1. Introduction
The global crisis of antibiotic resistance is multifaceted and has been linked to antibiotic overuse and misuse in humans and animals, improper prescribing practices and the widespread use of antibiotics as growth promoters in livestock production [
1,
2]. Pork production is widespread in South Africa, with the largest contributions coming from Limpopo and North West provinces, which account for 24% and 20% of the total production, respectively [
3].
In animals, antibiotics such as sulphonamides, tetracyclines, and fluoroquinolones are widely used for therapeutic, metaphylactic and prophylactic purposes or as growth promotors in animal feed [
4,
5,
6]. The Food and Agriculture Organization and the European Union have established tolerance or maximum residue limits (MRL) for antibiotic residues in animal-derived food products [
7]. In conjunction, the MRL for foodstuffs in South Africa are regulated in South Africa by the Foodstuffs, Cosmetics and Disinfectants Act No. 54 of 1972 [
8].
Previous studies have found that food products not only serve as a reservoir for antibiotic resistant bacteria and antibiotic resistance genes (ARG) but also act as a mediator, facilitating the transfer of antibiotic resistant bacteria and ARGs between the environment and humans through indirect contact through the consumption of contaminated foods [
2,
5,
9,
10]. Understanding the diversity and abundance of ARGs, virulence factor (VF) genes and antibiotic residues in food, especially retail pork meat, is important for controlling antibiotic resistance [
2,
4]. This involves implementing effective antibiotic stewardship programs, regulating the use of antibiotics in food production, improving hygiene and sanitation practices, and promoting responsible antibiotic usage in both human and veterinary medicine [
5]. The environment harbours a high prevalence of multidrug-resistant (MDR) strains, which affect both humans and animals, and facilitates the easy transmission of ARGs through horizontal gene transfer among different bacterial species (spp.) [
11]. These spp. include both commensal flora and pathogenic foodborne pathogens like
Campylobacter spp.,
Enterococcus spp.,
Escherichia coli and non-typhoidal
Salmonella spp. (NTS) that can cause disease in humans and animals [
11]. The aim of this study was to isolate and characterise four common indicator bacteria that overlap between humans and animals,
Campylobacter spp.,
Enterococcus spp.,
E. coli and NTS, to test for antibiotic residues and to assess the microbial community as well as the resistome present in purchased raw retail pork meat.
2. Materials and Methods
2.1. Ethical Clearance and Study Definitions
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Human Research Ethics Committee of the University of the Witwatersrand (M190244; 10/05/2019).
Supermarkets were defined as stores that sold raw meat and various other grocery items, while butcheries were defined as stores selling primarily raw meat commodities. Raw meat samples were defined as pork chops (alternative names included loin, rib, sirloin, top loin and blade chops). Indicator bacteria in this study were defined as Campylobacter spp., Enterococcus spp., E. coli and NTS spp.
2.2. Study Setting and Sampling
Raw meat samples (i.e. pork chops) were purchased once off from 10 randomly selected retail outlet stores in Johannesburg and Pretoria, Gauteng on the 4
th of January 2022. The stores selected for sample collection included both supermarkets (n=5) and butcheries (n=5). Pork chops were chosen as it is the most popular cut among consumers [
12]. Raw meat samples (containing at least two pork chops in the same container/packet) were randomly selected from the store shelves. All the raw meat samples were collected within the recommended dates for human consumption. Store demographics were captured by the study investigators on the day of sample collection (
Supplementary Table S1). A unique number was assigned to each raw meat sample as well as the sampled stores to maintain anonymity and to ensure that the results cannot be linked back to a specific store.
The purchased raw meat samples were transported on ice to the Centre for Healthcare-associated infections, Antimicrobial Resistance and Mycoses (CHARM), National Institute for Communicable Diseases (NICD), a division of the National Health Laboratory Service (NHLS). The outside of all the meat containers/packets was wiped with 70% ethanol before segregation and processing to avoid cross-contamination. The raw meat samples were separated aseptically. One raw meat sample from each store was sent on ice within 24 hours after collection to a subcontracted laboratory in Gauteng, South Africa for 1) the isolation of four selected indicator bacteria and 2) antibiotic residue testing using liquid chromatography tandem-mass spectrometry (LC-MS/MS). The remaining raw meat samples underwent metagenomics sequencing at the Sequencing Core Facility (SCF), NICD. All the raw meat samples were processed within 24 hours to 48 hours after sample collection.
2.3. Antibiotic Susceptibility Testing (AST) and Whole Genome Sequencing (WGS) of Isolated Indicator Bacteria from Raw Meat Samples
The bacteria isolated from the raw meat samples were transported at room temperature (20°C to 25°C) from the subcontracted laboratory to CHARM within 24 hours after isolation. Organism identification was reconfirmed at CHARM, NICD with the matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) (Microflex, Bruker Daltonics, Germany). AST was performed using the Microscan Walkaway System with the Gram-negative NM44 (Beckman Coulter, USA) and Gram-positive PM33 (Beckman Coulter, USA) cards. The AST results were interpreted using the 2023 European Committee on antimicrobial susceptibility testing (EUCAST) guidelines [
13].
The genomic DNA (gDNA) of all isolated organisms from the raw meat samples were extracted with the QIAamp mini kit (Qiagen, Germany) with the inclusion of lysozyme (10 mg/mL; Sigma-Aldrich, USA) to ensure sufficient lysis. The quantity and quality of the extracted gDNA were determined on Qubit 4.0 (Invitrogen, USA) with the high sensitivity assay kit (Invitrogen, USA). Multiplexed paired-end libraries were prepared using the Nextera DNA Prep kit, followed by sequencing (2×150 bp) on a NextSeq 550 instrument (Illumina Inc., USA) with 100x coverage at the SCF, NICD. Raw paired-end reads were analysed using the Jekesa pipeline (v1.0) [
14]. Briefly, Trim Galore! (v0.6.7) was used to filter the paired-end reads (Q >30 and length >50 bp) [
15].
De novo assembly was performed using SKESA (v2.3.0;
https://github.com/ncbi/SKESA) and the assembled contigs were polished using Shovill (v1.1.0;
https://github.com/tseemann/shovill) [
16]. Assembly metrics were calculated using QUAST (v5.0.2) [
17]. The multilocus sequence typing (MLST) profiles were determined using the MLST tool (version 2.16.4;
https://pubmlst.org/; https://github.com/tseemann/mlst) [
18]. VF genes and ARGs search was performed using ABRicate (version 1.0.1;
https://github.com/tseemann/abricate), against the Comprehensive Antibiotic Resistance Database (CARD), CARD-prevalence, Virulence Factor Database (VFDB) and ResFinder - Center for Genomic Epidemiology (CGE) database; with the gene alignment coverage cut-off of ≥95% and blastn sequence similarity of ≥95% [19-24]. The assembled genome files were deposited in the National Center for Biotechnology Information GenBank under BioProject number PRJNA1006163.
2.4. Metagenomics
Total gDNA was extracted using the QIAamp Fast DNA stool mini kit (Qiagen, Germany) and host depletion was done using the NEBNext Microbiome DNA enrichment kit (New England Biolabs, USA) with the inclusion of negative and positive controls (ZymoBIOMICS Gut Microbiome Standard). Sequencing was done on the NextSeq 550 (2×150 bp and 10 M reads) (Illumina, USA).
2.5. Resistome Gene Abundance Estimates
With the host-depleted metagenomic sequence reads, standard
de novo assembly of the metagenomic data were done using MEGAHIT (v1.2.9) [
26]. Using the assembled contigs, the ARG, toxin genes and VF genes were predicted using PathoFact (v1.0) pipeline [
27]. The predicted ARG, as well as the toxin genes and VF genes with high confident prediction (1: secreted toxin or 1: secreted VF) were selected for relative abundance analysis, using ShortBRED (v0.9.4) [
28] against the CARD database (v2023-06) and VFDB (database update: 06 August 2023). The predicted resistome’ relative abundance was quantified by ShortBRED-Quantify calls USEARCH, where reads with 95% identity to the resistome were counted and normalized by reads per kilobase of reference sequence per million sample reads (RPKMs). The antimicrobial resistance (AMR) category “multidrug” was defined as bacterial strains that have become resistant to multiple classes of antibacterial drugs or other agents (
https://card.mcmaster.ca/ontology/41472).
3. Results
3.1. Isolation of Indicator Bacteria
All 10 raw meat samples were tested for the selected four indicator bacteria (
Table 1).
E. coli was detected and isolated from one butchery raw meat sample (PC9-B4). Enterococci were detected and isolated from two supermarket raw meat samples (PC3-S3 and PC4-S4) and one butchery raw meat sample (PC10-B5).
Campylobacter spp. and
Salmonella spp. were not detected in any of the collected raw meat samples (
Supplementary Table S2).
3.2. AST and WGS of Isolated Indicator
A single
E. coli isolate was obtained from sample PC9-B4, while one
E. faecalis isolate each was obtained from samples PC3-S3, PC4-S4 and PC10-B5. The IDs of all the isolates were reconfirmed and AST was performed. All three
E. faecalis isolates had the same antibiotic susceptibility profile (
Table 2). There are no AST guidelines for isolates obtained from raw meat samples; thus, the AST results were interpreted using the 2023 EUCAST guidelines for human isolates [
13].
The isolated
E. coli (PC9-B4) and
E. faecalis (PC3-S3, PC4-S4 and PC10-B5) isolates were subjected to WGS (
Table 3). The
E. coli isolate exhibited ARGs conferring resistance to aminoglycoside (
aadA1), fluoroquinolone (
gyrA), tetracycline (
tetB), trimethoprim (
dfrA1) and sulphonamide (
sul2). All three
E. faecalis isolates exhibited ARGs conferring resistance to tetracycline (
tetM) and lincosamides (
isaA).
A total of eight VF genes were identified in E. coli, and nine VF genes were identified in E. faecalis. VF genes associated with E. coli included colicins (cba, cea, cia and cma), as well as the toxin gene (astA). VF genes associated with E. faecalis isolates included adhesins (ace and efaAfs), toxins (cyl-A, -L and -M), and genes associated with biofilm formation (ebp-A and -B) and pheromone production (cad, camE, cCF10 and cOB1).
The E. coli isolate harboured the IncB/O/K/Z and IncFII (pCoo) plasmids, while the E. faecalis isolates harboured the repUS43, repUS11 and rep9a plasmids.
3.3. Antibiotic Residue Testing
The average antibiotic residue concentration detected in all the raw meat samples were <50 µg/kg for all the majority of all the tested antibiotics (
Table 4). Sample PC5-S5 had an antibiotic residue concentration of 71,5 µg/kg for chlortetracycline. All tested residue levels were below the Codex/SA MRL acceptable limit [
7].
3.4. Metagenomics
3.4.1. Read Statistics
Approximately 7 million hosts reads (ranging from 5,797,550 – 8,900,728 sequencing reads) were removed from each sample prior to the metagenomic analysis (
Table 5). The remaining reads were subject to further bacterial community profiling and ARG prediction analyses.
3.4.2. Estimated Relative Abundance
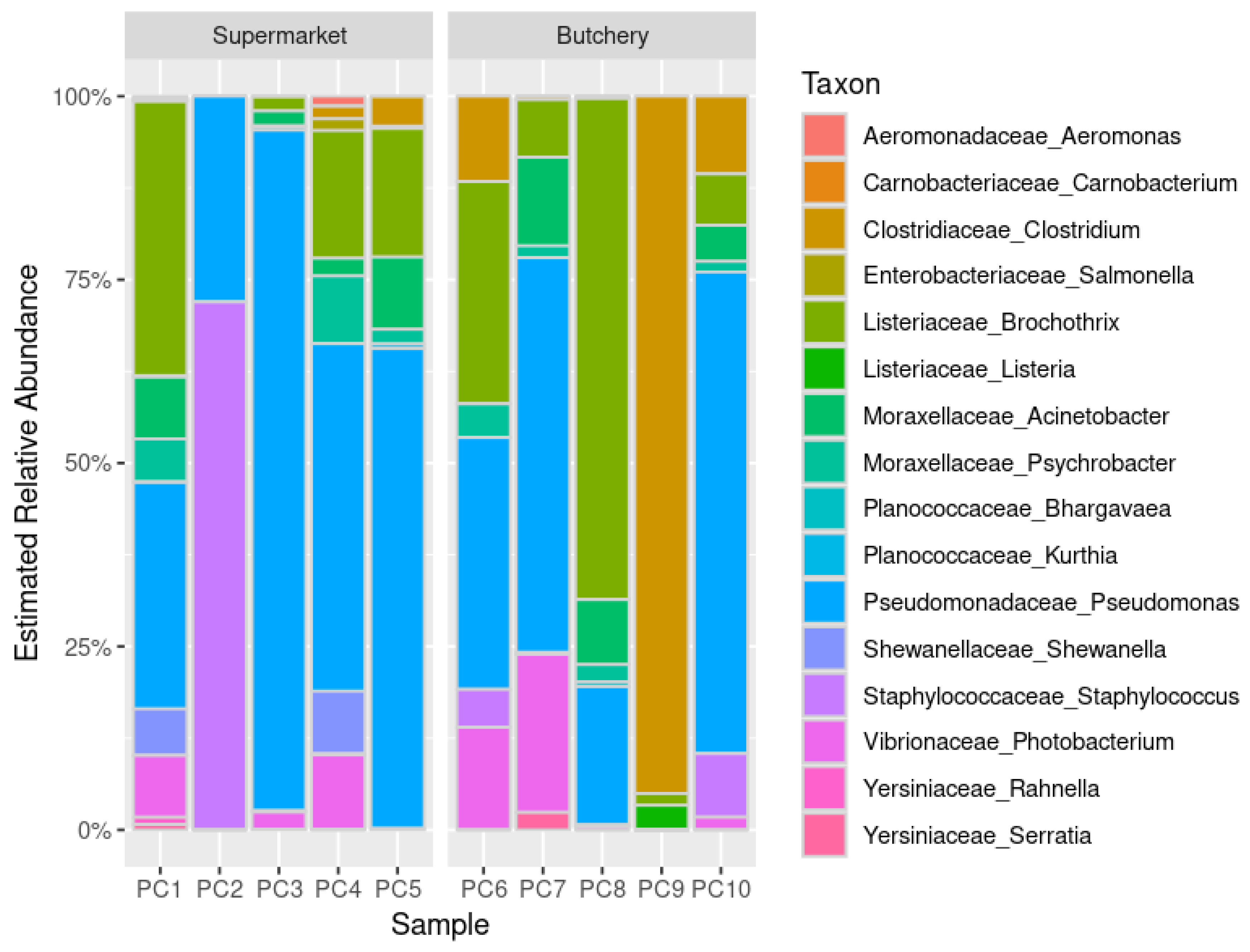
Distribution of the bacterial community structures in the raw meat samples reveals functional diversity among bacteria (
Figure 1). The relative abundance was similar between raw meat samples from supermarkets and butcheries, but PC9 was different.
Food and meat spoilage organisms such as
Pseudomonas (90%; 9/10),
Acinetobacter (80%; 8/10),
Brochothrix (80%; 8/10),
Psychrobacter (70%; 7/10),
Photobacterium (60; 6/10) and
Clostridium (50%; 5/10) were detected in the majority of raw meat samples [
29,
30,
31,
32,
33].
3.4.3. Resistome prediction
Using the assembled contigs generated from MEGAHIT from the 10 raw meat samples, 61665 ORFs were predicted by PathoFact pipeline, of which, 89, 138 and 40 ORFs are predicted to be involve with AMR, VF secretion and toxin secretion, respectively.
3.4.3.1. Antibiotic Resistance Gene Prediction
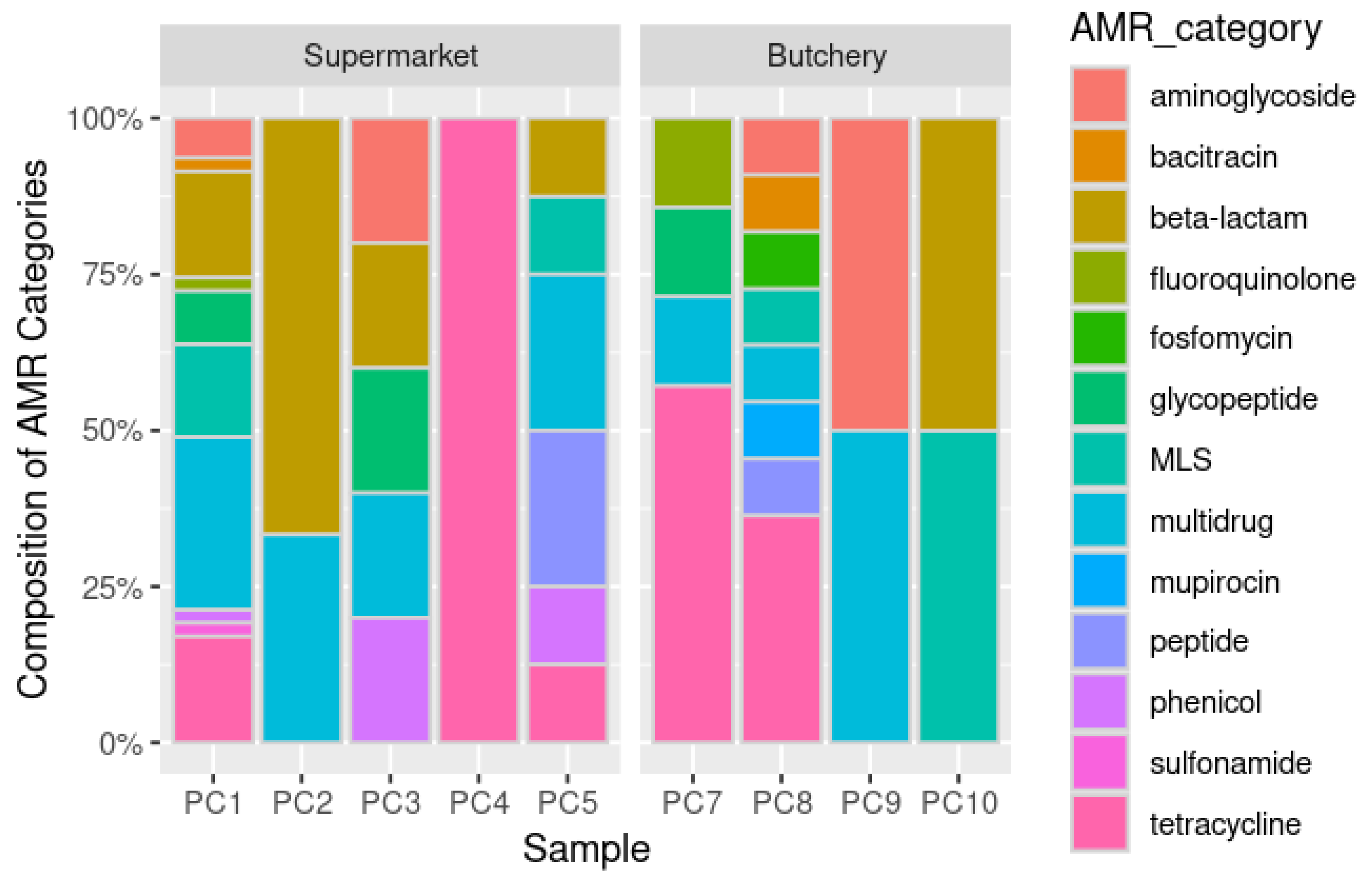
A total of 89 ORFs were predicted as ARGs (
Table 5;
Supplementary Table S3). Raw meat samples from supermarkets were dominated by multidrug (26.8%; 18/67), beta-lactam (20.9%; 14/67) and tetracycline (14.9%; 10/67) resistance genes. Raw meat samples from butcheries were dominated by tetracycline (36.4%; 8/22) and multidrug (13.6%; 3/22) (
Figure 2,
Figure 3A and
Figure 3C) resistance genes.
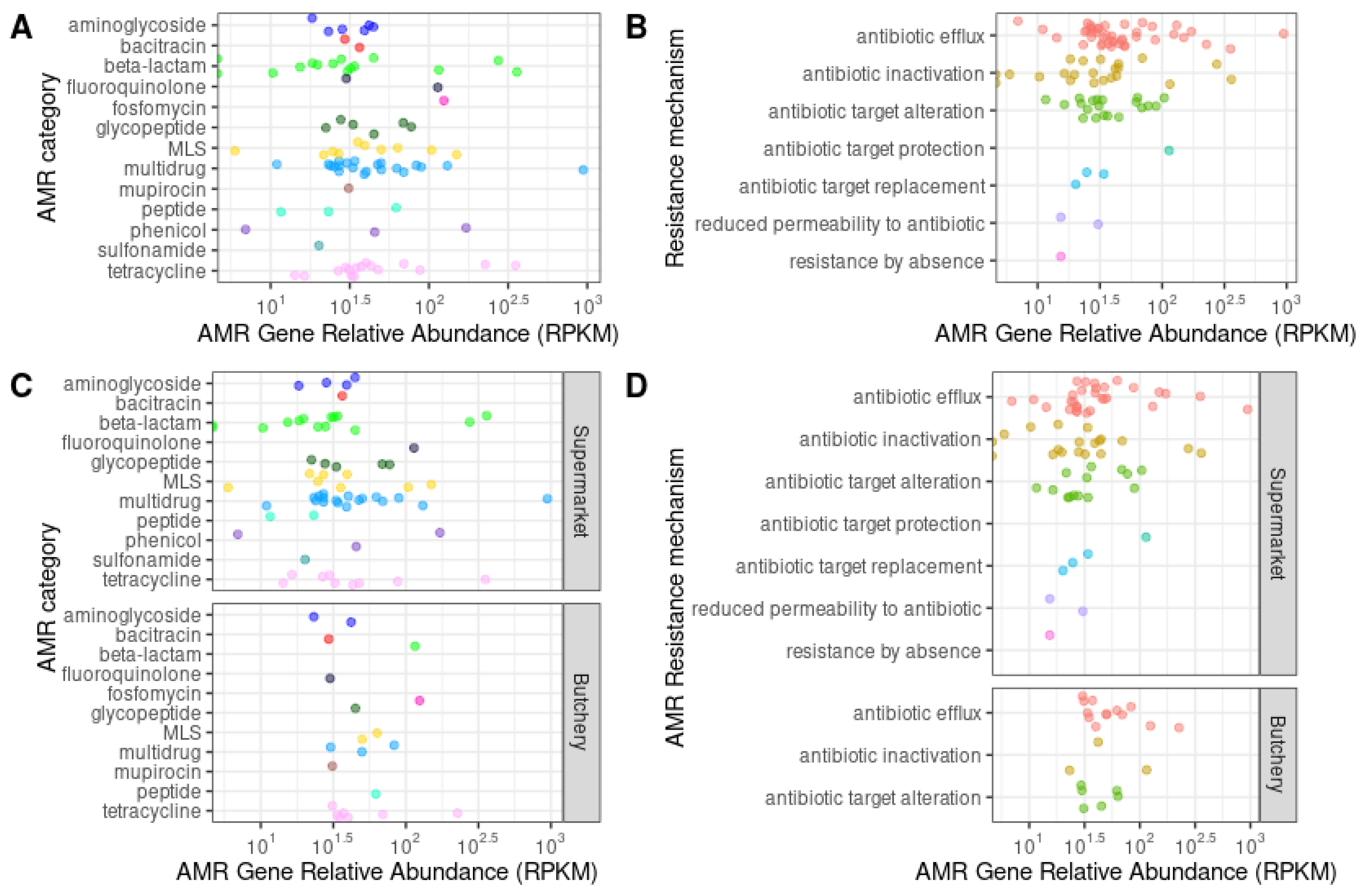
The majority of the ARGs were involved with resistance mechanisms such as antibiotic efflux (48.3%; 43/89), antibiotic inactivation (23.6%; 21/89), and antibiotic target alteration (12.36%; 11/89) (
Supplementary Table S4). No ARG were predicted in sample PC6 (butchery - B1) (
Figure 3B and
Figure 3D;
Supplementary Figure S1).
3.4.3.2 Virulence Factor and Toxin Gene Prediction
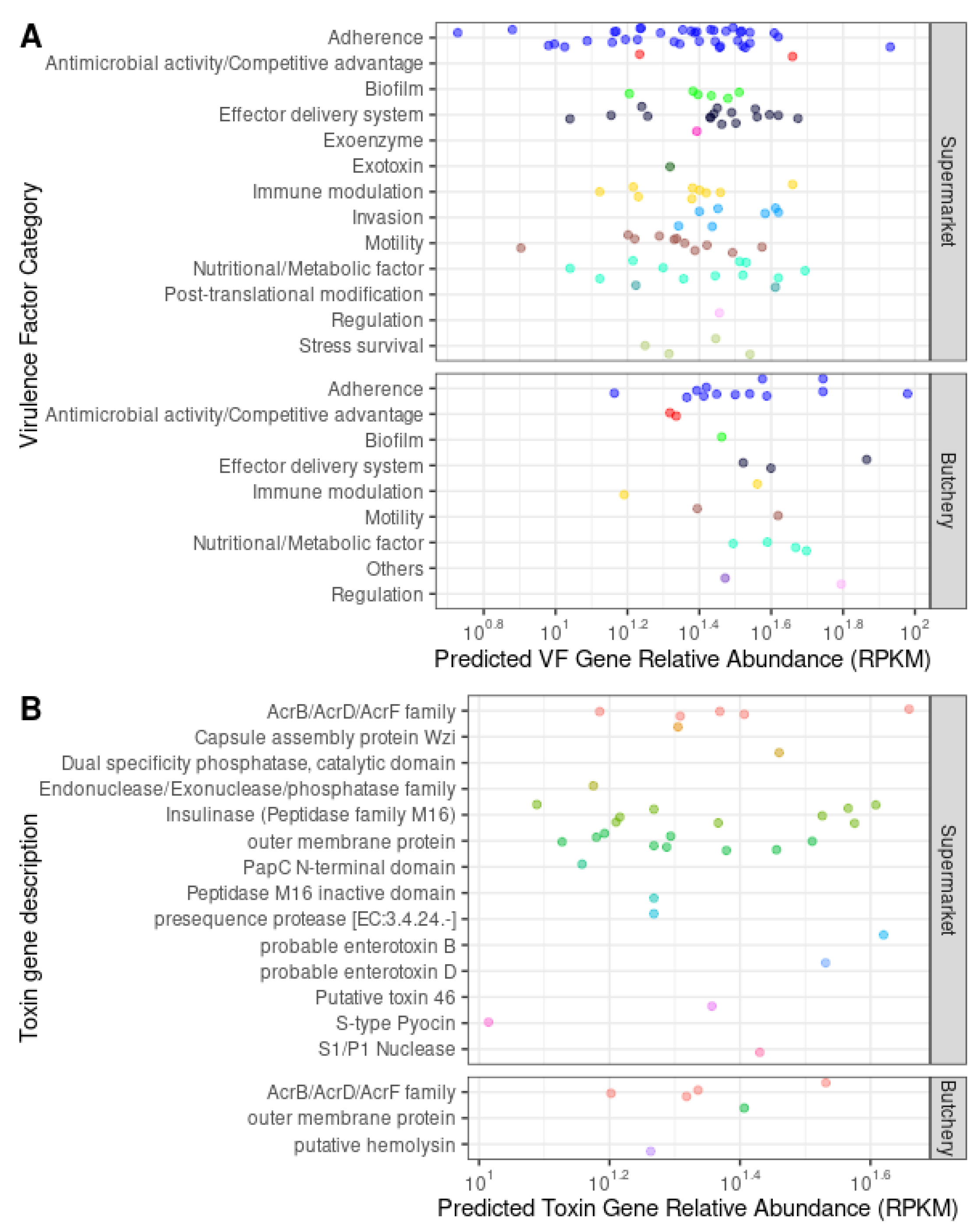
A total of 138 ORFs were predicted as secreted VF, and they are categorised into adherence (36.9%; 51/138), antimicrobial activity/competitive advantage (2.9%; 4/138), biofilm (5.1%; 7/138), effector delivery system (13.8%; 19/138), exoenzyme (0.7%; 1/138), exotoxin (0.7%; 1/138), immune modulation (8%; 11/138), invasion (5.1%; 7/138), motility (9.4%; 13/138), nutritional/metabolic factor (10.9%; 15/138), post-translational modification (1.4%; 2/138), regulation (1.4%; 2/138) and stress survival (3%; 4/138) VF categories (
Table 5;
Figure 4A;
Supplementary Table S5;
Supplementary Figure S1).
VF genes from all categories were present in the raw meat samples collected from supermarkets (
Supplementary Table S6). No exotoxin, invasion, post-translational modification and stress survival VF genes were detected in the raw meat samples collected from butcheries. Fifteen types of toxin genes were detected in the raw meat samples collected from supermarkets and butcheries (
Figure 4B).
4. Discussion
The global crisis of antibiotic resistance is multifaceted and linked to factors such as antibiotic overuse and misuse [
1,
2]. In this study, four selected indicator bacteria (
Campylobacter spp.,
Enterococcus spp.,
E. coli and NTS spp.) that overlap between human and animals, the diversity and abundance of bacterial communities, ARGs and antibiotic residues in raw pork meat samples were investigated.
The majority of the four indicator organisms were absent from the collected raw meat samples. A single E. coli isolate was obtained from sample PC9-B4, while one E. faecalis isolate each was obtained from raw pork meat samples PC3-S3, PC4-S4 and PC10-B5.
The isolated
E. coli and
E. faecalis isolates showed minimal phenotypic resistance. Specifically, the
E. coli isolate showed resistance to tigecycline and trimethoprim/sulfamethoxazole, while all three
E. faecalis isolates showed resistance to moxifloxacin only. WGS data for the
E. coli (PC9-B4) and
E. faecalis isolates (PC3-S3, PC4-S4 and PC10-B5) showed mainly the presence of tetracycline resistance genes. International studies have shown that
E. faecalis was the most dominant
Enterococcus spp. isolated (>80%) from pork samples [
34,
35]. High tetracycline resistance and associated resistance genes were reported in these studies, corresponding with the findings of this study [
34,
35]. In contrast, Aslam
et al. (2012) in Canada found additional ARGs in enterococci isolated from retail meats, including aminoglycosides (
aac,
aphA3,
aad-A and -E and sat4), macrolides (
erm-B and -A), streptogramin (
vatE), bacitracin (
bcrR) and lincosamide (
linB) [
34]. Hart
et al. (2004)
in Australia also reported widespread resistance to tetracycline in
E. coli isolated from pigs [
36]. Tetracycline, a broad-spectrum antibiotic used to treat various infections and as a growth promoter in animals, has contributed to the high tetracycline resistance rates reported in this study from one
E. coli and three
E. faecalis isolates [4,5,37-39]. The WGS data further revealed eight and nine different VF gene categories associated with the sequenced
E. coli (PC9-B4) and
E. faecalis PC3-S3, PC4-S4 and PC10-B5) isolates, respectively. VF genes enable pathogenic bacteria to colonise host niches and establish infections, contributing both directly and indirectly to disease processes [
27]. Aslam
et al. (2012) reported that cytolysin (
cyl-A, -B, -L and -
M), specifically
cyl-A, -L and -M found in this study, as well as aggregation substances (
agg), which were not detected in this study, were unconditionally associated the ARGs
tetM, linB and
bcrR in retail meats [
34]. Another study conducted in Iran has also reported
ace, ebp and
efaA VF genes in
E. faecalis isolates isolated from hospitalised patients [
41]. The
E. coli ST10 detected in this study has been identified as a reservoir for ARG and mobile genetic elements, such as class 1 integrons and plasmids [
39,
42,
43]. The IncB/O/K/Z and
IncFII (pCoo) plasmids detected in the
E. coli isolates have also been reported in
E. coli from hospitalised patients in South Africa and in carbapenem-resistant Enterobacterales from hospitalised patients in Thailand [
44,
45]. Therefore, sequencing, especially plasmid monitoring, is essential for future surveillance studies to track the spread and evolution of these genetic elements in humans, animals and food products.
The antibiotic residue concentrations in the 10 tested raw meat samples were below the acceptable MRL (µg/kg) detection rate according to the published guidelines [
7]. Ramatla
et al. (2017) reported higher antibiotic residue concentrations in raw chicken, pork and beef samples (muscle, liver, or kidney) collected in North West, South Africa [
4]. Possible explanation for the difference in findings between their study and the current one could be attributed to the sampling frequency (monthly sample collection over a period of five months
vs one sampling event) and sample types (muscle, liver, and kidney samples
vs solely muscle samples).
In the context of metagenomics, the majority of the identified bacteria were of food/meat spoilage and environmental origin [29-33]. However, their pathogenicity or ability to cause disease has not been confirmed in this study. Additionally, the resistome analysis revealed the presence of beta-lactam, tetracycline and multidrug resistance genes in the 10 raw meat samples sampled from supermarkets and butcheries. The resistance genes found in this study corresponds to previous local and internal studies [
38,
46,
47].
Further research and analysis are needed to fully understand the potential implications of these findings in terms of environmental health and antibiotic resistance.
This study has several limitations. Firstly, a high number of host reads (i.e. pig DNA) were detected in the metagenomic data, indicating an insufficient number of reads available for the recovery of bacterial metagenome-assembled genomes (MAGs) within the raw meat samples. Consequently, this limited our ability to validate the predications of antibiotic resistance, VF and toxin genes. Using the MAGs approach could have provided more comprehensive information about the bacterial species in the meat samples, potentially identifying novel microorganisms carrying ARGs and uncovering new putative, unculturable pathogens. Secondly, it is recommended to consider increasing the sequencing depth or using alternative microbiome enrichment methods. Thirdly, the study's limited sample size could affect the generalisability of the findings to other supermarkets and butcheries across South Africa.
5. Conclusions
In conclusion, this study highlights the importance of evidence-based investigation and laboratory testing for significant bacterial organisms, as well as antibiotic susceptibility testing in food production systems. While none of the indicator organisms were detected in significant colony-forming units in the collected raw meat samples, isolated E. coli and E. faecalis strains exhibited minimal phenotypic resistance, with notable resistance to tigecycline, trimethoprim/sulfamethoxazole, and moxifloxacin. WGS data highlighted prevalent tetracycline resistance genes, aligning with international studies on tetracycline use in pork production. The presence of various ARG and VF genes in supermarket and butchery meat samples highlights the need for effective antimicrobial use in pork production. Although antibiotic residue concentrations are within acceptable limits, further research is needed to understand the broader implications of these findings on environmental health and antibiotic resistance. This study emphasises the need for vigilance and comprehensive strategies to combat antibiotic resistance in food production systems.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org. Table S1: Demographic data collected from the sampled stores (five supermarkets and five butcheries); Table S2: Captured demographic data collected from the sampled stores five supermarkets and five butcheries); Table S3: Summary of antimicrobial resistance categories by number of open reading frame genes from 10 raw retail meat samples; Table S4: Summary of antimicrobial resistance mechanisms by number of ORF genes from 10 raw retail meat samples; Table S5: Summary of virulence factor category by number of ORF genes from 10 raw retail meat samples; Table S6: Summary of toxin gene description by number of ORF genes from 10 raw retail meat samples; Figure S1: Mobile genetic element prediction of ARGs of all raw meat samples from both supermarket and butchery groups; Figure S2: Mobile genetic element prediction of VF and toxin genes of all raw meat samples from both supermarket and butchery groups.
Author Contributions
Conceptualisation, O.P. and W.S.; methodology, M.L., and W.S.; software, M.L. and W.Y.C.; validation, M.L. and W.S.; formal analysis, M.L. and W.Y.C.; investigation, M.L. and W.S.; resources, O.P.; data curation, M.L., W.S. and W.Y.C.; writing—original draft preparation, M.L.; writing—review and editing, M.L., W.S., W.Y.C. and O.P.; visualisation, M.L. and W.Y.C.; supervision, O.P.; project administration, M.L., W.S. and O.P.; funding acquisition, O.P. All authors have read and agreed to the published version of the manuscript.
Funding
Research reported in this publication was supported by the South African Medical Research Council (SAMRC) as a sub-grant received from the Bill and Melinda Gates Foundation (Grant number: 96086). The content and findings reported/illustrated are the sole deduction, view and responsibility of the authors and do not reflect the official position and sentiments of the SAMRC or the Bill and Melinda Gates Foundation. WS was also supported by a Fogarty International Center Global Infectious Disease research-training grant, National Institutes of Health, to the University of Pittsburgh and the National Institute for Communicable Diseases (D43TW011255).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Human Research Ethics Committee of the University of the Witwatersrand (Protocol No: M190244).
Informed Consent Statement
Not applicable.
Data Availability Statement
The assembled genome files were submitted deposited in the National Center for Biotechnology Information GenBank and are available under the following BioProject numbers: PRJNA1006163. The supplementary materials contain additional data.
Acknowledgments
We would like to thank Prof Arshad Ismail and the rest of the staff members of the Sequencing Core Facility (SCF) at the National Institute for Communicable Diseases (NICD) for guidance and for the sequencing of our isolate and retail pork meat selection.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Eckstrom, K.; Barlow, J.W. Resistome metagenomics from plate to farm: the resistome and microbial composition during food waste feeding and composting on a Vermont poultry farm. PLoS One 2019, 14, e0219807. [CrossRef]
- Li, Y.; Cao, W.; Liang, S.; Yamasaki, S.; Chen, X.; Shi, L.; Ye, L. Metagenomic characterization of bacterial community and antibiotic resistance genes in representative ready-to-eat food in southern China. Sci Rep 2020, 10, 15175. [CrossRef]
- Department of Agriculture Land Reform and Rural Development. A profile of the South African pork market value chain. Available online: http://webapps1.daff.gov.za/AmisAdmin/upload/Pork%20Market%20Value%20Chain%20Profile%202021.pdf (accessed on 26 October 2023).
- Ramatla, T.; Ngoma, L.; Adetunji, M.; Mwanza, M. Evaluation of antibiotic residues in raw meat using different analytical methods. Antibiotics (Basel) 2017, 6, 34. [CrossRef]
- Patel, S.J.; Wellington, M.; Shah, R.M.; Ferreira, M.J. Antibiotic stewardship in food-producing animals: challenges, progress, and opportunities. Clin Ther 2020, 42, 1649-1658. [CrossRef]
- World Health Organization. WHO guidelines on use of medically important antimicrobials in food-producing animals. Available online: https://www.who.int/publications/i/item/9789241550130 (accessed on 31 October 2023).
- Food and Agriculture Organization of the United Nations, F.W.H.O., WHO. CX/MRL 2-2021: Maximum residue limits (MRLs) and risk management recommendations (RMRs) for residues of veterinary drugs in foods. Available online: https://www.fao.org/fao-who-codexalimentarius/codex-texts/maximum-residue-limits/en/ (accessed on 20 July 2023).
- 2007, F.C.a.D.A.A.o. Available online: https://www.gov.za/sites/default/files/gcis_document/201409/a39-07.pdf (accessed on 26 October 2023).
- Bezanson, G.S.; MacInnis, R.; Potter, G.; Hughes, T. Presence and potential for horizontal transfer of antibiotic resistance in oxidase-positive bacteria populating raw salad vegetables. Int J Food Microbiol 2008, 127, 37-42. [CrossRef]
- Sultana, F.; Kamrunnahar; Afroz, H.; Jahan, A.; Fakruddin, M.; Datta, S. Multi-antibiotic resistant bacteria in frozen food (ready to cook food) of animal origin sold in Dhaka, Bangladesh. Asian Pac J Trop Biomed 2014, 4, S268-271. [CrossRef]
- Urban-Chmiel, R.; Marek, A.; Stepien-Pysniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic resistance in bacteria-A review. Antibiotics (Basel) 2022, 11. [CrossRef]
- Schulz, L. Consumers respond to meat price differences. Available online: https://www.extension.iastate.edu/agdm/articles/schulz/SchApr22.html (accessed on 31 October 2023).
- EUCAST. EUCAST Breakpoint tables for interpretation of MICs and zone diameters (v. 13.1). 2023.
- Kwenda SAM, K.Z., Mtshali S, Mnyameni F, Ismail A. Jekesa: an automated easy-to-use pipeline for bacterial whole genome typing. 2020.
- Felix Krueger, F.J., Phil Ewels, Ebrahim Afyounian, Benjamin Schuster-Boeckler. FelixKrueger/TrimGalore: v0.6.7 - DOI via Zenodo (0.6.7). Zenodo 2021. [CrossRef]
- Souvorov, A.; Agarwala, R.; Lipman, D.J. SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol 2018, 19, 153. [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072-1075. [CrossRef]
- Jolley, K.A.; Maiden, M.C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 2010, 11, 595. [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2017, 45, D566-D573. [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: hierarchical and refined dataset for big data analysis--10 years on. Nucleic Acids Res 2016, 44, D694-697. [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012, 67, 2640-2644. [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 2020, 75, 3491-3500. [CrossRef]
- Zankari, E.; Allesoe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 2017, 72, 2764-2768. [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: architecture and applications. BMC Bioinformatics 2009, 10, 421. [CrossRef]
- Lu, J.; Rincon, N.; Wood, D.E.; Breitwieser, F.P.; Pockrandt, C.; Langmead, B.; Salzberg, S.L.; Steinegger, M. Metagenome analysis using the Kraken software suite. Nat Protoc 2022, 17, 2815-2839. [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674-1676. [CrossRef]
- de Nies, L.; Lopes, S.; Busi, S.B.; Galata, V.; Heintz-Buschart, A.; Laczny, C.C.; May, P.; Wilmes, P. PathoFact: a pipeline for the prediction of virulence factors and antimicrobial resistance genes in metagenomic data. Microbiome 2021, 9, 49. [CrossRef]
- Kaminski, J.; Gibson, M.K.; Franzosa, E.A.; Segata, N.; Dantas, G.; Huttenhower, C. High-specificity targeted functional profiling in microbial communities with ShortBRED. PLoS Comput Biol 2015, 11, e1004557. [CrossRef]
- Stellato, G.; La Storia, A.; De Filippis, F.; Borriello, G.; Villani, F.; Ercolini, D. Overlap of spoilage-associated microbiota between meat and the meat processing environment in small-scale and large-scale retail distributions. Appl Environ Microbiol 2016, 82, 4045-4054. [CrossRef]
- Stellato, G.; Utter, D.R.; Voorhis, A.; De Angelis, M.; Eren, A.M.; Ercolini, D. A few Pseudomonas oligotypes dominate in the meat and dairy processing environment. Front Microbiol 2017, 8, 264. [CrossRef]
- Fuertes-Perez, S.; Hauschild, P.; Hilgarth, M.; Vogel, R.F. Biodiversity of Photobacterium spp. isolated from meats. Front Microbiol 2019, 10, 2399. [CrossRef]
- lulietto, M.F.; Sechi, P.; Borgogni, E.; Cenci-Goga, B.T. Meat spoilage: A critical review of a neglected alteration due to ropy slime producing bacteria. Italian Journal of Animal Science 2016, 14. [CrossRef]
- Cha, M.H.; Kim, S.H.; Kim, S.; Lee, W.; Kwak, H.S.; Chi, Y.M.; Woo, G.J. Antimicrobial resistance profile of Acinetobacter spp. isolates from retail meat samples under Campylobacter-selective conditions. J Microbiol Biotechnol 2021, 31, 733-739. [CrossRef]
- Aslam, M.; Diarra, M.S.; Checkley, S.; Bohaychuk, V.; Masson, L. Characterization of antimicrobial resistance and virulence genes in Enterococcus spp. isolated from retail meats in Alberta, Canada. Int J Food Microbiol 2012, 156, 222-230. [CrossRef]
- Tyson, G.H.; Nyirabahizi, E.; Crarey, E.; Kabera, C.; Lam, C.; Rice-Trujillo, C.; McDermott, P.F.; Tate, H. Prevalence and antimicrobial resistance of Enterococci isolated from retail meats in the United States, 2002 to 2014. Appl Environ Microbiol 2018, 84. [CrossRef]
- Hart, W.S.; Heuzenroeder, M.W.; Barton, M.D. Antimicrobial resistance in Campylobacter spp., Escherichia coli and Enterococci associated with pigs in Australia. J Vet Med B Infect Dis Vet Public Health 2004, 51, 216-221. [CrossRef]
- Strasheim, W.; Etter, E.M.C.; Lowe, M.; Perovic, O. Method to Assess Farm-Level Vaccine and Antibiotic Usage Utilizing Financial Documentation: A Pilot Study in a Commercial Pig Farm in South Africa From 2016 to 2018. Front Vet Sci 2022, 9, 856729. [CrossRef]
- Liu, Z.; Klumper, U.; Shi, L.; Ye, L.; Li, M. From pig breeding environment to subsequently produced pork: comparative analysis of antibiotic resistance denes and cacterial community composition. Front Microbiol 2019, 10, 43. [CrossRef]
- Peng, Z.; Hu, Z.; Li, Z.; Zhang, X.; Jia, C.; Li, T.; Dai, M.; Tan, C.; Xu, Z.; Wu, B.; et al. Antimicrobial resistance and population genomics of multidrug-resistant Escherichia coli in pig farms in mainland China. Nat Commun 2022, 13, 1116. [CrossRef]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; van Nimwegen, E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Molecular Biology and Evolution 2014, 31, 1077-1088. [CrossRef]
- Kafil, H.S.; Mobarez, A.M.; Moghadam, M.F. Adhesion and virulence factor properties of Enterococci isolated from clinical samples in Iran. Indian J Pathol Microbiol 2013, 56, 238-242. [CrossRef]
- Reid, C.J.; Wyrsch, E.R.; Roy Chowdhury, P.; Zingali, T.; Liu, M.; Darling, A.E.; Chapman, T.A.; Djordjevic, S.P. Porcine commensal Escherichia coli: a reservoir for class 1 integrons associated with IS26. Microb Genom 2017, 3. [CrossRef]
- Reid, C.J.; DeMaere, M.Z.; Djordjevic, S.P. Australian porcine clonal complex 10 (CC10) Escherichia coli belong to multiple sublineages of a highly diverse global CC10 phylogeny. Microb Genom 2019, 5. [CrossRef]
- Perovic, O.; Singh-Moodley, A.; Lowe, M. In vitro activity of ceftolozane-tazobactam against Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa obtained from blood cultures from sentinel public hospitals in South Africa. Antibiotics 2023, 12, 453. [CrossRef]
- Paveenkittiporn, W.; Kamjumphol, W.; Ungcharoen, R.; Kerdsin, A. Whole-genome sequencing of clinically isolated carbapenem-resistant Enterobacterales harboring mcr genes in Thailand, 2016-2019. Front Microbiol 2020, 11, 586368. [CrossRef]
- Abdalla, S.E.; Abia, A.L.K.; Amoako, D.G.; Perrett, K.; Bester, L.A.; Essack, S.Y. From farm-to-fork: E. coli from an intensive pig production system in South Africa shows high resistance to critically important antibiotics for human and animal use. Antibiotics 2021, 10. [CrossRef]
- Barroga, T.R.M.; Morales, R.G.; Benigno, C.C.; Castro, S.J.M.; Caniban, M.M.; Cabullo, M.F.B.; Agunos, A.; de Balogh, K.; Dorado-Garcia, A. Antimicrobials used in backyard and commercial poultry and swine farms in the Philippines: a qualitative pilot study. Front Vet Sci 2020, 7, 329. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).