Submitted:
29 June 2024
Posted:
03 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Cultivated Crustacean Meat
1.2. Stem Cells and Myogenic Factors
1.3. Regenerating Limb Bud Model
1.4. Molecular Factors Relevant to CM and CCM Research
2. Results
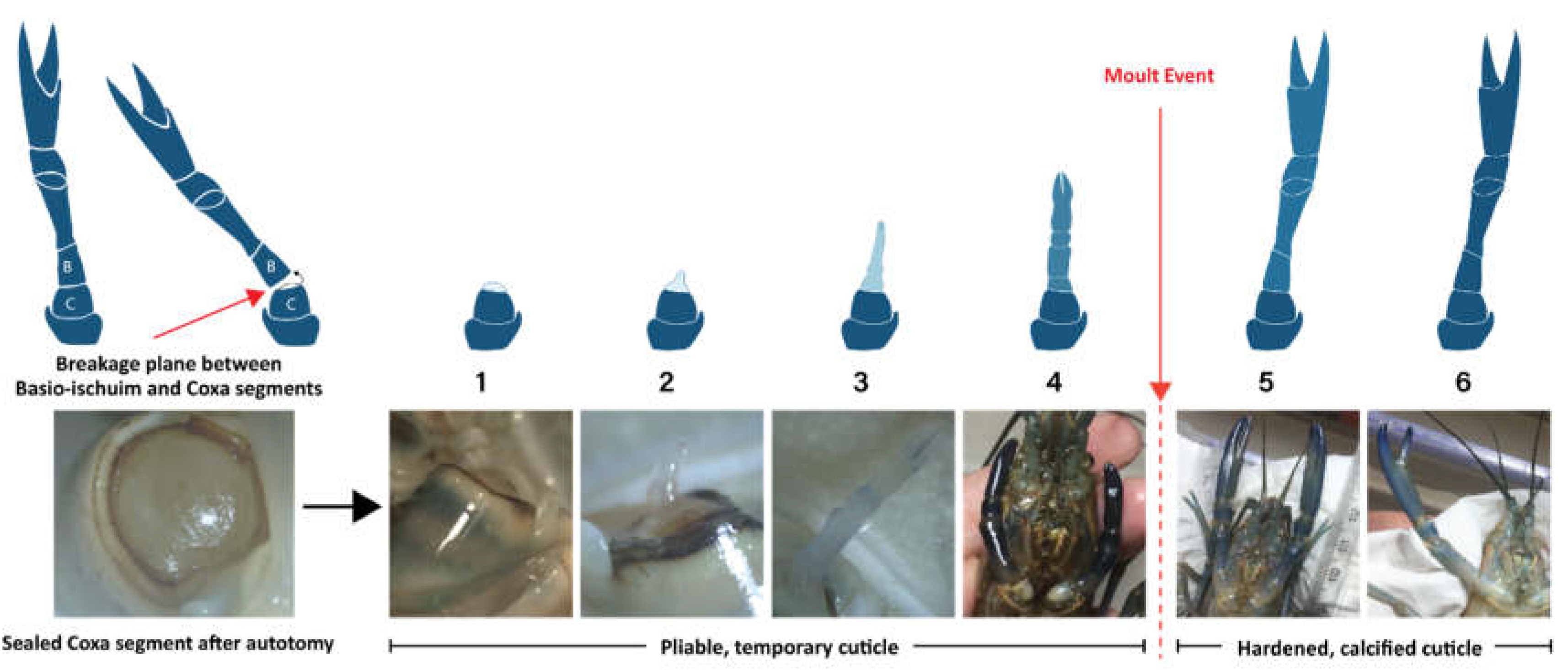
2.1. Stage Allocation and Sample Preparation
2.2. Transcriptome Sequencing, Assembly and Quantification
2.3. Annotation of the Reference Transcriptome
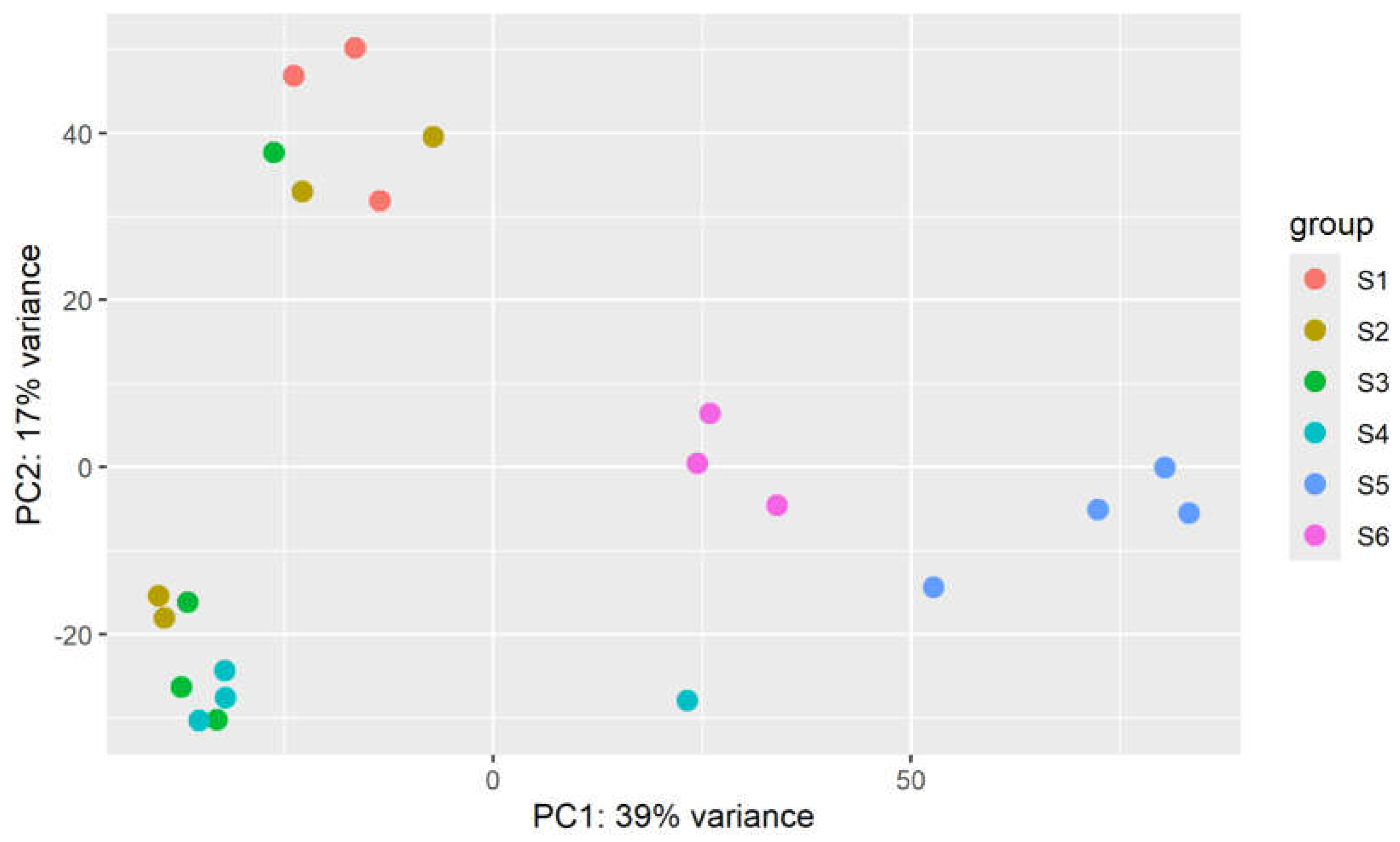
2.4. Principal Component Analyis
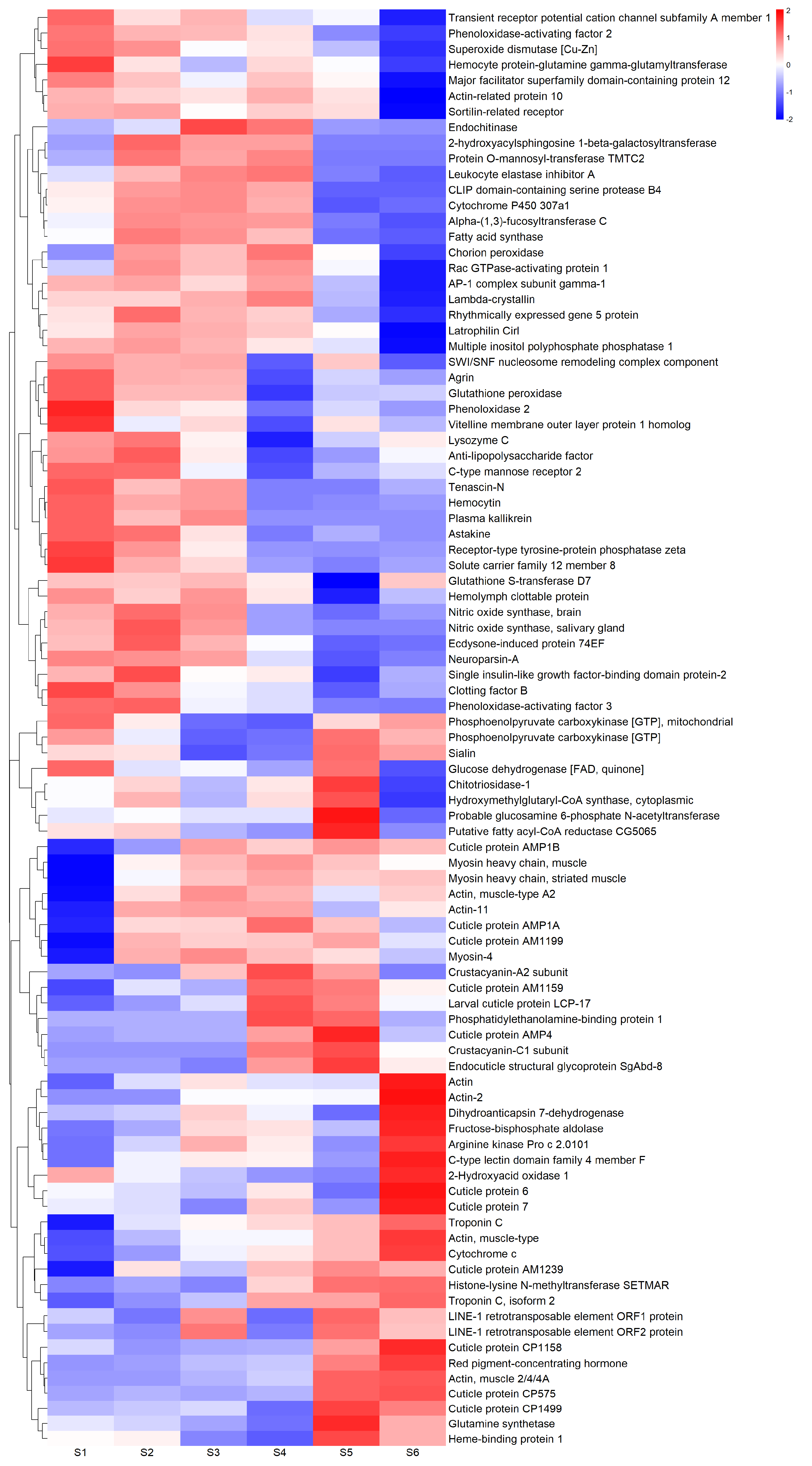
2.5. Selection of Differentially Expressed Genes
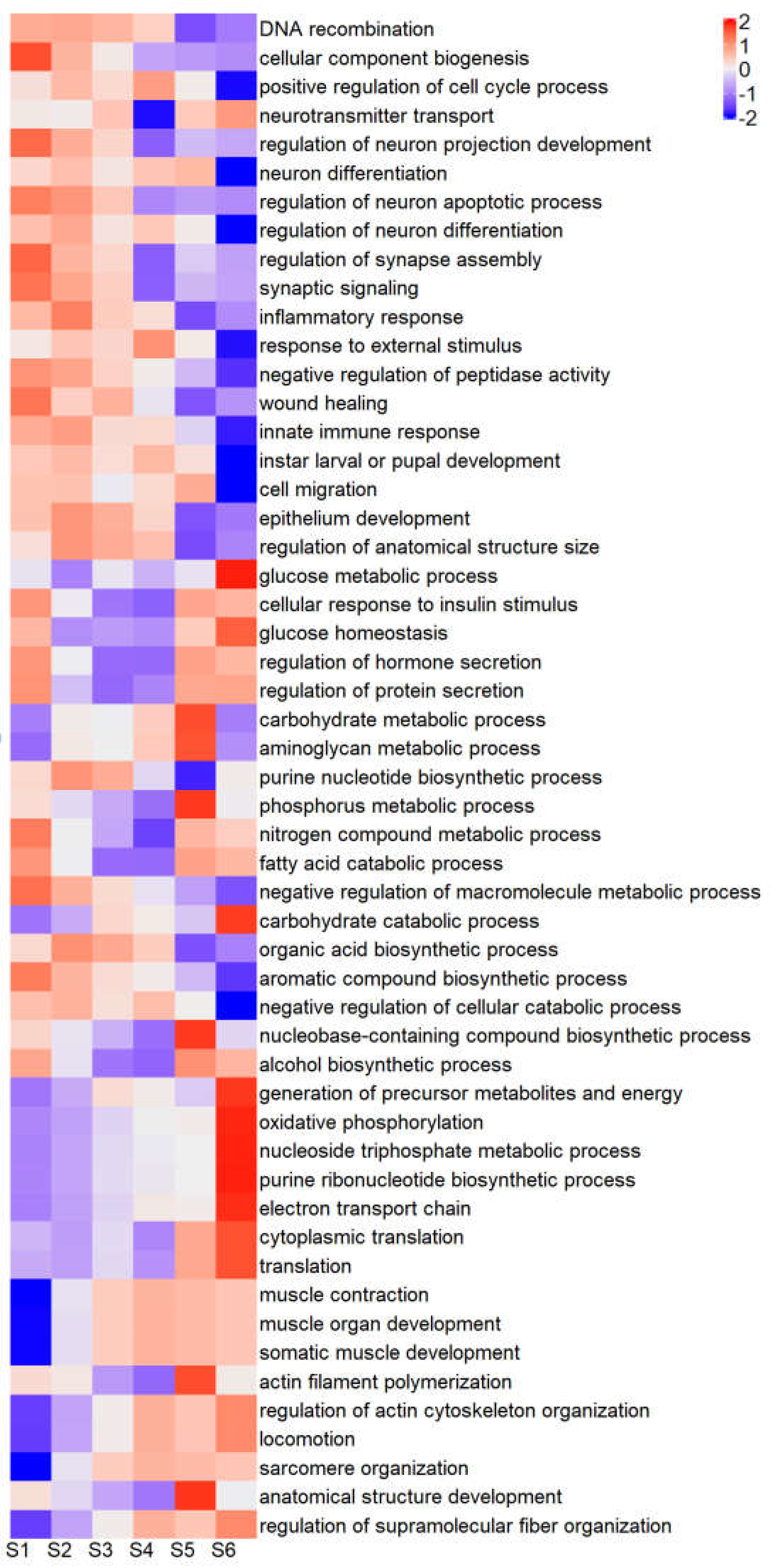
2.6. GO Enrichment of Differentially Expressed Genes
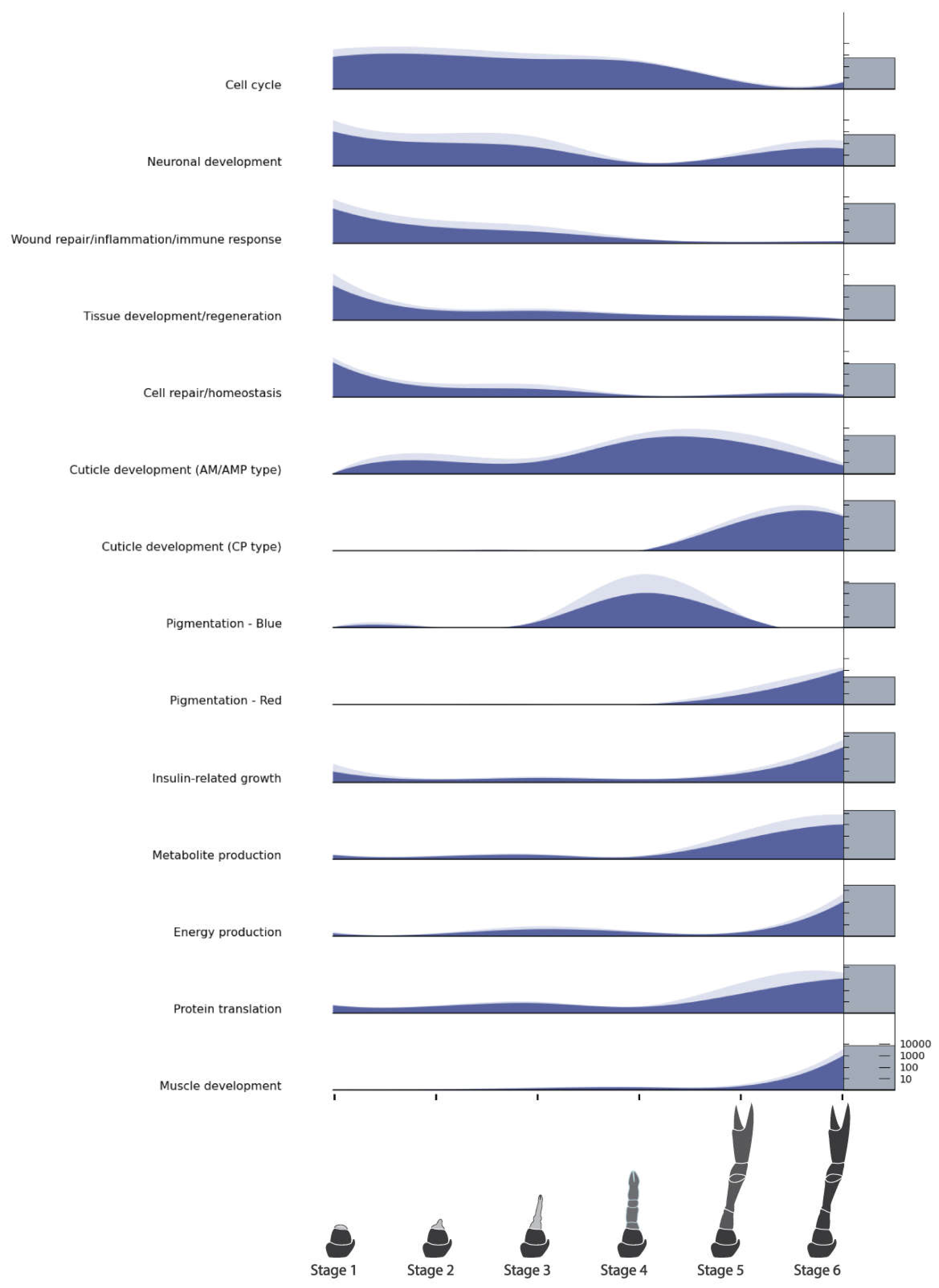
2.7. Functional Characterization of Differentially Expressed Genes and GO Terms
2.8. Identification and Relative Expression Analysis of Target Homologs
2.8.1. Endocrine Factors
2.8.2. Myogenic Factors
2.8.3. Cell Cycle Genes
2.8.4. Pluripotency Factors
2.9. Stage 4 vs Stage 6 Tissues as Potential Stem Cells Sources for CCM Development
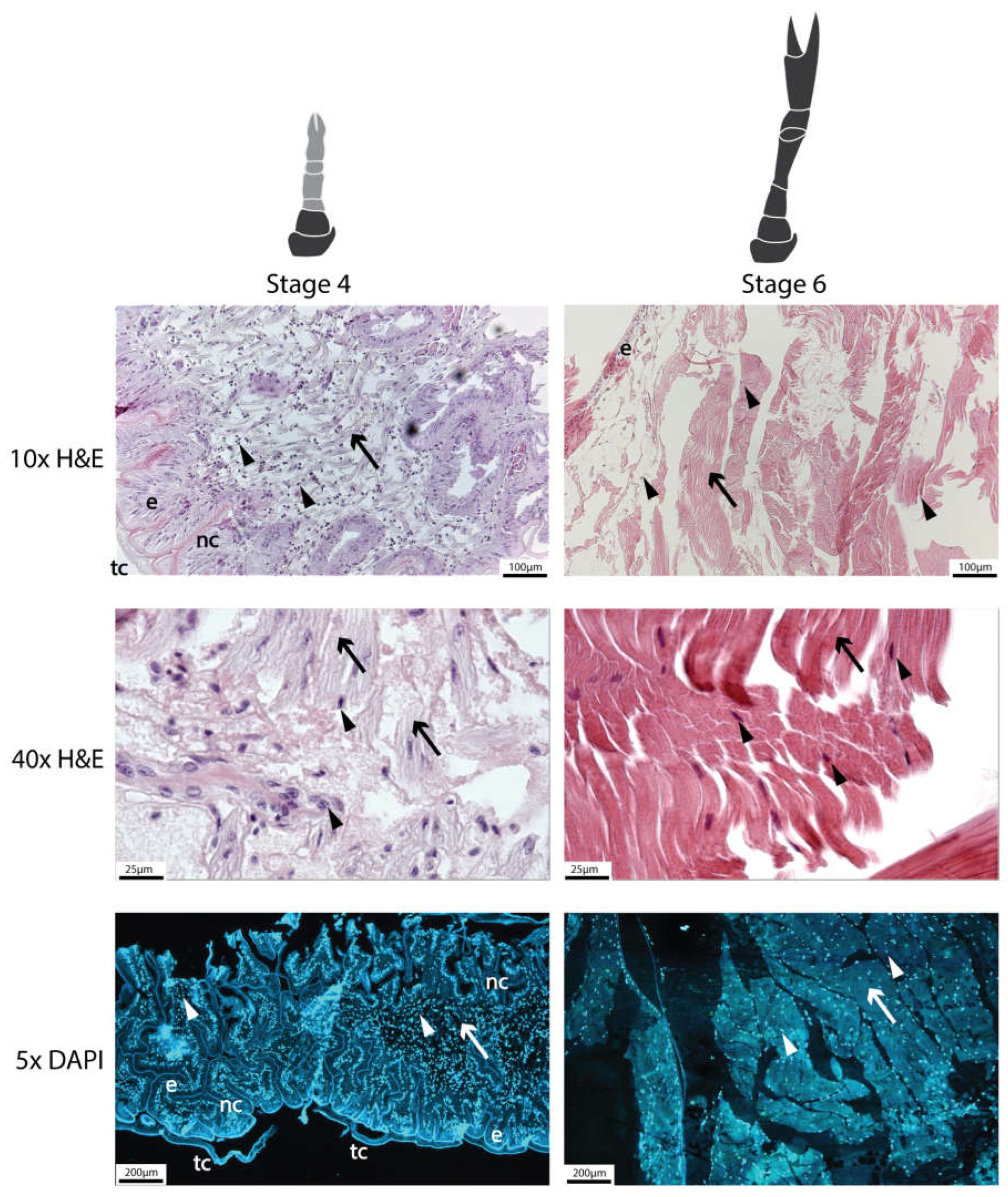
2.9.1. Histological Analysis
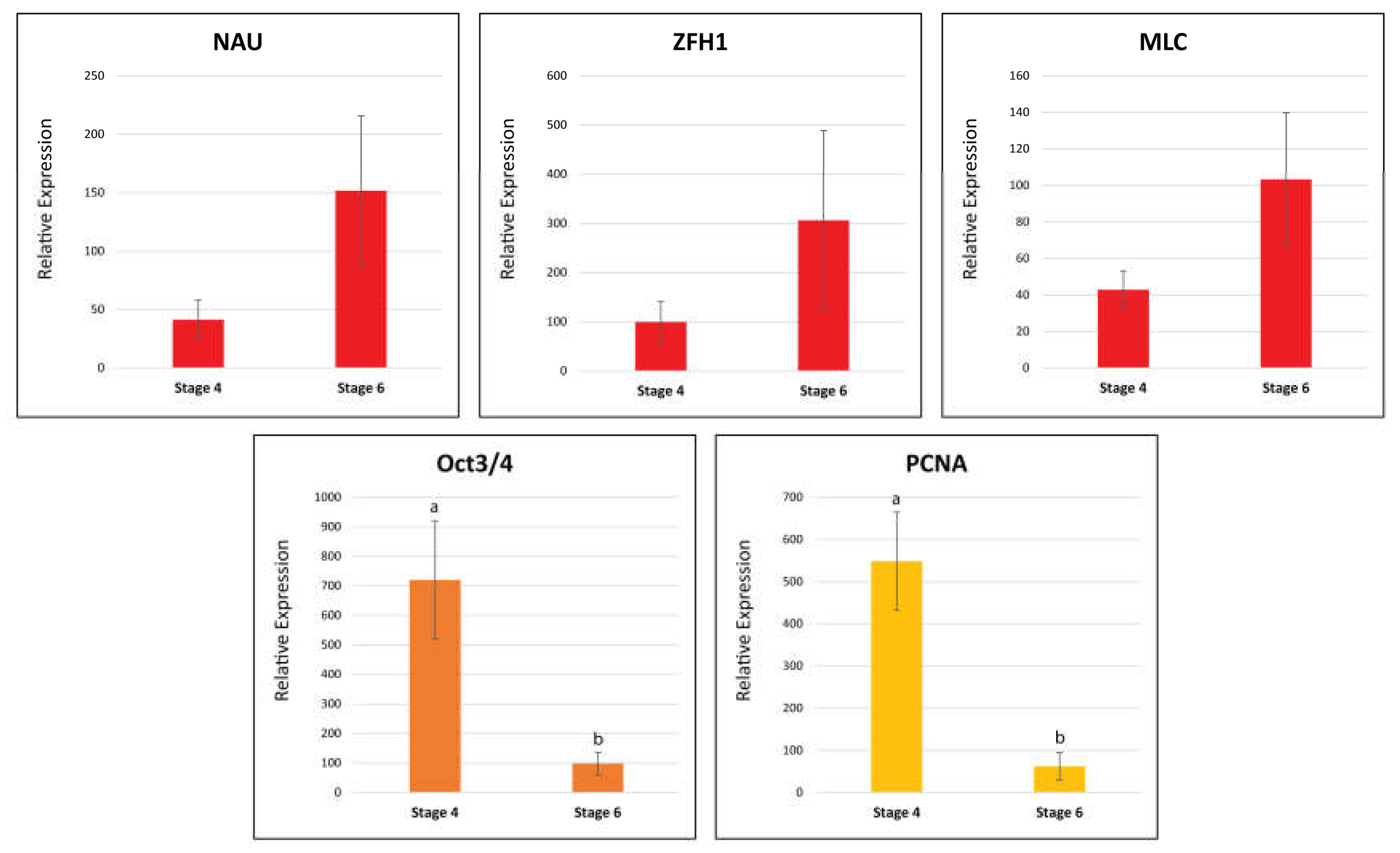
2.9.2. Real-Time Quantitative PCR
3. Discussion
3.1. Differentially Expressed Genes (DEG) Analysis
3.2. Target Gene Analysis
3.2.1. Endocrine Factors
3.2.2. Myogenic Factors
3.2.3. Cell Cycle and Pluripotency Factors
3.3. Limitations
4. Materials and Methods
4.1. Animal Handling
4.2. Regeneration Assessment
4.3. Anesthetic
4.4. Sample Preparation
4.5. RNA Extraction, Quantification, and Sequencing
4.6. Transcriptome Assembly and Quantification
4.7. Data Upload into CrustyBase.org
4.8. Differential Expression Analysis
4.9. Reference Assembly Annotation
4.10. Gene Ontology Enrichment Analysis
4.11. Graphical Representation of the Functional DEG and GO Term Categories
4.12. Target gene Analysis
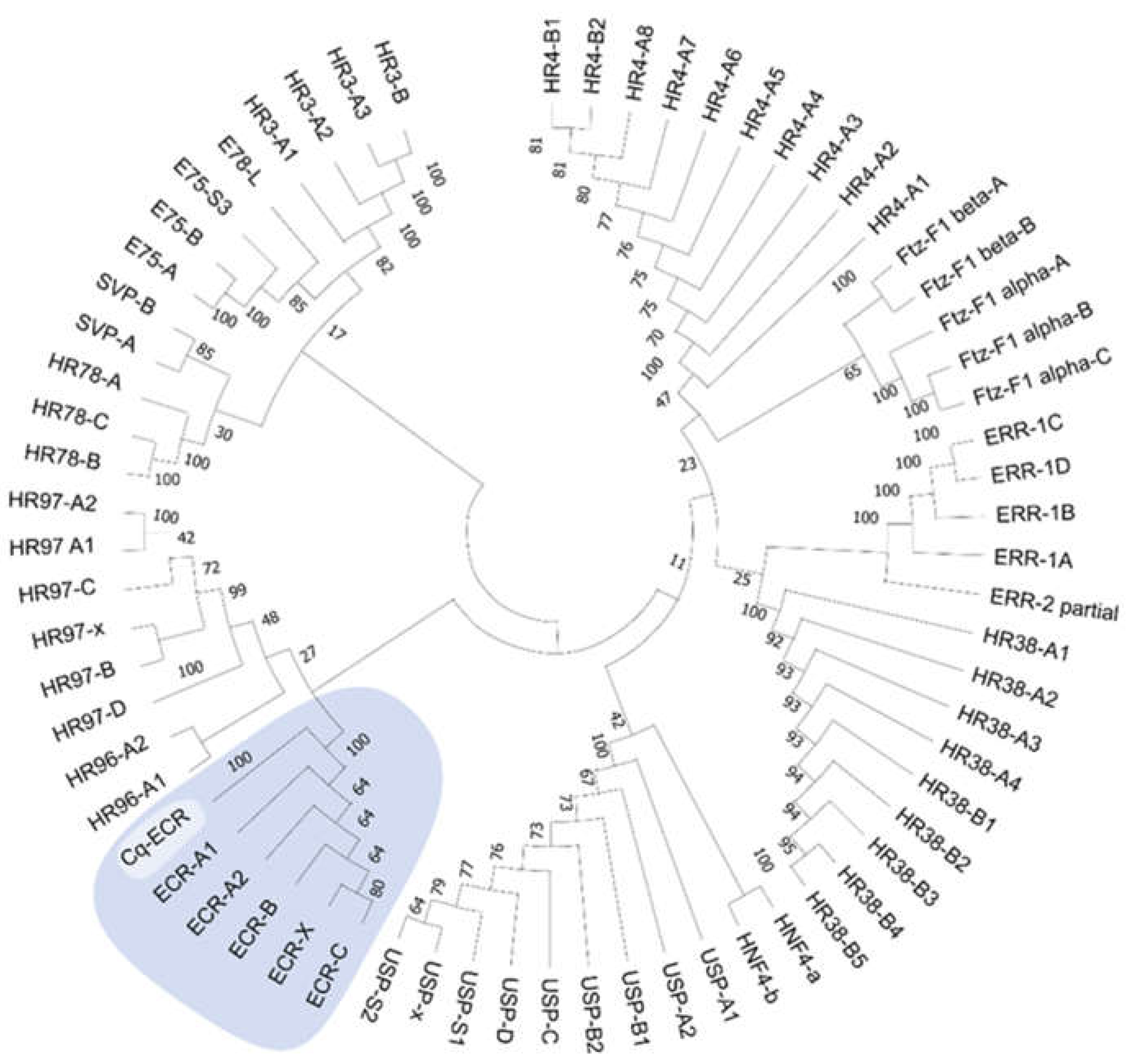
4.13. Phylogeny
4.14. Histology
4.15. Real-Time Quantitative PCR
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Animal Ethics
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2020; 978-92-5-132692-3; United Nations Food and Agriculture Organisation: Rome, Italy, 2020 2020. [Google Scholar]
- Baechler, B.R.; Stienbarger, C.D.; Horn, D.A.; Joseph, J.; Taylor, A.R.; Granek, E.F.; Brander, S.M. Microplastic occurrence and effects in commercially harvested North American finfish and shellfish: Current knowledge and future directions. Limnology & Oceanography Letters 2020, 5, 113–136. [Google Scholar] [CrossRef]
- Baki, M.A.; Hossain, M.M.; Akter, J.; Quraishi, S.B.; Haque Shojib, M.F.; Atique Ullah, A.K.M.; Khan, M.F. Concentration of heavy metals in seafood (fishes, shrimp, lobster and crabs) and human health assessment in Saint Martin Island, Bangladesh. Ecotoxicology & Environmental Safety 2018, 159, 153–163. [Google Scholar] [CrossRef]
- Fatema, U.K.; Faruque, H.; Salam, M.A.; Matsuda, H. Vulnerability assessment of target shrimps and bycatch species from industrial shrimp trawl fishery in the bay of Bengal, Bangladesh. Sust. 2022, 14. [Google Scholar] [CrossRef]
- Lira, A.S.; Le Loc'h, F.; Andrade, H.A.; Lucena-Frédou, F.; Zhou, S. Vulnerability of marine resources affected by a small-scale tropical shrimp fishery in northeast Brazil. ICES Journal of Marine Science 2022, 79, 633–647. [Google Scholar] [CrossRef]
- Ahmed, N.; Cheung, W.W.L.; Thompson, S.; Glaser, M. Solutions to blue carbon emissions: Shrimp cultivation, mangrove deforestation and climate change in coastal Bangladesh. Marine Policy 2017, 82, 68–75. [Google Scholar] [CrossRef]
- Barraza-Guardado, R.H.; Arreola-Lizarraga, J.A.; Lopez-Torres, M.A.; Casillas-Hernandez, R.; Miranda-Baeza, A.; Magallon-Barrajas, F.; Ibarra-Gamez, C. Effluents of shrimp farms and its influence on the coastal ecosystems of Bahia de Kino, Mexico. The Scientific World Journal 2013, 2013, 306370. [Google Scholar] [CrossRef]
- Boone Kauffman, J.; Arifanti, V.B.; Hernández Trejo, H.; Carmen Jesús García, M.; Norfolk, J.; Cifuentes, M.; Hadriyanto, D.; Murdiyarso, D. The jumbo carbon footprint of a shrimp: Carbon losses from mangrove deforestation. Frontiers in Ecology & the Environment 2017, 15, 183–188. [Google Scholar] [CrossRef]
- Macusi, E.D.; Estor, D.E.P.; Borazon, E.Q.; Clapano, M.B.; Santos, M.D. Environmental and socioeconomic impacts of shrimp farming in the Philippines: A critical analysis using PRISMA. Sust. 2022, 14. [Google Scholar] [CrossRef]
- Rubio, N.; Datar, I.; Stachura, D.; Kaplan, D.; Krueger, K. Cell-Based Fish: A novel approach to seafood production and an opportunity for cellular agriculture. Frontiers in Sustainable Food Systems 2019, 3, 43. [Google Scholar] [CrossRef]
- Albalat, A.; Zacarias, S.; Coates, C.J.; Neil, D.M.; Planellas, S.R. Welfare in farmed decapod crustaceans, with particular reference to Penaeus vannamei. Frontiers in Marine Science 2022, 9, 886024. [Google Scholar] [CrossRef]
- Elwood, R.W.; Barr, S.; Patterson, L. Pain and stress in crustaceans? Applied Animal Behaviour Science 2009, 118, 128–136. [Google Scholar] [CrossRef]
- Passantino, A.; Elwood, R.W.; Coluccio, P. Why Protect Decapod Crustaceans Used as Models in Biomedical Research and in Ecotoxicology? Ethical and Legislative Considerations. Animals 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Reiss, J.; Robertson, S.; Suzuki, M. Cell sources for cultivated meat: Applications and considerations throughout the production workflow. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Musgrove, L.; Russell, F.D.; Ventura, T. Considerations for cultivated crustacean meat: Potential cell sources, potential differentiation and immortalization strategies, and lessons from crustacean and other animal models. Critical Reviews in Food Science and Nutrition 2024, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Post, M.J.; Levenberg, S.; Kaplan, D.L.; Genovese, N.; Fu, J.; Bryant, C.J.; Negowetti, N.; Verzijden, K.; Moutsatsou, P. Scientific, sustainability and regulatory challenges of cultured meat. Nature Food 2020, 1, 403–415. [Google Scholar] [CrossRef]
- Bomkamp, C.; Musgrove, L.; Marques, D.M.C.; Fernando, G.F.; Ferreira, F.C.; Specht, E.A. Differentiation and maturation of muscle and fat cells in cultivated seafood: Lessons from developmental biology. Marine Biotechnology 2023, 25, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Vogt, G. Cytology, function and dynamics of stem and progenitor cells in decapod crustaceans. Biological Reviews 2022, 97, 817–850. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, P.M.; Chung, A.C.K.; Durica, D.S. Limb regeneration in the fiddler crab, Uca pugilator: Histological, physiological and molecular considerations. American Zoologist 1999, 39, 513–526. [Google Scholar] [CrossRef]
- Hopkins, P.M.; Das, S. Ch6 Regeneration in Crustaceans. In Physiology: Crustacea, Chang, E.S., Thiel, M., Eds.; Oxford University Press: Oxford, England, 2015; pp. 168–198. [Google Scholar]
- Uhrík, B.; Rýdlová, K.; Zacharová, D. The roles of haemocytes during degeneration and regeneration of crayfish muscle fibres. Cell Tissue Res. 1989, 255, 443–449. [Google Scholar] [CrossRef]
- Das, S. Morphological, molecular, and hormonal basis of limb regeneration across Pancrustacea. Integrative & Comparative Biology 2015, 55, 869–877. [Google Scholar] [CrossRef]
- Read, A.T.; Govind, C.K. Cell types in regenerating claws of the snapping shrimp, Alpheus heterochelis. Canadian Journal of Zoology 1998, 76, 1080–1090. [Google Scholar] [CrossRef]
- Sinigaglia, C.; Almazan, A.; Lebel, M.; Semon, M.; Gillet, B.; Hughes, S.; Edsinger, E.; Averof, M.; Paris, M. Distinct gene expression dynamics in developing and regenerating crustacean limbs. Proc. Natl. Acad. Sci. USA 2022, 119, e2119297119. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, P.M. Limb regeneration in the fiddler crab, Uca pugilator: Hormonal and growth factor control. American Zoologist 2001, 41, 389–398. [Google Scholar] [CrossRef]
- White, R.B.; Lamey, T.M.; Ziman, M.; Koenders, A. Isolation and expression analysis of a Pax group III gene from the crustacean Cherax destructor. Development Genes & Evolution 2005, 215, 306–312. [Google Scholar] [CrossRef]
- Andersen, S.O. Exoskeletal proteins from the crab, Cancer pagurus. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 1999, 123, 203–211. [Google Scholar] [CrossRef]
- Hyde, C.J.; Fitzgibbon, Q.P.; Elizur, A.; Smith, G.G.; Ventura, T. Transcriptional profiling of spiny lobster metamorphosis reveals three new additions to the nuclear receptor superfamily. BMC Genomics 2019, 20, 531. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Sriram, S.; Ling, K.Y. Isolation and cultivation of muscle and fat cells from crustaceans. WO2020149791A1, Jan 13, 2020 2020.
- Babu, D.E. Histological and histochemical studies on regeneration and tissue differentiation in the crab Menippe rumphii (Fabricius) (Crustacea: Brachyura). Journal of Experimental Marine Biology & Ecology 1987, 111, 213–230. [Google Scholar] [CrossRef]
- Novotová, M.; Uhrik, B. Structural characteristics and distribution of satellite cells along crayfish muscle fibers. Experientia 1992, 48, 593–596. [Google Scholar] [CrossRef]
- O'Neill, E.N.; Cosenza, Z.A.; Baar, K.; Block, D.E. Considerations for the development of cost-effective cell culture media for cultivated meat production. Comprehensive Reviews In Food Science & Food Safety 2021, 20, 686–709. [Google Scholar] [CrossRef]
- Jiwlawat, N.; Lynch, E.; Jeffrey, J.; Van Dyke, J.M.; Suzuki, M. Current progress and challenges for skeletal muscle differentiation from human pluripotent stem cells using transgene-free approaches. Stem Cells International 2018, 2018, 6241681. [Google Scholar] [CrossRef] [PubMed]
- Kolkmann, A.M.; Van Essen, A.; Post, M.J.; Moutsatsou, P. Development of a Chemically Defined Medium for in vitro Expansion of Primary Bovine Satellite Cells. Front Bioeng Biotechnol 2022, 10, 895289. [Google Scholar] [CrossRef] [PubMed]
- Chaulet, A.; Medesani, D.A.; Freitas, J.; Cervino, A.; Cervino, N.; Rodríguez, E.M. Induction of somatic growth in juvenile crayfish Cherax quadricarinatus (Decapoda, Parastacidae), by ecdysone and insulin growth factor. Aquaculture 2012, 370-371, 1–6. [Google Scholar] [CrossRef]
- Jayesh, P.; Philip, R.; Bright Singh, I.S. Multifactorial interaction of growth factors on Penaeus monodon lymphoid cells and the impact of IGFs in DNA synthesis and metabolic activity in vitro. Cytotechnology 2015, 67, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Shinji, J.; Gotoh, H.; Miyanishi, H.; Lavine, M.D.; Lavine, L.C. The activin signaling transcription factor Smox is an essential regulator of appendage size during regeneration after autotomy in the crayfish. Evol Dev 2019, 21, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Morris, B.J. Control of receptor sensitivity at the mRNA level. Molecular Neurobiology 1993, 7, 189–205. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.; Gandhi, N.; Mancera, R.; Smith, G.; Elizur, A.; Ventura, T. Understanding insulin endocrinology in decapod crustacea: Molecular modelling characterization of an insulin-binding protein and insulin-like peptides in the eastern spiny lobster, Sagmariasus verreauxi. Int. J. Mol. Sci. 2017, 18, 1832. [Google Scholar] [CrossRef] [PubMed]
- Rosen, O.; Weil, S.; Manor, R.; Roth, Z.; Khalaila, I.; Sagi, A. A crayfish insulin-like-binding protein: Another piece in the androgenic gland insulin-like hormone puzzle is revealed. Journal of Biological Chemistry 2013, 288, 22289–22298. [Google Scholar] [CrossRef] [PubMed]
- Manor, R.; Weil, S.; Oren, S.; Glazer, L.; Aflalo, E.D.; Ventura, T.; Chalifa-Caspi, V.; Lapidot, M.; Sagi, A. Insulin and gender: an insulin-like gene expressed exclusively in the androgenic gland of the male crayfish. General and Comparative Endocrinology 2007, 150, 326–336. [Google Scholar] [CrossRef]
- Mykles, D.L. Signaling pathways that regulate the crustacean molting gland. Frontiers in Endocrinology 2021, 12. [Google Scholar] [CrossRef]
- Genovese, N.J.; Domeier, T.L.; Telugu, B.P.V.L.; Roberts, R.M. Enhanced development of skeletal myotubes from porcine induced pluripotent stem cells. Scientific Reports 2017, 7, 41833. [Google Scholar] [CrossRef] [PubMed]
- Chal, J.; Pourquié, O. Making muscle: Skeletal myogenesis in vivo and in vitro. Development 2017, 144, 2104–2122. [Google Scholar] [CrossRef] [PubMed]
- Kodaka, Y.; Rabu, G.; Asakura, A. Skeletal Muscle Cell Induction from Pluripotent Stem Cells. Stem Cells International 2017, 2017, 1376151. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.V. Comparison of muscle development in Drosophila and vertebrates. Madame Curie Bioscience Database [Internet], 2006. [Google Scholar]
- Baylies, M.K.; Bate, M.; Gomez, M.R. Myogenesis: A view from Drosophila. Cell 1998, 93, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Patel, N.H. Investigating divergent mechanisms of mesoderm development in arthropods: The expression of Ph-twist and Ph-mef2 in Parhyale hawaiensis. Journal of Experimental Zoology Part B: Molecular & Developmental Evolution 2008, 310, 24–40. [Google Scholar] [CrossRef]
- Miskiewicz, P.; Morrissey, D.; Lan, Y.; Raj, L.; Kessler, S.; Fujioka, M.; Goto, T.; Weir, M. Both the paired domain and homeodomain are required for in vivo function of Drosophila Paired. Development 1996, 122, 2709–2718. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Li, X.; Noll, M. Multiple protein functions of paired in Drosophila development and their conservation in the Gooseberry and Pax3 homologs. Development 2001, 128, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Boukhatmi, H.; Bray, S. A population of adult satellite-like cells in Drosophila is maintained through a switch in RNA-isoforms. eLife 2018, 7, e35954. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, D.; Reichert, H.; Gunage, R.D.; VijayRaghavan, K. Identification and functional characterization of muscle satellite cells in Drosophila. eLife 2017, 6, e30107. [Google Scholar] [CrossRef]
- Konstantinides, N.; Averof, M. A common cellular basis for muscle regeneration in arthropods and vertebrates. Science 2014, 343, 788–791. [Google Scholar] [CrossRef]
- Zammit, P.S. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev Biol 2017, 72, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Ghanawi, J.; Saoud, G.; Zakher, C.; Monzer, S.; Saoud, I.P. Clove oil as an anaesthetic for Australian redclaw crayfish Cherax quadricarinatus. Aquaculture Research 2019, 50, 3628–3632. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Suvorov, V.; Prjibelski, A.D. rnaQUAST: a quality assessment tool for de novo transcriptome assemblies. Bioinformatics 2016, 32, 2210–2212. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kopylova, E.; Noe, L.; Touzet, H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Hyde, C.J.; Fitzgibbon, Q.P.; Elizur, A.; Smith, G.G.; Ventura, T. CrustyBase: An interactive online database for crustacean transcriptomes. BMC Genomics 2020, 21, 637. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 2015, 4, 1521. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Stephens, M. False discovery rates: a new deal. Biostatistics 2017, 18, 275–294. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Vicens, R.E.; Garcia-Escudero, C.A.; Conci, N.; Eitel, M.; Worheide, G. TransPi-a comprehensive TRanscriptome ANalysiS PIpeline for de novo transcriptome assembly. Mol Ecol Resour 2022, 22, 2070–2086. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology., Bioconductor version: Release (3.18) R package version 2.54.0; 2023.
- Gu, Z. Complex heatmap visualization. Imeta 2022, 1, e43. [Google Scholar] [CrossRef] [PubMed]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Smith, G.; Glendinning, S.; Ventura, T. Transcriptomic changes following induced de-masculinisation of Australian red claw crayfish Cherax quadricarinatus. Internatinoal Journal of Molecular Sciences 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Ventura, T.; Fitzgibbon, Q.P.; Battaglene, S.C.; Elizur, A. Redefining metamorphosis in spiny lobsters: molecular analysis of the phyllosoma to puerulus transition in Sagmariasus verreauxi. Scientific Reports 2015, 5. [Google Scholar] [CrossRef]
- Glendinning, S.; Fitzgibbon, Q.P.; Smith, G.G.; Ventura, T. Unravelling the neuropeptidome of the ornate spiny lobster Panulirus ornatus: A focus on peptide hormones and their processing enzymes expressed in the reproductive tissues. Gen Comp Endocrinol 2023, 332, 114183. [Google Scholar] [CrossRef]
| Stage | Sample | R-value* | Avg Rv | Stage | Sample | R-value* | Avg Rv |
| Stage 1 | S1.1 | 1.21 | 0.89 | Stage 4 | S4.1 | 29.95 | 49.49 |
| S1.2 | 1.17 | S4.2 | 68.31 | ||||
| S1.3 | 0.23 | S4.3 | 36.24 | ||||
| S1.4 | 0.97 | S4.4 | 26.45 | ||||
| Stage 2 | S2.1 | 2.37 | 3.41 | Stage 5 | S5.1 | 138.59 | 136.36(1-3 dpm) |
| S2.2 | 4.81 | S5.2 | 145.60 | ||||
| S2.3 | 3.62 | S5.3 | 158.20 | ||||
| S2.4 | 2.85 | S5.4 | 103.05 | ||||
| Stage 3 | S3.1 | 24.83 | 22.85 | Stage 6 | S6.1 | NA | NA (18-21 dpm) |
| S3.2 | 20.93 | S6.2 | NA | ||||
| S3.3 | 21.47 | S6.3 | NA | ||||
| S3.4 | 24.19 | S6.4 | NA |
| Complete | 85.1% |
| Single and complete BUSCO | 44.1% |
| Duplicated and complete BUSCO | 41.0% |
| Fragmented BUSCO | 6.9% |
| Missing BUSCO | 8.0% |
| Transcripts | 55 018 |
| Transcripts > 500 bp | 44 584 |
| Transcripts > 100 bp | 25 516 |
| Average length of assembled transcripts | 1 488.461 |
| Longest transcript | 22 480 |
| Total length | 81 892 142 |
| Transcript N50 | 2 242 |
| Number of transcripts | 55 018 |
| Total number of ORFs | 43 515 |
| ORFs with annotations | 39 360 |
| ORFs without annotations | 4 155 |
| ORFs complete | 17 773 |
| ORFs complete with annotations | 16 223 |
| Transcripts with eggNOG COG | 79 |
| Transcripts with GO | 23 105 |
| Transcripts with KEGG | 18 749 |
| Transcripts with PFAM | 23 635 |
| Transcripts with BLAST | 21 444 |
| DEG Categories | GO Term Categories | Combined Categories |
|---|---|---|
| Neuronal development | Cell cycle | Cell cycle |
| Wound repair/inflammation/immune response | Neuronal development | Neuronal development |
| Tissue development/regeneration | Wound repair/inflammation/immune response | Wound repair/inflammation/immune response |
| Cell repair/homeostasis | Tissue development/regeneration | Tissue development/regeneration |
| Cuticle development (AM/AMP type) | Insulin-regulated growth | Cell repair/homeostasis |
| Cuticle development (CP type) | Metabolite production | Cuticle development (AM/AMP type) |
| Pigmentation - Blue | Energy production | Cuticle development (CP type) |
| Pigmentation - Red | Protein translation | Pigmentation - Blue |
| Metabolite production | Muscle development | Pigmentation - Red |
| Energy production | Insulin-regulated growth | |
| Muscle development | Metabolite production | |
| Energy production | ||
| Protein translation | ||
| Muscle development |
| Gene Category | Gene/Factor | Transcript_ID | Expression |
|---|---|---|---|
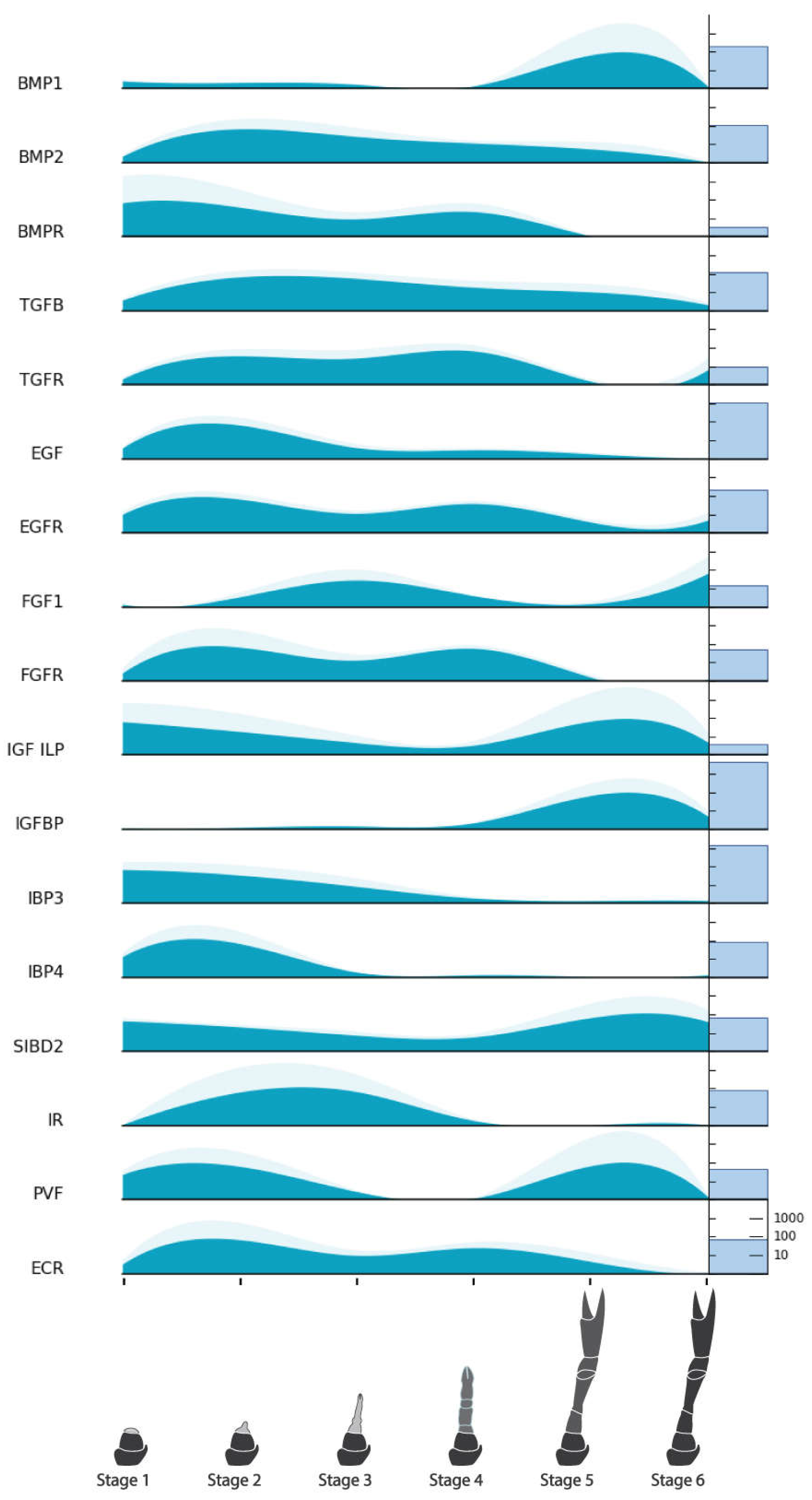
| Endocrine Factors | Bone Morphogenetic Protein 1 (BMP1) | NonamEVm007201t1 | Figure 6 |
| Bone Morphogenetic Protein 2 (BMP2) | NonamEVm003174t1 | ||
| Bone Morphogenetic Protein Receptor (BMPR) | NonamEVm006724t1 | ||
| Transforming Growth Factor Beta (TGFβ) | NonamEVm004397t1 | ||
| Transforming Growth Factor Beta (TGFβR) | NonamEVm003982t1 | ||
| Ecdysone Receptor (ECR) | NonamEVm002506t1 | ||
| Epidermal Growth Factor (EGF) | NonamEVm000011t1 | ||
| Epidermal Growth Factor Receptor (EGFR) | NonamEVm000307t1 | ||
| Fibroblast Growth Factor 1 (FGF1) | NonamEVm009793t1 | ||
| Fibroblast Growth Factor Receptor (FGFR) | NonamEVm002596t1 | ||
| Insulin-like Growth Factor/Peptide (IGF/ILP) | NonamEVm015416t1 | ||
| Insulin-like Growth Factor Binding Protein (IGFBP) | NonamEVm006292t1 | ||
| Insulin-like Growth Factor Binding Protein 3 (IBP3) Insulin-like Growth Factor Binding Protein 4 (IBP4) Insulin-like growth factor 2 mRNA-binding protein (IGF2B) |
NonamEVm007418t1 NonamEVm018337t1 NonamEVm002726t1 |
||
| Single insulin-like growth factor-binding domain protein-2 (SBD2) | NonamEVm018319t1 | ||
| Insulin Receptor (IR) | NonamEVm000244t1 | ||
| Platelet Derived/Vascular Endothelial Factor (PVF) | NonamEVm006397t1 | ||
| Platelet Derived/Vascular Endothelial Factor Receptor (PVR) | Not found | ||
| Hepatocyte Growth Factor & Receptor (HGF/HGFR) | Not found | ||
| Myostatin (MSTN) | Not found | ||
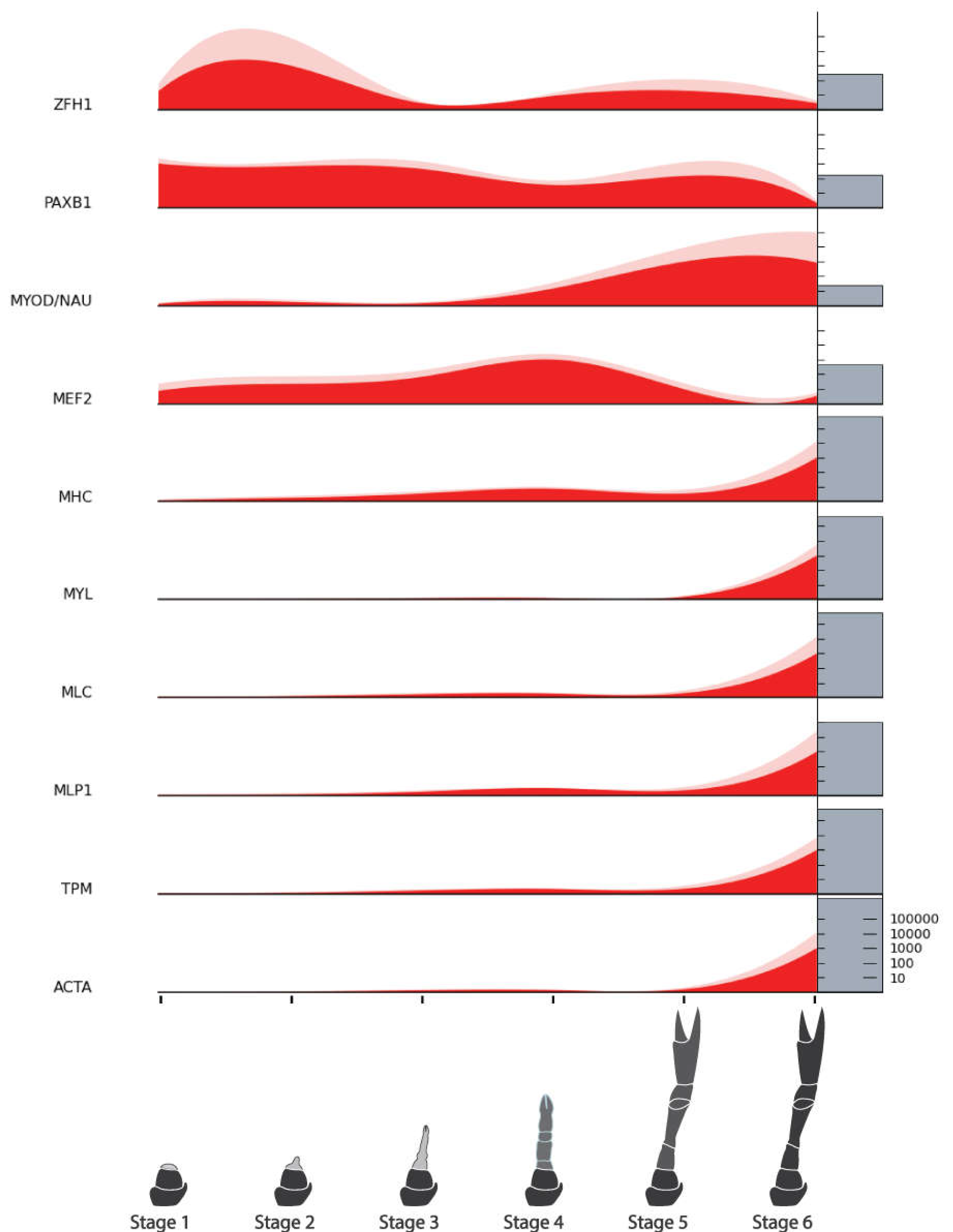
| Myogenic Factors | Zinc Finger Homeodomain 1 (ZFH1) | NonamEVm000901t1 | Figure 8 |
| Pax 3 and Pax 7 Binding Protein 1 (PAXB1) | NonamEVm001208t1 | ||
| Myogenic Determination Protein/Nautilus (MYOD/NAU) | NonamEVm006620t1 | ||
| Myocyte Enhancer Factor 2 (MEF2) | NonamEVm004258t1 | ||
| Myosin Heavy Chain, Muscle (MHC) | NonamEVm000136t1 | ||
| Myosin Regulatory Light Chain (MYL) | NonamEVm011187t1 | ||
| Myosin Light Chain (MLC) | NonamEVm013270t1 | ||
| Muscle Lim Protein (MLP) | NonamEVm002170t1 | ||
| Tropomyosin (TMP) | NonamEVm005681t1 | ||
| Actin, Muscle-Related (ACTA) | NonamEVm004870t1 | ||
| Myogenic Determination Protein/Twist (MYOD/TWIST) | Not found | ||
| Paired Box Protein 3 (PAX3) | Not found | ||
| Paired Box Protein 7 (PAX7) | Not found | ||
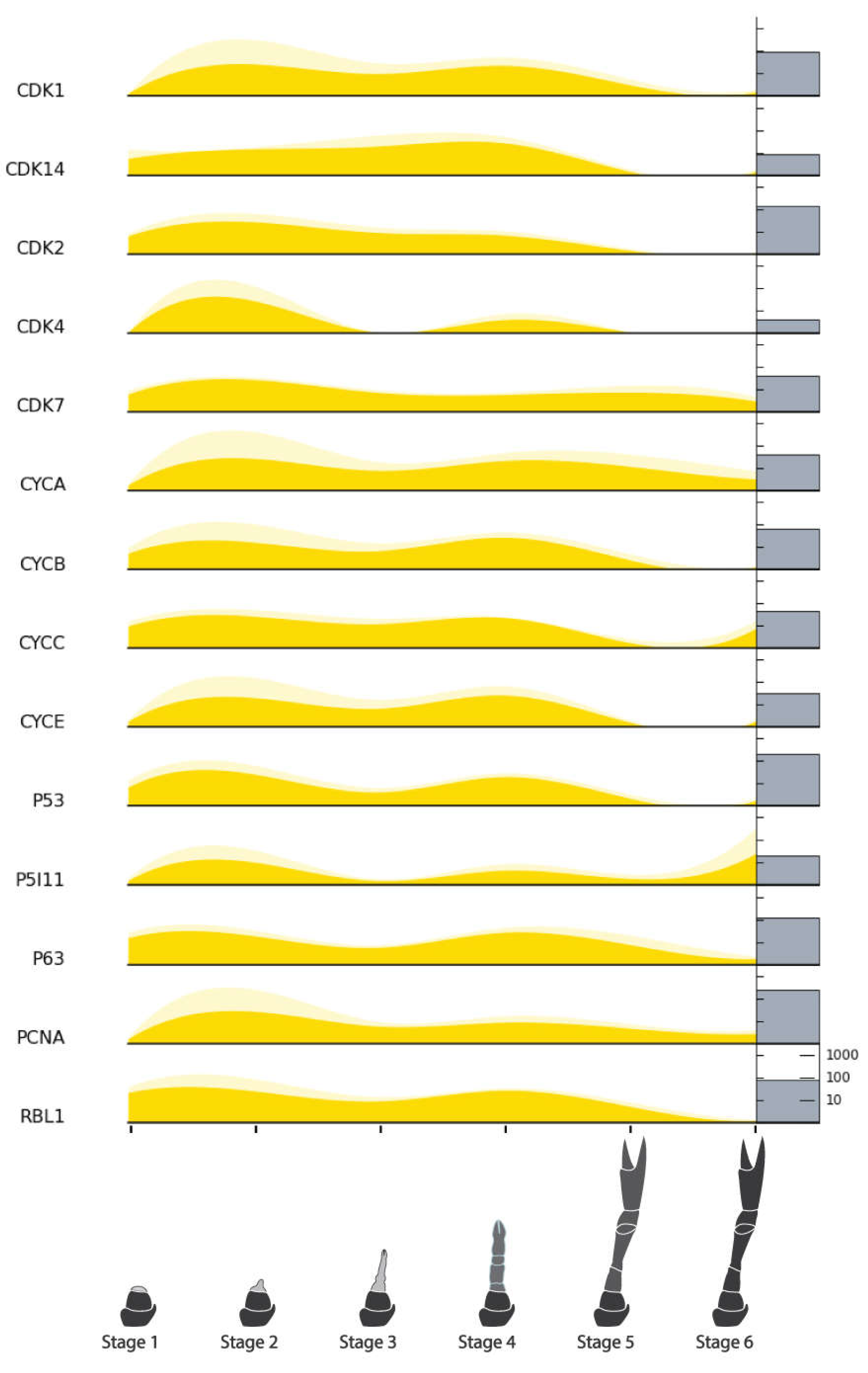
| Cell Cycle Factors | Cyclin-dependent Kinase 1 (CDK1) | NonamEVm006446t1 | Figure 9 |
| Cyclin-dependent Kinase 14 (CDK14) | NonamEVm007728t1 | ||
| Cyclin-dependent Kinase 2 (CDK2) | NonamEVm006000t1 | ||
| Cyclin-dependent Kinase 4 (CDK4) | NonamEVm010173t1 | ||
| Cyclin-dependent Kinase 7 (CDK7) | NonamEVm005270t1 | ||
| Cyclin A (CYC-A) | NonamEVm003889t1 | ||
| Cyclin B (CYC-B) | NonamEVm004508t1 | ||
| Cyclin C (CYC-C) | NonamEVm007180t1 | ||
| Cyclin E (CYC-E) | NonamEVm006398t1 | ||
| Cellular Tumor Antigen p53 (P53) | NonamEVm004102t1 | ||
| Tumor Protein p53-inducible Protein 11 (P5I11) | NonamEVm007863t1 | ||
| Cellular Tumor Antigen p53 (P63) | NonamEVm001352t1 | ||
| Proliferating Cell Nuclear Antigen (PCNA) | NonamEVm007483t1 | ||
| Retinoblastoma-like Associated Protein (RBL1) | NonamEVm000671t1 | ||
| Cyclin-dependent Kinase 3 (CDK3) | Not found | ||
| Cyclin-dependent Kinase 6 (CDK6) | Not found | ||
| Cyclin D (CYC-D) | Not found | ||
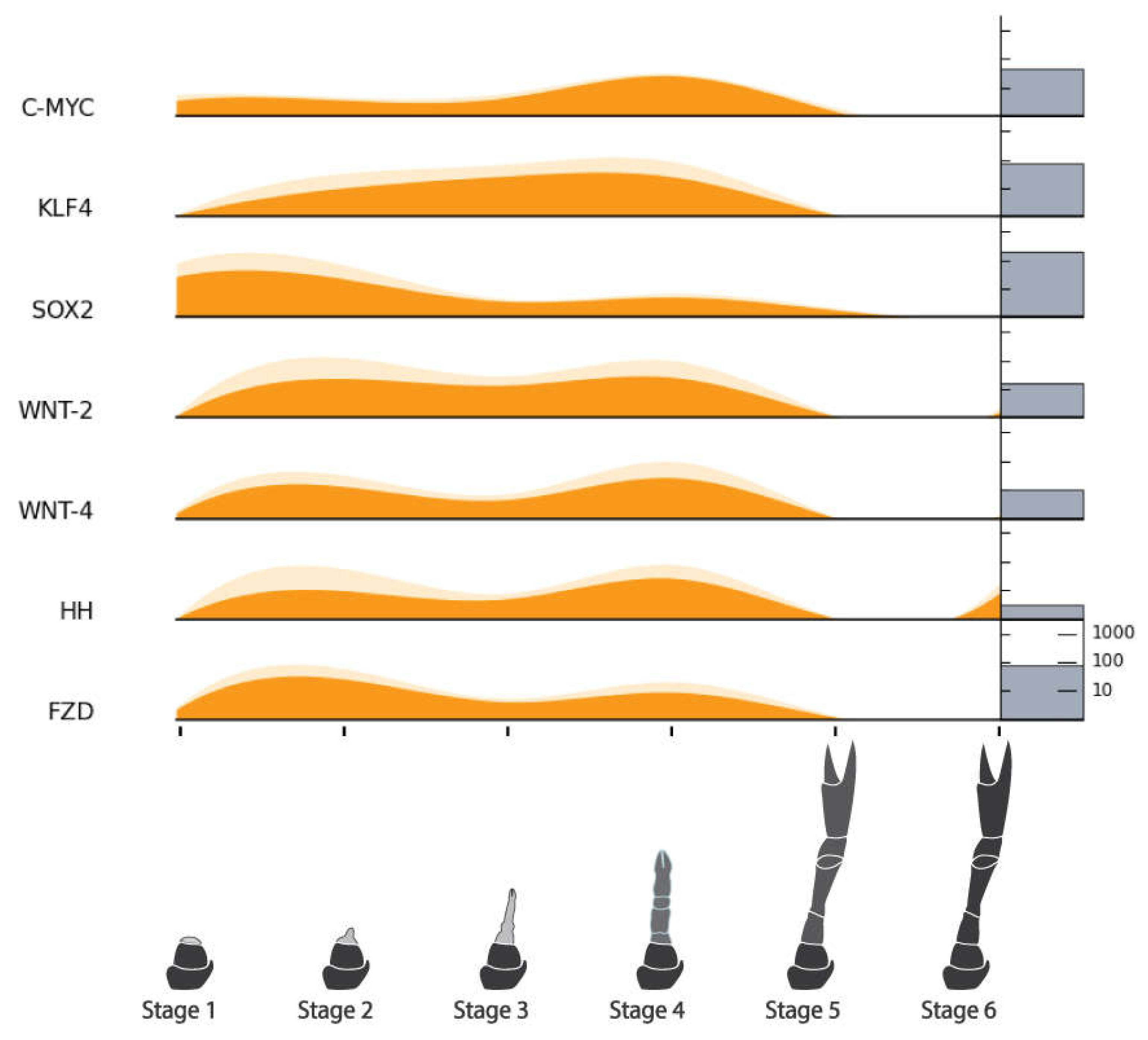
| Pluripotency Factors | MYC Proto-oncogene Protein (C-MYC) | NonamEVm003188t1 | Figure 10 |
| Krueppel-like Factor 4 (KLF4) | NonamEVm004452t1 | ||
| SRY-Box Transcription Factor 2 (SOX2) | NonamEVm004482t1 | ||
| POU Domain, Class 5, Transcription Factor 1 (OCT3/4) | NonamEVm004528t1 | ||
| Int/Wingless Family Protein 2 (WNT-2) | NonamEVm005041t1 | ||
| Int/Wingless Family Protein 4 (WNT-4) | NonamEVm003744t1 | ||
| Hedgehog (HH) | NonamEVm003556t1 | ||
| Frizzled (FZD) | NonamEVm005809t1 | ||
| Nanog (NANOG) | Not found | ||
| Int/Wingless Family Protein 1 (WNT-1) | Not found |
| Primer Name | UPL Probe # | Forward Primer | Reverse Primer |
| qCq18S | 133 | GCGCTACACTGAAGGGATCA | AGGGGTTTGAACGGGTTACC |
| qCqMLC | 135 | CGTGTGCTGGGAATCACTGA | CGGGCTGGACCTTCGTTATT |
| qCqZfh1 | 10 | AGAAGTCCCCCTCATCTCCC | CAACGCAACTATTAGCCCGC |
| qCqPCNA | 42 | GGCTCGTCTTGTCCAAGGAA | TGACGCTTCATTCAGCAGCT |
| qCqOct3/4 | 85 | CGAAGTCGAAGAGGAGACCC | CCAGCTTGATACGCCTCTGT |
| cCqNautilus | 5 | CGTGCAAGAGAAAGAGCGTC | TCACCTTCCGTAGTCTCCGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).