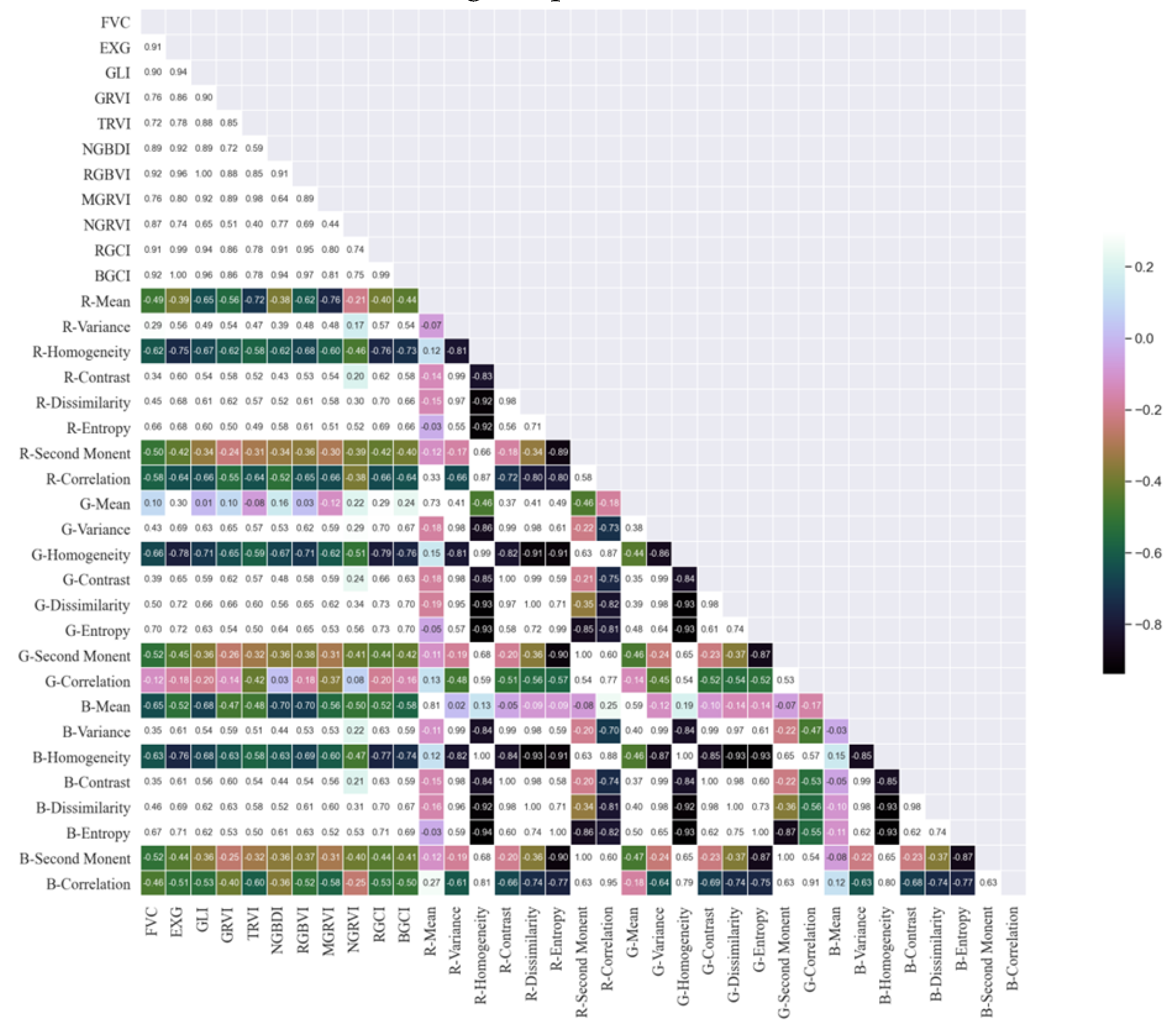
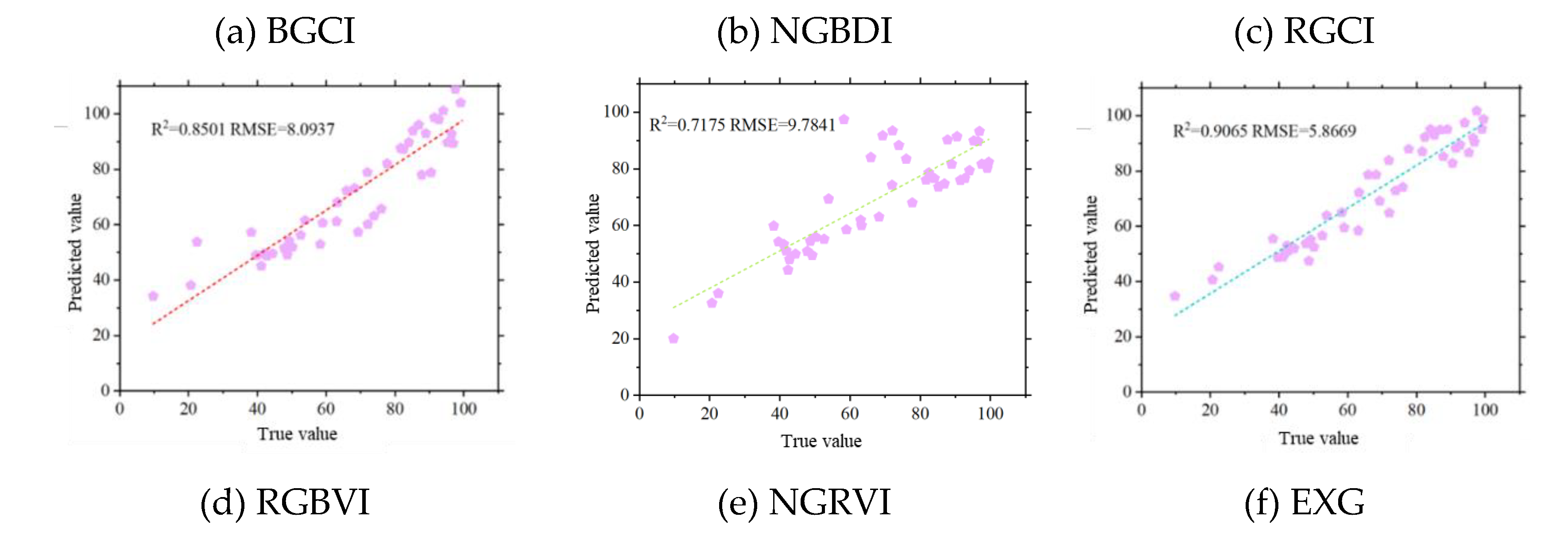
Figure 11, the Pearson correlation coefficient method over the entire potato growth period revealed that the BGCI and RGBVI exhibited the highest correlation with FVC (R=0.92), followed by the EXG and RGCI, with correlation coefficients of 0.91 for both indices. Next in line were the NGRVI (R=0.90) and NGBDI (R=0.89).
Among the 24 texture features, B-correlation was the most highly correlated feature with FVC (R=0.73), but its correlation coefficient was still less than 0.75. Therefore, the results from the Pearson correlation coefficient method indicated that texture features are not suitable for establishing the FVC estimation model for the tuber formation stage of potatoes.
The results of the random forest feature selection also showed that the BGCI obtained the highest score, with an importance of 0.1969. Next in line were the NGBDI, RGCI, RGBVI, NGRVI, and EXG, with importances of 0.1241, 0.0894, 0.0554, 0.0483, and 0.0332, respectively. These scores accounted for less than 0.1 of the total importance. Although the feature importance ranking from random forest selection was not entirely consistent with that of the Pearson correlation coefficient method, both methods identified the MGRVI, BGCI, TRVI, NGRVI, RGCI, and NGRVI as the six remote sensing feature factors with relatively high correlations to potato FVC. These selected vegetation indices were used to estimate potato vegetation coverage during the tuber formation stage.
Using the six selected vegetation indices, the estimation of the vegetation coverage during the potato tuber formation period was carried out, and the estimation results were analysed and evaluated. Evaluating the accuracy of FVC estimation of the model using R
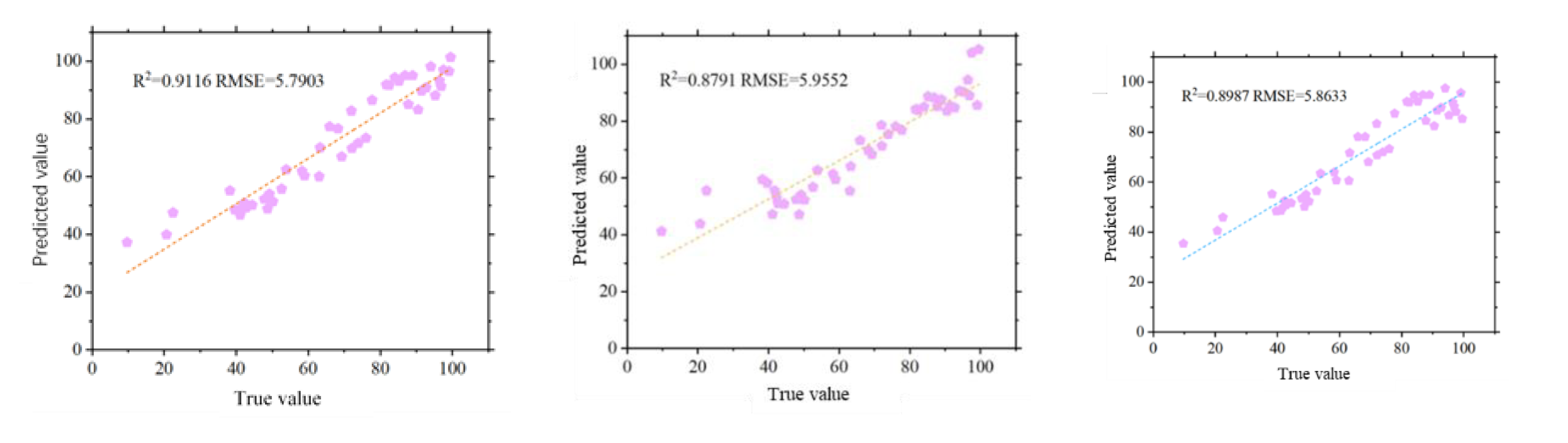
2 and RMSE values, it was observed that with the increase in vegetation coverage data volume throughout the potato growth period, the BGCI and RGCI achieved better accuracy in FVC estimation. As shown in
Figure 12, the BGCI obtained the highest FVC estimation accuracy during the entire potato growth period (R
2=0.9116, RMSE=5.7903), followed by the EXG (R
2=0.9065, RMSE=5.8669) and RGCI (R
2=0.8987, RMSE=5.8633). The NGRVI had the lowest FVC estimation accuracy (R
2=0.7175, RMSE=9.7841).
4. Discussion
4.1. Comparison of Different Threshold Extraction Methods
Common methods for estimating vegetation coverage include machine learning and thresholding methods [
25]. Threshold-based methods are known for their simplicity, efficiency, and accuracy and have been successfully applied in crop vegetation coverage extraction studies based on UAV remote sensing images [
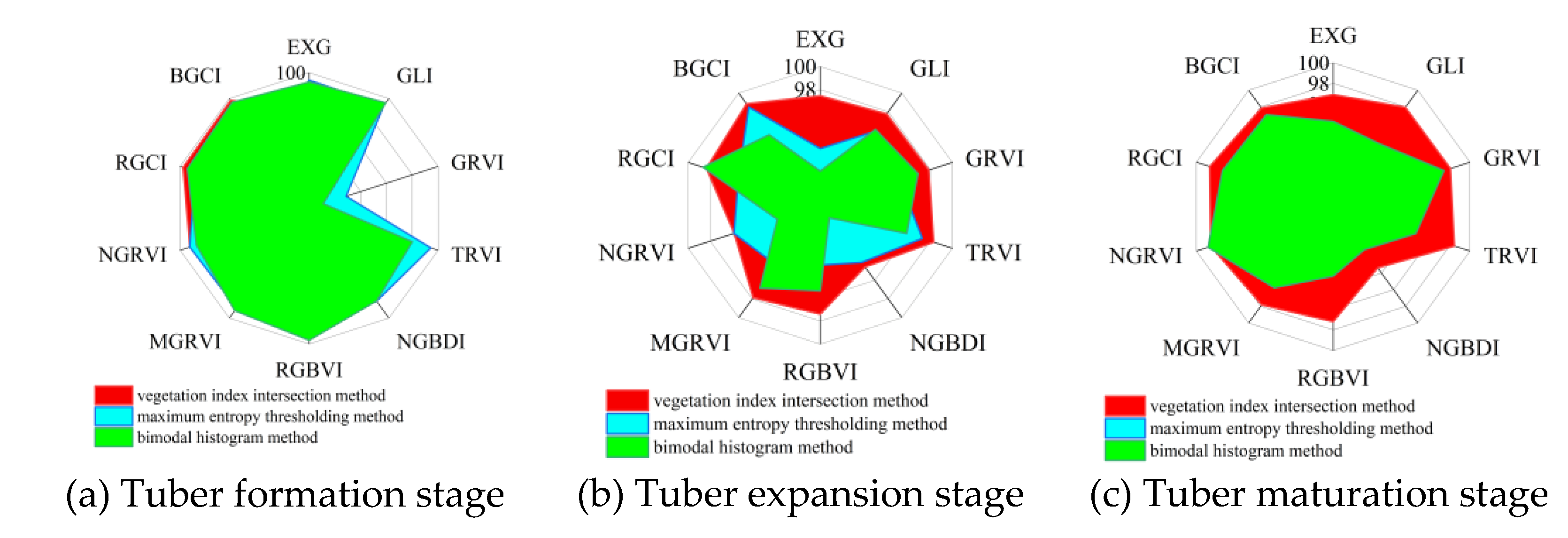
26]. In crop vegetation coverage extraction, the maximum entropy thresholding method and the bimodal histogram method are two widely used methods for FVC extraction. To better elucidate the effectiveness of the two vegetation indices constructed in this study under different methods, 10 vegetation indices were combined with the maximum entropy thresholding method and the bimodal histogram method to extract potato vegetation coverage during three growth periods. By comparing the FVC extraction accuracy of the 10 vegetation indices under the vegetation index intersection method, the maximum entropy thresholding method, and the bimodal histogram method, the most suitable vegetation index and extraction method for vegetation coverage extraction were explored. To more intuitively compare the FVC extraction accuracy of the 10 vegetation indices under the three methods, the data were visualized, as shown in
Figure 13.
As shown in
Figure 13(a), during the potato tuber formation stage, the BGCI combined with the vegetation index intersection method achieved the best vegetation coverage accuracy. Compared with the maximum entropy thresholding method and the bimodal histogram method, the RGCI combined with the vegetation index intersection method also achieved greater vegetation coverage accuracy.
Figure 13(b) shows that during the potato tuber expansion stage, the BGCI combined with the vegetation index intersection method achieved the best vegetation coverage extraction accuracy. As shown in
Figure 13(c), during the potato maturation stage, the RGCI combined with the vegetation index intersection method achieved the highest vegetation coverage estimation accuracy. However, the BGCI combined with the vegetation index intersection method did not achieve the best vegetation coverage extraction accuracy during this growth stage, as it did in the previous two growth stages.
Based on the analysis results of the 10 vegetation indices combined with the three vegetation coverage extraction methods, it can be concluded that during the potato tuber formation and expansion stages, the BGCI combined with the vegetation index intersection method achieved the best vegetation coverage extraction accuracy. During the potato maturation stage, the RGCI combined with the vegetation index intersection method achieved the highest vegetation coverage extraction accuracy.
4.2. Validation of the General Applicability of the Vegetation Index Intersection Method
Through the potato vegetation coverage extraction results of Experiment B, it is evident that the vegetation index combined with the vegetation index intersection method achieved the highest accuracy. To further explore whether this method has transferability, data from Experiment A were selected to validate the general applicability of the vegetation index intersection method.
As shown in
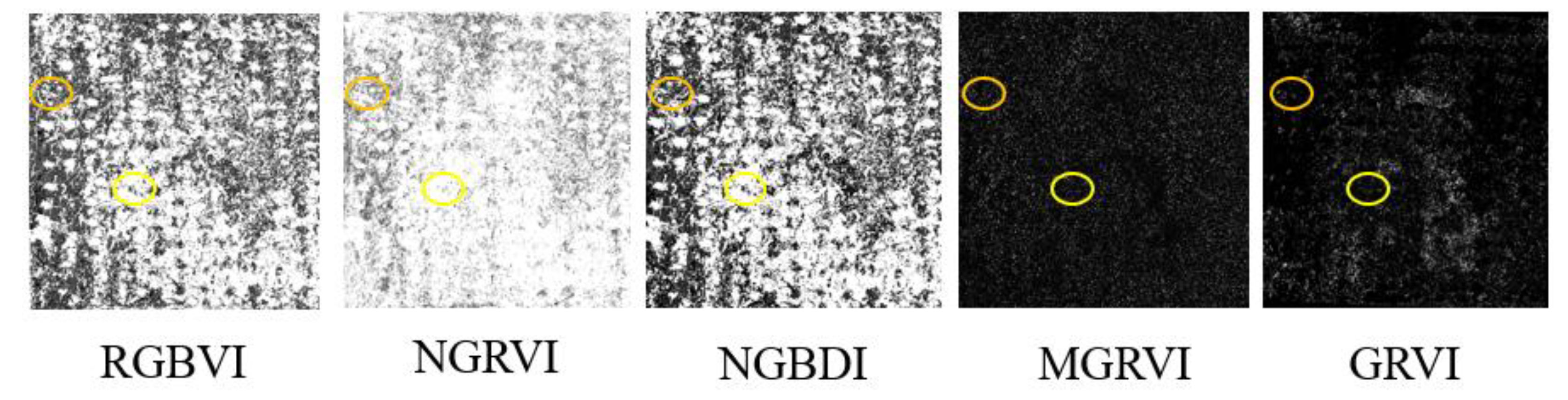
Figure 14, after irrigating the potatoes, some of the soil on the surface of the fields in Experiment A had not dried and appeared darker in colour. This caused the grayscale images of the RGBVI, NGRVI, MGBDI, NGBDI, MGRVI, and GRVI to show less distinct differences between the soil and potato vegetation. The brighter portions of the soil had higher grayscale values, similar to those of the potato plants. This phenomenon can lead to lower vegetation coverage extraction accuracy [
27].
In the grayscale images of the RGCI, BGCI, EXG, TRVI, and GLI, the potato plants had higher grayscale values and appeared white, while the soil areas had lower grayscale values and appeared black (
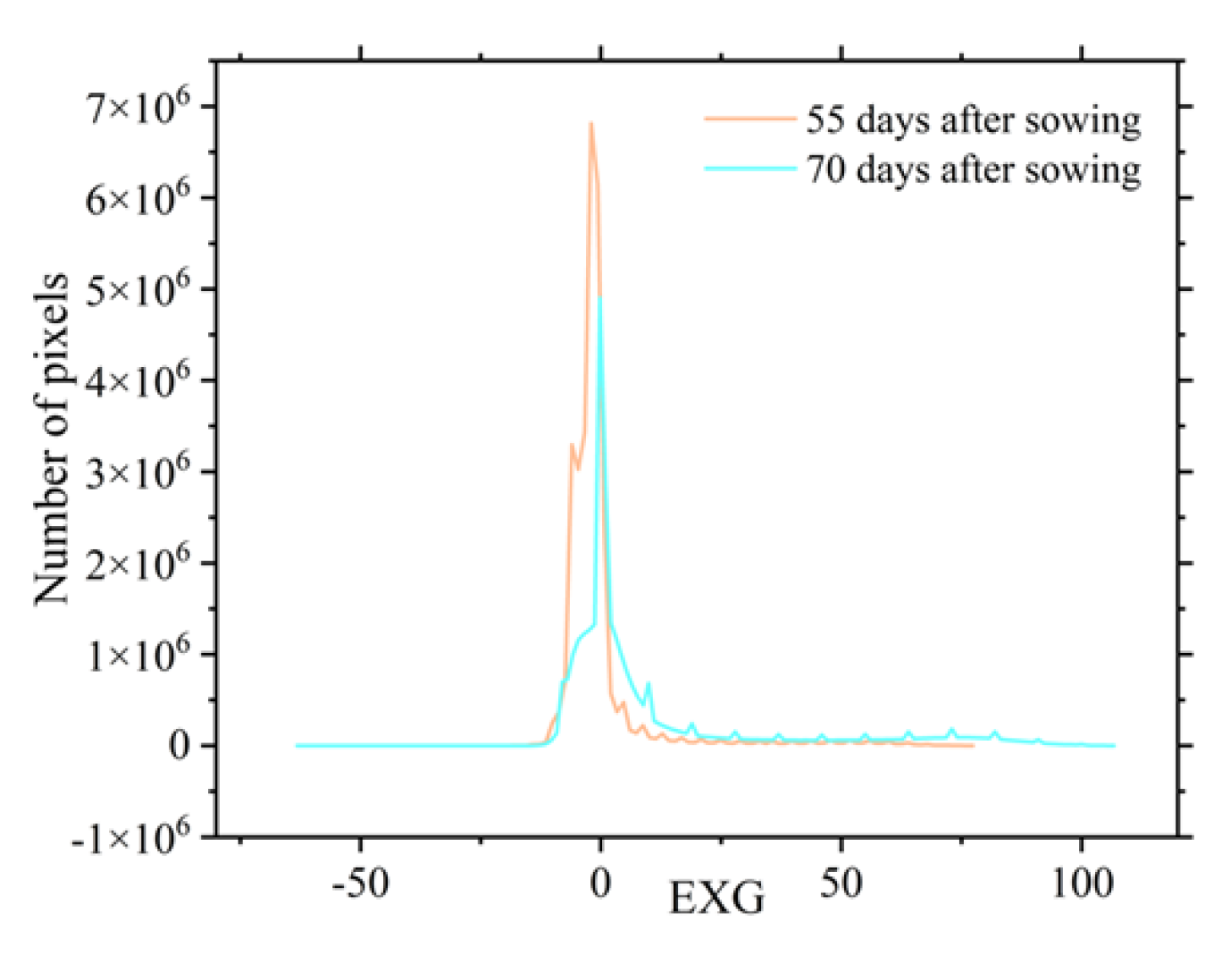
Figure 15); this allowed for a clear distinction between the plants and the soil. However, the EXG histogram exhibited irregularities (
Figure 16), making it difficult to determine the vegetation coverage extraction threshold using the vegetation index intersection method. Therefore, the RGCI, BGCI, TRVI, and GLI were selected to extract potato vegetation coverage using the vegetation index intersection method.
By visual interpretation, 300 regions of interest of potato plants and soil were selected on the orthoimages of the experimental field to verify the potato vegetation coverage extracted using the vegetation index intersection method with a confusion matrix. As shown in
Table 4, during the first two growth stages of the potatoes, the BGCI achieved the highest vegetation coverage extraction accuracy. In the tuber formation stage, the overall classification accuracy and kappa coefficient were 99.97% and 0.9974, respectively. In the tuber expansion stage, the overall accuracy and kappa coefficient were 99.81% and 0.9999, respectively. The overall classification accuracy and kappa coefficient in the tuber formation stage were 99.77% and 0.9952, respectively, and those in the tuber expansion stage were 99.77% and 0.9952, respectively; this is consistent with the conclusions drawn from Experiment B, demonstrating that the two vegetation indices proposed in this study combined with the vegetation index intersection method achieved an ideal accuracy and general applicability for vegetation coverage extraction.
4.3. The Effect of Fitting Parameter Selection on Estimation Accuracy
Experiment A and Experiment B involved different potato varieties, soil types, and growth environments. To validate the generality of the potato FVC estimation model developed in Experiment B, FVC data extracted using the BGCI combined with the vegetation index threshold method from Experiment A were used as independent data to validate the estimation model established in Experiment B.
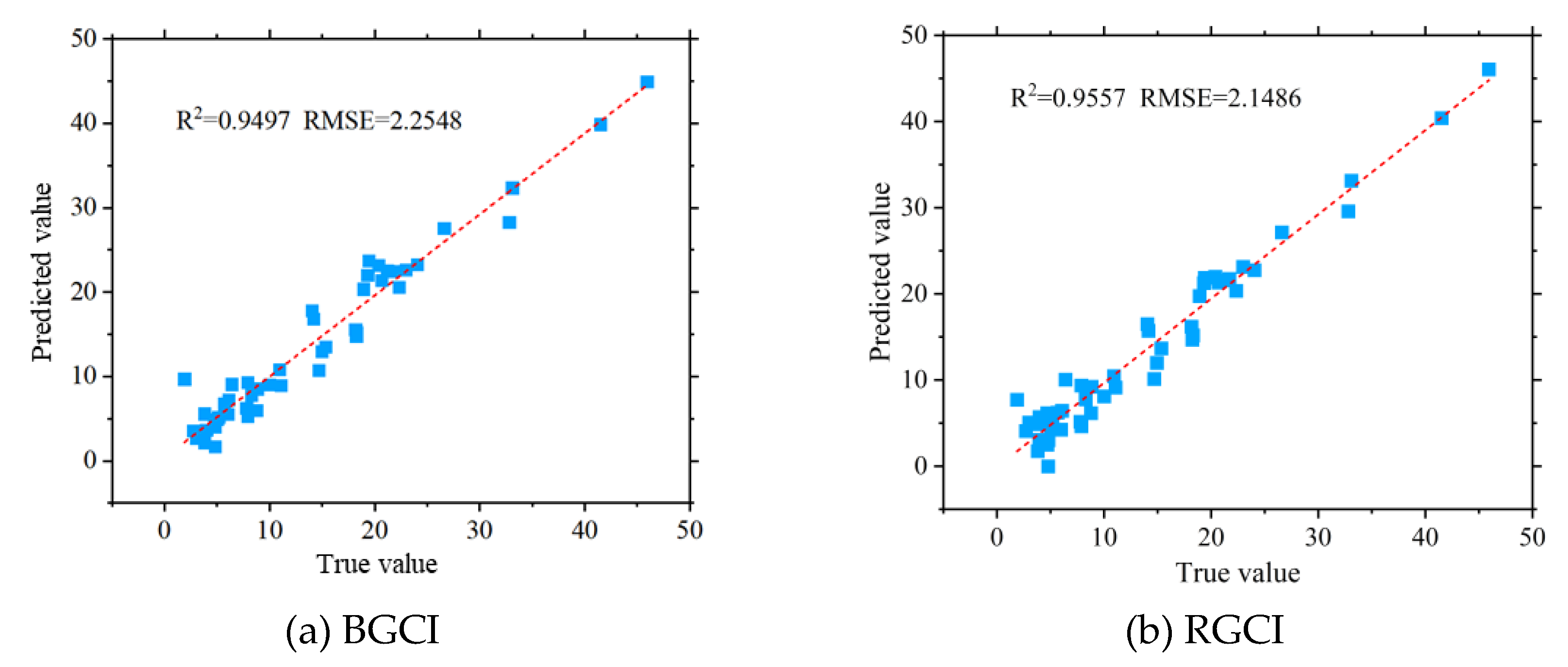
Figure 17 shows the accuracy validation results of the potato FVC model based on the BGCI and RGCI..
The results indicate that the potato FVC models based on the BGCI and RGCI achieved an ideal potato FVC estimation accuracy, with R2 values of both the RGCI and BGCI models exceeding 0.94 and RMSE values below 2.30. Compared to Experiment B, both the RGCI and BGCI models showed greater accuracy in estimating FVC for Experiment A. The main factor contributing to the difference in potato FVC estimation accuracy was the presence of three potato varieties in Experiment B, making the field background more complex and leading to a decrease in the accuracy of potato FVC estimation. Both the BGCI models in Experiment A and Experiment B achieved R2 values greater than 0.90, while the RGCI model's FVC estimation accuracy exceeded an R2 value of 0.89. The validation results demonstrate that the FVC estimation models constructed using the BGCI and RGCI proposed in this study have high accuracy and strong generality. These models effectively improve the accuracy of FVC estimation using traditional parameters based on UAV visible light images and perform well in different planting environments and growth stages.
5. Conclusions
This study presents an analysis of the differences in grayscale values between potato plants and the background in the visible light spectrum in the field via the construction of RGCI and BGCI vegetation indices based on the relationships in the red‒green and blue‒green bands. The computation processes of eight common visible light spectrum vegetation indices and 24 texture features were described. The potato FVC was extracted using the vegetation index intersection method, maximum entropy thresholding method, and bimodal histogram method. The accuracy of 10 vegetation indices combined with three FVC extraction methods was validated using a confusion matrix, and the best vegetation index and potato FVC extraction method were further selected. The Pearson correlation coefficient method and random forest feature selection were used to screen the aforementioned vegetation indices and 24 texture features. Based on the top 6 selected features, potato FVC estimation models were established. The main conclusions are as follows.
(1) This study newly constructed two vegetation indices, BGCI and RGCI, and successfully obtained classification thresholds using the SVM combined with vegetation indices, which can effectively differentiate between the background and potato plants. The extraction thresholds for the BGCI during the three growth periods were -13.0583, 10.1801, and -4.3000, respectively. For the RGCI, the extraction thresholds during the three growth periods were 2.5892, 23.0584, and 16.9357, respectively. The BGCI and RGCI could effectively distinguish potato plants from the background under the above thresholds.
(2) The BGCI and RGCI combined with the vegetation index intersection method both achieved excellent results in the extraction of potato vegetation coverage throughout the entire growth period. During the potato tuber formation and expansion stages, the BGCI combined with the vegetation index intersection method achieved the highest vegetation coverage extraction accuracy, with overall accuracies of 99.61% and 98.84%, respectively. The RGCI combined with the vegetation index intersection method achieved the highest accuracy, 98.63%, in terms of vegetation coverage extraction during the maturation stage. Overall, the RGCI combined with the vegetation index intersection method obtained the most ideal vegetation coverage extraction results throughout the entire potato growth period.
(3) This study screened multiple vegetation indices and texture features and successfully established a highly accurate potato vegetation coverage estimation model. Using the Pearson correlation coefficient method and random forest feature selection, six vegetation indices highly correlated with potato FVC (BGCI, NGBDI, RGCI, RGBVI, NGRVI, and EXG) were selected, and corresponding vegetation coverage estimation models were constructed. Among these models, the vegetation coverage estimation model based on the BGCI exhibited the highest accuracy (R2=0.9116, RMSE=5.7903). In the validation of model generality, FVC estimation models based on the newly constructed BGCI and RGCI also achieved good accuracy. The RGCI had an R2 value of 0.9497 and an RMSE of 2.2548, while BGCI had an R2 value of 0.9557 and an RMSE of 2.1486. This study proposed two novel vegetation information indices, and the potato vegetation coverage models based on these indices demonstrated good model accuracy and generality.
Author Contributions
Methodology, X.S. and H.Y.; formal analysis, X.S. and T.G.; data curation, T.G. and R.L.; writing—original draft X.S. and H.Y..; writing—review and editing, H.Y., R.L., L.Y. and Y.H.; visualization, Y.C.; investigation, Y.C.; supervision, L.Y. and Y.H.; project administration, Y.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (32171894 (C 0043619), 31971787(C 0043628)).
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank Chao Wang and Shaoxiang Wang for their work on field data collection.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Huang, X.; Cheng, F.; Wang, J.L.; Duan, P.; Wang, J.S. Forest Canopy Height Extraction Method Based on ICESat-2/ATLAS Data. IEEE Trans. Geosci. Remote Sensing 2023, 61, 14. [Google Scholar] [CrossRef]
- Teng, Y.J.; Ren, H.Z.; Zhu, J.S.; Jiang, C.C.; Ye, X.; Zeng, H. A practical method for angular normalization on land surface temperature using space between thermal radiance and fraction of vegetation cover. Remote Sens. Environ. 2023, 291, 20. [Google Scholar] [CrossRef]
- Neyns, R.; Canters, F. Mapping of Urban Vegetation with High-Resolution Remote Sensing: A Review. Remote Sens. 2022, 14, 27. [Google Scholar] [CrossRef]
- Hu, J.Y.; Feng, H.; Wang, Q.L.; Shen, J.N.; Wang, J.; Liu, Y.; Feng, H.K.; Yang, H.; Guo, W.; Qiao, H.B.; et al. Pretrained Deep Learning Networks and Multispectral Imagery Enhance Maize LCC, FVC, and Maturity Estimation. Remote Sens. 2024, 16, 23. [Google Scholar] [CrossRef]
- Niu, Y.X.; Han, W.T.; Zhang, H.H.; Zhang, L.Y.; Chen, H.P. Estimating fractional vegetation cover of maize under water stress from UAV multispectral imagery using machine learning algorithms. Comput. Electron. Agric. 2021, 189, 11. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, W.; Xing, J.H.; Zhang, X.P.; Tian, H.J.; Tang, H.Z.; Bi, P.S.; Li, G.C.; Zhang, F.J. Extracting vegetation information from high dynamic range images with shadows: A comparison between deep learning and threshold methods. Comput. Electron. Agric. 2023, 208, 12. [Google Scholar] [CrossRef]
- Yu, Y.; Bao, Y.D.; Wang, J.C.; Chu, H.J.; Zhao, N.; He, Y.; Liu, Y.F. Crop Row Segmentation and Detection in Paddy Fields Based on Treble-Classification Otsu and Double-Dimensional Clustering Method. Remote Sens. 2021, 13, 24. [Google Scholar] [CrossRef]
- Ding, F.; Li, C.C.; Zhai, W.G.; Fei, S.P.; Cheng, Q.; Chen, Z. Estimation of Nitrogen Content in Winter Wheat Based on Multi-Source Data Fusion and Machine Learning. Agriculture-Basel 2022, 12, 16. [Google Scholar] [CrossRef]
- Chen, H.; Li, W.; Zhu, Y.Y. Improved window adaptive gray level co-occurrence matrix for extraction and analysis of texture characteristics of pulmonary nodules. Comput. Meth. Programs Biomed. 2021, 208, 6. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.M.; Denoeux, T.; Liu, Z.G.; Pan, Q. EGMM: An evidential version of the Gaussian mixture model for clustering. Appl. Soft. Comput. 2022, 129, 17. [Google Scholar] [CrossRef]
- Yu, Y.; Meng, L.; Luo, C.; Qi, B.; Zhang, X.; Liu, H. Early Mapping Method for Different Planting Types of Rice Based on Planet and Sentinel-2 Satellite Images. Agronomy 2024, 14. [Google Scholar] [CrossRef]
- Yang, H.B.; Hu, Y.H.; Zheng, Z.Z.; Qiao, Y.C.; Hou, B.R.; Chen, J. A New Approach for Nitrogen Status Monitoring in Potato Plants by Combining RGB Images and SPAD Measurements. Remote Sens. 2022, 14, 15. [Google Scholar] [CrossRef]
- Madec, S.; Irfan, K.; Velumani, K.; Baret, F.; David, E.; Daubige, G.; Samatan, L.B.; Serouart, M.; Smith, D.; James, C.; et al. VegAnn, Vegetation Annotation of multi-crop RGB images acquired under diverse conditions for segmentation. Sci. Data 2023, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Li, D.L.; Li, C.; Yao, Y.; Li, M.D.; Liu, L.C. Modern imaging techniques in plant nutrition analysis: A review. Comput. Electron. Agric. 2020, 174, 14. [Google Scholar] [CrossRef]
- Jiang, J.; Atkinson, P.M.; Chen, C.S.; Cao, Q.; Tian, Y.C.; Zhu, Y.; Liu, X.J.; Cao, W.X. Combining UAV and Sentinel-2 satellite multi-spectral images to diagnose crop growth and N status in winter wheat at the county scale. Field Crop. Res. 2023, 294, 13. [Google Scholar] [CrossRef]
- Yang, N.; Zhang, Z.T.; Zhang, J.R.; Guo, Y.H.; Yang, X.Z.; Yu, G.D.; Bai, X.Q.; Chen, J.Y.; Chen, Y.W.; Shi, L.S.; et al. Improving estimation of maize leaf area index by combining of UAV-based multispectral and thermal infrared data: The potential of new texture index. Comput. Electron. Agric. 2023, 214, 16. [Google Scholar] [CrossRef]
- Garofalo, S.P.; Giannico, V.; Costanza, L.; Alhajj Ali, S.; Camposeo, S.; Lopriore, G.; Pedrero Salcedo, F.; Vivaldi, G.A. Prediction of Stem Water Potential in Olive Orchards Using High-Resolution Planet Satellite Images and Machine Learning Techniques. Agronomy 2023, 14. [Google Scholar] [CrossRef]
- Shi, H.Z.; Guo, J.J.; An, J.Q.; Tang, Z.J.; Wang, X.; Li, W.Y.; Zhao, X.; Jin, L.; Xiang, Y.Z.; Li, Z.J.; et al. Estimation of Chlorophyll Content in Soybean Crop at Different Growth Stages Based on Optimal Spectral Index. Agronomy-Basel 2023, 13, 17. [Google Scholar] [CrossRef]
- Yang, H.B.; Hu, Y.H.; Zheng, Z.Z.; Qiao, Y.C.; Zhang, K.L.; Guo, T.F.; Chen, J. Estimation of Potato Chlorophyll Content from UAV Multispectral Images with Stacking Ensemble Algorithm. Agronomy-Basel 2022, 12, 16. [Google Scholar] [CrossRef]
- Zhang, M.Z.; Chen, T.E.; Gu, X.H.; Kuai, Y.; Wang, C.; Chen, D.; Zhao, C.J. UAV-borne hyperspectral estimation of nitrogen content in tobacco leaves based on ensemble learning methods. Comput. Electron. Agric. 2023, 211, 11. [Google Scholar] [CrossRef]
- Huang, L.S.; Li, T.K.; Ding, C.L.; Zhao, J.L.; Zhang, D.Y.; Yang, G.J. Diagnosis of the Severity of Fusarium Head Blight of Wheat Ears on the Basis of Image and Spectral Feature Fusion. Sensors 2020, 20, 17. [Google Scholar] [CrossRef] [PubMed]
- Hayit, T.; Erbay, H.; Varçin, F.; Hayit, F.; Akci, N. The classification of wheat yellow rust disease based on a combination of textural and deep features. Multimed. Tools Appl. 2023, 82, 47405–47423. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Wang, Y.; Zhu, D.Q.; Zou, Y.; Zhang, S.; Zhou, H.Y. Automatic Segmentation of Crop/Background Based on Luminance Partition Correction and Adaptive Threshold. IEEE Access 2020, 8, 202611–202622. [Google Scholar] [CrossRef]
- Zhang, H.S.; Huang, L.S.; Huang, W.J.; Dong, Y.Y.; Weng, S.Z.; Zhao, J.L.; Ma, H.Q.; Liu, L.Y. Detection of wheat Fusarium head blight using UAV-based spectral and image feature fusion. Front. Plant Sci. 2022, 13, 14. [Google Scholar] [CrossRef] [PubMed]
- Estévez, J.; Salinero-Delgado, M.; Berger, K.; Pipia, L.; Rivera-Caicedo, J.P.; Wocher, M.; Reyes-Muñoz, P.; Tagliabue, G.; Boschetti, M.; Verrelst, J. Gaussian processes retrieval of crop traits in Google Earth Engine based on Sentinel-2 top-of-atmosphere data. Remote Sens. Environ. 2022, 273, 19. [Google Scholar] [CrossRef] [PubMed]
- Aslan, M.F.; Durdu, A.; Sabanci, K.; Ropelewska, E.; Gueltekin, S.S. A Comprehensive Survey of the Recent Studies with UAV for Precision Agriculture in Open Fields and Greenhouses. Appl. Sci.-Basel 2022, 12, 29. [Google Scholar] [CrossRef]
- Li, L.Y.; Mu, X.H.; Jiang, H.L.; Chianucci, F.; Hu, R.H.; Song, W.J.; Qi, J.B.; Liu, S.Y.; Zhou, J.X.; Chen, L.; et al. Review of ground and aerial methods for vegetation cover fraction (fCover) and related quantities estimation: definitions, advances, challenges, and future perspectives. ISPRS-J. Photogramm. Remote Sens. 2023, 199, 133–156. [Google Scholar] [CrossRef]
Figure 1.
Location and division of the two research fields and UAV-based field observations. (a) Location of the experimental areas in Shaanxi Province. (b) and (c) Aerial views of Experiment Area A and Experiment Area B, respectively.
Figure 1.
Location and division of the two research fields and UAV-based field observations. (a) Location of the experimental areas in Shaanxi Province. (b) and (c) Aerial views of Experiment Area A and Experiment Area B, respectively.
Figure 2.
Scatter plots of potato plants, soil, shadows, and drip irrigation strips.
Figure 2.
Scatter plots of potato plants, soil, shadows, and drip irrigation strips.
Figure 3.
Fitting results of BGCI and RGCI vegetation indices.
Figure 3.
Fitting results of BGCI and RGCI vegetation indices.
Figure 4.
Validation results of the confusion matrix for FVC extraction thresholds at different growth stages of potatoes.
Figure 4.
Validation results of the confusion matrix for FVC extraction thresholds at different growth stages of potatoes.
Figure 5.
Process of potato plant threshold extraction based on the vegetation index threshold method during the tuber formation period.
Figure 5.
Process of potato plant threshold extraction based on the vegetation index threshold method during the tuber formation period.
Figure 6.
Process of potato plant threshold extraction based on the vegetation index threshold method during the tuber expansion period.
Figure 6.
Process of potato plant threshold extraction based on the vegetation index threshold method during the tuber expansion period.
Figure 7.
Process of potato plant threshold extraction based on the vegetation index threshold method during the maturation stage.
Figure 7.
Process of potato plant threshold extraction based on the vegetation index threshold method during the maturation stage.
Figure 8.
Extraction results of potato vegetation coverage based on the vegetation index intersection method.
Figure 8.
Extraction results of potato vegetation coverage based on the vegetation index intersection method.
Figure 9.
Confusion matrix verification results of potato vegetation coverage extraction accuracy based on the vegetation index intersection method.
Figure 9.
Confusion matrix verification results of potato vegetation coverage extraction accuracy based on the vegetation index intersection method.
Figure 10.
Extraction results of FVC based on the BGCI and RGCI vegetation index intersection methods.
Figure 10.
Extraction results of FVC based on the BGCI and RGCI vegetation index intersection methods.
Figure 11.
Results of feature selection using the Pearson correlation coefficient method in the entire stage.
Figure 11.
Results of feature selection using the Pearson correlation coefficient method in the entire stage.
Figure 12.
Results of model accuracy in the entire stage.
Figure 12.
Results of model accuracy in the entire stage.
Figure 13.
Comparison of the extraction accuracy of potato FVC using the three threshold extraction methods.
Figure 13.
Comparison of the extraction accuracy of potato FVC using the three threshold extraction methods.
Figure 14.
Comparison of the vegetation index misclassification results.
Figure 14.
Comparison of the vegetation index misclassification results.
Figure 15.
Grayscale images of the 5 vegetation indices.
Figure 15.
Grayscale images of the 5 vegetation indices.
Figure 16.
Histogram of the EXG.
Figure 16.
Histogram of the EXG.
Figure 17.
Validation of the accuracy of the FVC estimation model established by the RGCI and BGCI.
Figure 17.
Validation of the accuracy of the FVC estimation model established by the RGCI and BGCI.
Table 1.
Statistical results of the pixel values of potato vegetation, soil, drip irrigation belts, and shadows in the blue, green, and red bands.
Table 1.
Statistical results of the pixel values of potato vegetation, soil, drip irrigation belts, and shadows in the blue, green, and red bands.
| Typical objects |
Blue band pixel values |
Green band pixel values |
Red band pixel values |
| Average |
Standard deviation |
Average |
Standard deviation |
Average |
Standard deviation |
| Potato |
84.010 |
20.734 |
135.858 |
21.962 |
79.523 |
20.165 |
| Soil |
219.783 |
12.291 |
204.621 |
12.597 |
173.848 |
16.144 |
| Shadow |
83.114 |
33.934 |
75.344 |
30.681 |
52.283 |
27.783 |
| Drip irrigation tape |
146.108 |
37.974 |
136.684 |
37.058 |
121.778 |
38.578 |
Table 2.
Names of 24 texture features.
Table 2.
Names of 24 texture features.
| Texture feature name |
Red band |
Green band |
Blue band |
| Mean |
R-Mean |
G-Mean |
B-Mean |
| Variance |
R-Variance |
G-Variance |
B-Variance |
| Homogeneity |
R-Homogeneity |
G-Homogeneity |
B-Homogeneity |
| Contrast |
R-Contrast |
G-Contrast |
B-Contrast |
| Dissimilarity |
R-Dissimilarity |
G-Dissimilarity |
B-Dissimilarity |
| Entropy |
R-Entropy |
G-Entropy |
B-Entropy |
| Second moment |
R-Second moment |
G-Second moment |
B-Second moment |
| Correlation |
R-Correlation |
G-Correlation |
B-Correlation |
Table 3.
Threshold extraction results based on the vegetation index threshold method.
Table 3.
Threshold extraction results based on the vegetation index threshold method.
| Vegetation index |
Threshold extraction results |
| Tuber formation stage |
Tuber expansion Stage |
Tuber maturation Stage |
| EXG |
29.567 |
51.642 |
30.342 |
| GLI |
0.071 |
0.111 |
0.075 |
| GRVI |
0.002 |
0.053 |
0.004 |
| TRVI |
-13.987 |
-10.125 |
-20.482 |
| NGBDI |
0.098 |
0.163 |
0.149 |
| RGBVI |
0.143 |
0.222 |
0.169 |
| RGFI |
-6.825 |
16.943 |
-6.698 |
| MGRVI |
0.074 |
0.111 |
0.014 |
| NGRVI |
-3.951 |
-5.209 |
-1.759 |
| RGCI |
2.589 |
23.058 |
16.936 |
| BGCI |
-13.058 |
10.180 |
-4.300 |
Table 4.
Threshold extraction results based on the vegetation index threshold method.
Table 4.
Threshold extraction results based on the vegetation index threshold method.
| Vegetation index |
Image acquisition period |
Overall accuracy(%) |
Kappa coefficient |
| GLI |
Tuber formation period |
99.12 |
0.9210 |
| Tuber expansion period |
99.76 |
0.9951 |
| TRVI |
Tuber formation period |
96.81 |
0.7591 |
| Tuber expansion period |
97.67 |
0.9525 |
| RGCI |
Tuber formation period |
99.79 |
0.9798 |
| Tuber expansion period |
99.77 |
0.9952 |
| BGCI |
Tuber formation period |
99.97 |
0.9974 |
| Tuber expansion period |
99.81 |
0.9999 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).