Submitted:
02 July 2024
Posted:
03 July 2024
You are already at the latest version
Abstract
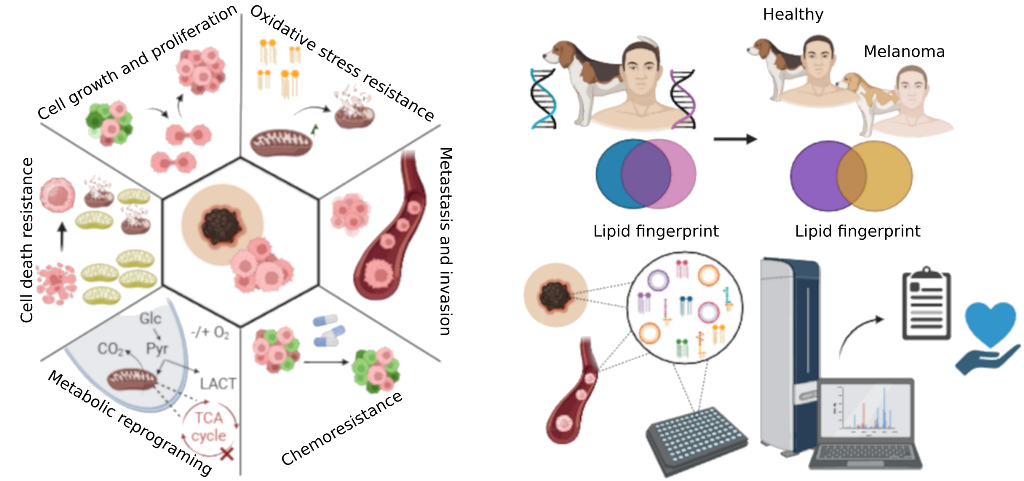
Keywords:
1. Introduction
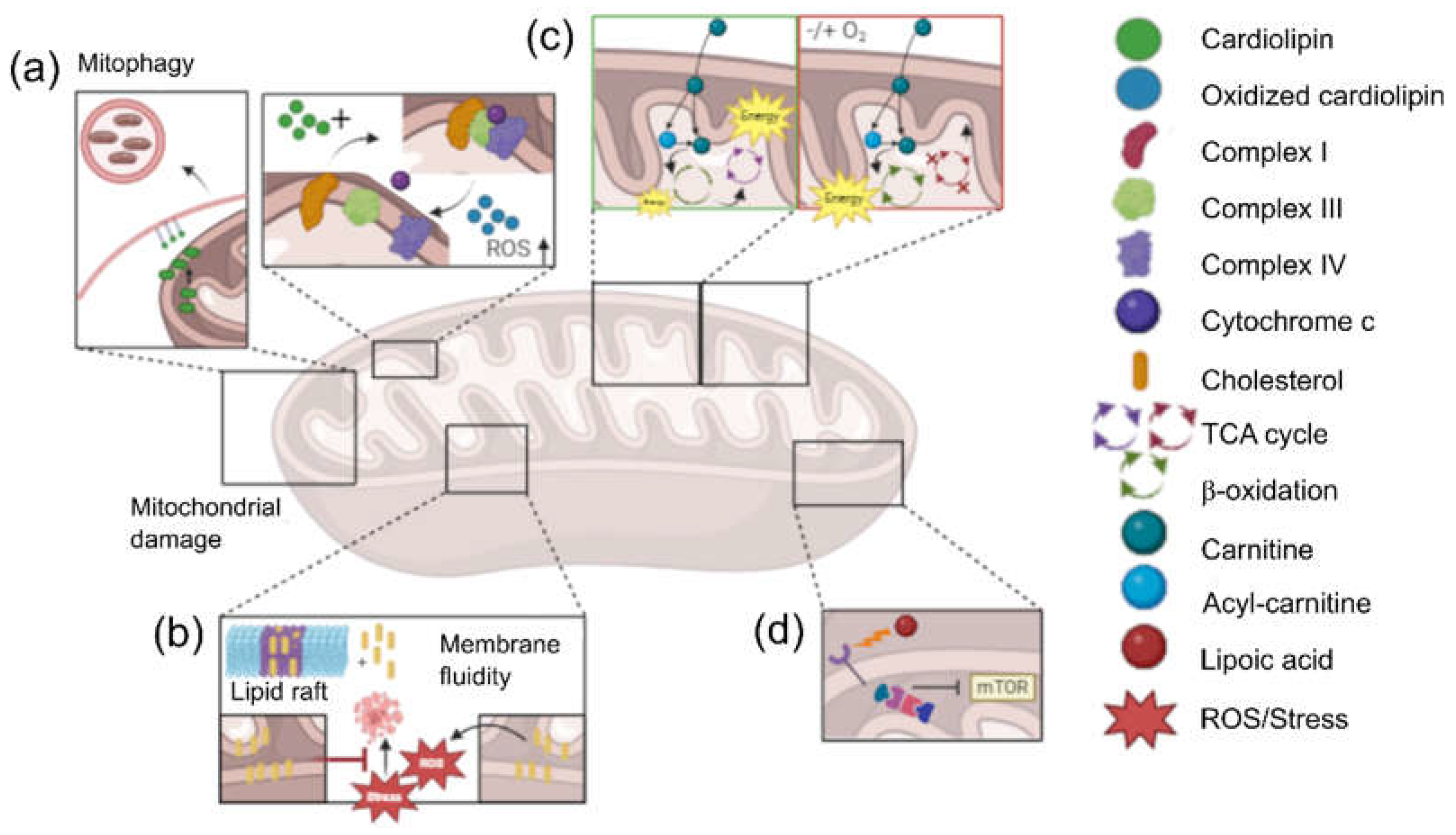
2. Interplay Between Lipid Metabolism and Mitochondrial Function in Cancer
3. Lipid Profile in Melanoma
4. Canine Models in Comparative Melanoma Research
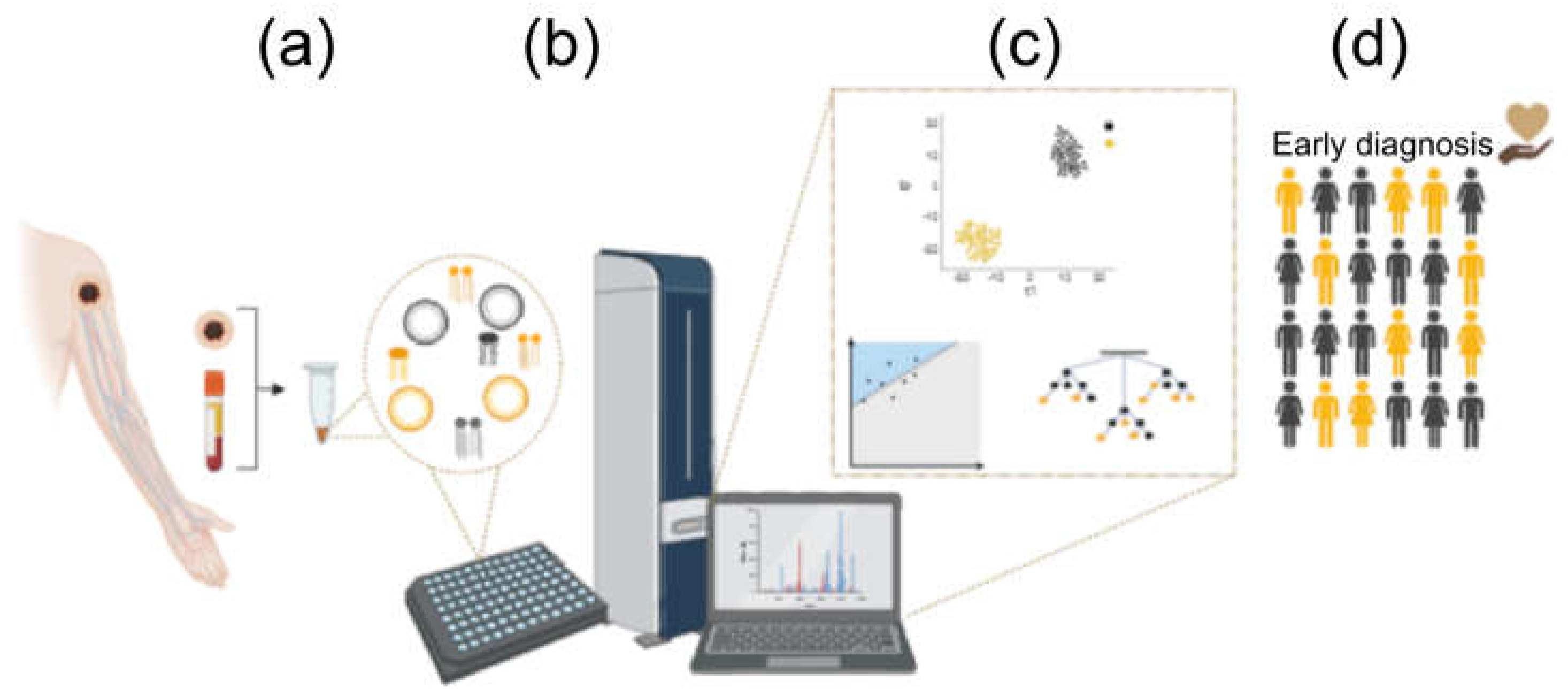
5. Lipidomics and Machine Learning
6. Perspectives
References
- Curtin, S.C.; Tejada-Vera, B.; Bastian, B.A. Deaths: Leading Causes for 2021. National Vital Statistics Reports 2024, 73, 1–116. [Google Scholar]
- Zheng, R.; Zeng, H.; Zhang, S.; Chen, T.; Chen, W. National Estimates of Cancer Prevalence in China, 2011. Cancer Lett 2016, 370, 33–38. [Google Scholar] [CrossRef]
- Yao, C.; Billette, J.M. Short-Term Cancer Prevalence in Canada, 2018. Health Rep 2022, 33, 15–21. [Google Scholar] [CrossRef]
- De Angelis, R.; Demuru, E.; Baili, P.; Troussard, X.; Katalinic, A.; Chirlaque Lopez, M.D.; Innos, K.; Santaquilani, M.; Blum, M.; Ventura, L.; et al. Complete Cancer Prevalence in Europe in 2020 by Disease Duration and Country (EUROCARE-6): A Population-Based Study. Lancet Oncol 2024, 25, 293–307. [Google Scholar] [CrossRef]
- Rahib, L.; Wehner, M.R.; Matrisian, L.M.; Nead, K.T. Estimated Projection of US Cancer Incidence and Death to 2040. JAMA Netw Open 2021, 4. [Google Scholar] [CrossRef]
- Sleeckx, N.; de Rooster, H.; Veldhuis Kroeze, E.J.B.; van Ginneken, C.; van Brantegem, L. Canine Mammary Tumours, an Overview. Reprod Domest Anim 2011, 46, 1112–1131. [Google Scholar] [CrossRef]
- Misdorp, W.; Misdorp1, W. Veterinary Cancer Epidemiology. Veterinary Quarterly 1996, 18, 32–36. [Google Scholar] [CrossRef]
- Blackwood, L. Cats with Cancer: Where to Start. J Feline Med Surg 2013, 15, 366–377. [Google Scholar] [CrossRef]
- Grüntzig, K.; Graf, R.; Hässig, M.; Welle, M.; Meier, D.; Lott, G.; Erni, D.; Schenker, N.S.; Guscetti, F.; Boo, G.; et al. The Swiss Canine Cancer Registry: A Retrospective Study on the Occurrence of Tumours in Dogs in Switzerland from 1955 to 2008. J Comp Pathol 2015, 152, 161–171. [Google Scholar] [CrossRef]
- Brunsgaard, E.K.; Wu, Y.P.; Grossman, D. Melanoma in Skin of Color: Part I. Epidemiology and Clinical Presentation. J Am Acad Dermatol 2023, 89, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Wohlmuth, C.; Wohlmuth-Wieser, I. Vulvar Melanoma: Molecular Characteristics, Diagnosis, Surgical Management, and Medical Treatment. Am J Clin Dermatol 2021, 22, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Abbas, O.; Miller, D.D.; Bhawan, J. Cutaneous Malignant Melanoma: Update on Diagnostic and Prognostic Biomarkers. Am J Dermatopathol 2014, 36, 363–379. [Google Scholar] [CrossRef] [PubMed]
- Pavri, S.N.; Clune, J.; Ariyan, S.; Narayan, D. Malignant Melanoma: Beyond the Basics. Plast Reconstr Surg 2016, 138, 330e–340e. [Google Scholar] [CrossRef] [PubMed]
- de Matos, L.L.; Trufelli, D.C.; de Matos, M.G.L.; Pinhal, M.A. da S. Immunohistochemistry as an Important Tool in Biomarkers Detection and Clinical Practice. Biomark Insights 2010, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Goyal, L.; Hingmire, S.; Parikh, P.M. Newer Diagnostic Methods in Oncology. Med J Armed Forces India 2006, 62, 162. [Google Scholar] [CrossRef]
- Whelehan, P.; Evans, A.; Wells, M.; MacGillivray, S. The Effect of Mammography Pain on Repeat Participation in Breast Cancer Screening: A Systematic Review. Breast 2013, 22, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Morton, C.A.; Mackie, R.M. Clinical Accuracy of the Diagnosis of Cutaneous Malignant Melanoma. Br J Dermatol 1998, 138, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Badu-Peprah, A.; Adu-Sarkodie, Y. Accuracy of Clinical Diagnosis, Mammography and Ultrasonography in Preoperative Assessment of Breast Cancer. Ghana Med J 2018, 52, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Schaffter, T.; Buist, D.S.M.; Lee, C.I.; Nikulin, Y.; Ribli, D.; Guan, Y.; Lotter, W.; Jie, Z.; Du, H.; Wang, S.; et al. Evaluation of Combined Artificial Intelligence and Radiologist Assessment to Interpret Screening Mammograms. JAMA Netw Open 2020, 3, E200265. [Google Scholar] [CrossRef]
- Marghoob, A.A.; Changchien, L.; DeFazio, J.; Dessio, W.C.; Malvehy, J.; Zalaudek, I.; Halpern, A.C.; Scope, A. The Most Common Challenges in Melanoma Diagnosis and How to Avoid Them. Australas J Dermatol 2009, 50, 1–13. [Google Scholar] [CrossRef]
- Poulet, G.; Massias, J.; Taly, V. Liquid Biopsy: General Concepts. Acta Cytol 2019, 63, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Copeland, C.; Le, A. Glutamine Metabolism in Cancer. Adv Exp Med Biol 2021, 1311, 17–38. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P.; Schmidberger, H.; Mayer, A. The Warburg Effect: Essential Part of Metabolic Reprogramming and Central Contributor to Cancer Progression. Int J Radiat Biol 2019, 95, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Schiliro, C.; Firestein, B.L. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, R.M.; Galli, C.; Ferro-Luzzi, A.; Iacono, J.M. Lipid and Phospholipid Fatty Acid Composition of Plasma, Red Blood Cells, and Platelets and How They Are Affected by Dietary Lipids: A Study of Normal Subjects from Italy, Finland, and the USA. Am J Clin Nutr 1987, 45, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Halpern, Z.; Rubin, M.; Harach, G.; Grotto, I.; Mosor, A.; Dvir, A.; Lichtenberg, D.; Gilat, T. Bile and Plasma Lipid Composition in Non-Obese Normolipidemic Subjects with and without Cholesterol Gallstones. Liver 1993, 13, 246–252. [Google Scholar] [CrossRef]
- Maldonado, E.N.; Romero, J.R.; Ochoa, B.; Aveldao, M.I. Lipid and Fatty Acid Composition of Canine Lipoproteins. Comparative Biochemistry and Physiology - B Biochemistry and Molecular Biology 2001, 128, 719–729. [Google Scholar] [CrossRef]
- Meikle, P.J.; Barlow, C.K.; Mellett, N.A.; Mundra, P.A.; Bonham, M.P.; Larsen, A.; Cameron-Smith, D.; Sinclair, A.; Nestel, P.J.; Wong, G. Postprandial Plasma Phospholipids in Men Are Influenced by the Source of Dietary Fat. J Nutr 2015, 145, 2012–2018. [Google Scholar] [CrossRef]
- Feingold, K.R. Lipid and Lipoprotein Metabolism. Endocrinol Metab Clin North Am 2022, 51, 437–458. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu Rev Biochem 2019, 88, 487–514. [Google Scholar] [CrossRef] [PubMed]
- Hessvik, N.P.; Llorente, A. Current Knowledge on Exosome Biogenesis and Release. Cell Mol Life Sci 2018, 75, 193–208. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.F. A Niche Role for Cancer Exosomes in Metastasis. Nat Cell Biol 2015, 17, 709–711. [Google Scholar] [CrossRef] [PubMed]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic Cancer Exosomes Initiate Pre-Metastatic Niche Formation in the Liver. Nat Cell Biol 2015, 17, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.K.; Enderle, D.; Noerholm, M.; Breakefield, X.O.; Skog, J.K. Exosome-Based Liquid Biopsies in Cancer: Opportunities and Challenges. Ann Oncol 2021, 32, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.K.L.; Jose, J.; Wahba, M.; Bernaus-Esqué, M.; Hoy, A.J.; Enrich, C.; Rentero, C.; Grewal, T. Linking Late Endosomal Cholesterol with Cancer Progression and Anticancer Drug Resistance. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Munir, R.; Usman, H.; Hasnain, S.; Smans, K.; Kalbacher, H.; Zaidi, N. Atypical Plasma Lipid Profile in Cancer Patients: Cause or Consequence? Biochimie 2014, 102, 9–18. [Google Scholar] [CrossRef]
- Irshad, R.; Tabassum, S.; Husain, M. Aberrant Lipid Metabolism in Cancer: Current Status and Emerging Therapeutic Perspectives. Curr Top Med Chem 2023, 23, 1090–1103. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Zhao, H.; Zeng, Y. Lipidomics: A Promising Cancer Biomarker. Clin Transl Med 2018, 7. [Google Scholar] [CrossRef]
- Di Gregorio, J.; Petricca, S.; Iorio, R.; Toniato, E.; Flati, V. Mitochondrial and Metabolic Alterations in Cancer Cells. Eur J Cell Biol 2022, 101. [Google Scholar] [CrossRef]
- Caino, M.C.; Altieri, D.C. Molecular Pathways: Mitochondrial Reprogramming in Tumor Progression and Therapy. Clin Cancer Res 2016, 22, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Srivastava, P.; Mathur, S.; Abbas, S.; Rai, N.; Tiwari, S.; Tiwari, M.; Sharma, L. Lipid Metabolism and Mitochondria: Cross Talk in Cancer. Curr Drug Targets 2022, 23, 606–627. [Google Scholar] [CrossRef]
- Horvath, S.E.; Daum, G. Lipids of Mitochondria. Prog Lipid Res 2013, 52, 590–614. [Google Scholar] [CrossRef] [PubMed]
- Poulaki, A.; Giannouli, S. Mitochondrial Lipids: From Membrane Organization to Apoptotic Facilitation. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef]
- Paradies, G.; Paradies, V.; Ruggiero, F.M.; Petrosillo, G. Role of Cardiolipin in Mitochondrial Function and Dynamics in Health and Disease: Molecular and Pharmacological Aspects. Cells 2019, 8. [Google Scholar] [CrossRef]
- Ahmadpour, S.T.; Mahéo, K.; Servais, S.; Brisson, L.; Dumas, J.F. Cardiolipin, the Mitochondrial Signature Lipid: Implication in Cancer. Int J Mol Sci 2020, 21, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Paradies, G.; Paradies, V.; De Benedictis, V.; Ruggiero, F.M.; Petrosillo, G. Functional Role of Cardiolipin in Mitochondrial Bioenergetics. Biochim Biophys Acta 2014, 1837, 408–417. [Google Scholar] [CrossRef]
- Praharaj, P.P.; Naik, P.P.; Panigrahi, D.P.; Bhol, C.S.; Mahapatra, K.K.; Patra, S.; Sethi, G.; Bhutia, S.K. Intricate Role of Mitochondrial Lipid in Mitophagy and Mitochondrial Apoptosis: Its Implication in Cancer Therapeutics. Cell Mol Life Sci 2019, 76, 1641–1652. [Google Scholar] [CrossRef]
- Goicoechea, L.; Conde de la Rosa, L.; Torres, S.; García-Ruiz, C.; Fernández-Checa, J.C. Mitochondrial Cholesterol: Metabolism and Impact on Redox Biology and Disease. Redox Biol 2023, 61. [Google Scholar] [CrossRef]
- Ribas, V.; García-Ruiz, C.; Fernández-Checa, J.C. Mitochondria, Cholesterol and Cancer Cell Metabolism. Clin Transl Med 2016, 5. [Google Scholar] [CrossRef]
- Garcia-Ruiz, C.; Conde de la Rosa, L.; Ribas, V.; Fernandez-Checa, J.C. MITOCHONDRIAL CHOLESTEROL AND CANCER. Semin Cancer Biol 2021, 73, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Qu, Q.; Zeng, F.; Liu, X.; Wang, Q.J.; Deng, F. Fatty Acid Oxidation and Carnitine Palmitoyltransferase I: Emerging Therapeutic Targets in Cancer. Cell Death Dis 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Melone, M.A.B.; Valentino, A.; Margarucci, S.; Galderisi, U.; Giordano, A.; Peluso, G. The Carnitine System and Cancer Metabolic Plasticity. Cell Death Dis 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Console, L.; Scalise, M.; Mazza, T.; Pochini, L.; Galluccio, M.; Giangregorio, N.; Tonazzi, A.; Indiveri, C. Carnitine Traffic in Cells. Link With Cancer. Front Cell Dev Biol 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Farahzadi, R.; Hejazi, M.S.; Molavi, O.; Pishgahzadeh, E.; Montazersaheb, S.; Jafari, S. Clinical Significance of Carnitine in the Treatment of Cancer: From Traffic to the Regulation. Oxid Med Cell Longev 2023, 2023. [Google Scholar] [CrossRef] [PubMed]
- Dörsam, B.; Fahrer, J. The Disulfide Compound α-Lipoic Acid and Its Derivatives: A Novel Class of Anticancer Agents Targeting Mitochondria. Cancer Lett 2016, 371, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, U.; Nickel, A.; Daniel, H. Alpha-Lipoic Acid Induces Apoptosis in Human Colon Cancer Cells by Increasing Mitochondrial Respiration with a Concomitant O2-*-Generation. Apoptosis 2005, 10, 359–368. [Google Scholar] [CrossRef]
- Bosso, M.; Haddad, D.; Al Madhoun, A.; Al-Mulla, F. Targeting the Metabolic Paradigms in Cancer and Diabetes. Biomedicines 2024, 12. [Google Scholar] [CrossRef]
- Carrié, L.; Virazels, M.; Dufau, C.; Montfort, A.; Levade, T.; Ségui, B.; Andrieu-Abadie, N. New Insights into the Role of Sphingolipid Metabolism in Melanoma. Cells 2020, 9, 1–29. [Google Scholar] [CrossRef]
- Yesmin, F.; Bhuiyan, R.H.; Ohmi, Y.; Yamamoto, S.; Kaneko, K.; Ohkawa, Y.; Zhang, P.; Hamamura, K.; Cheung, N.K. V.; Kotani, N.; et al. Ganglioside GD2 Enhances the Malignant Phenotypes of Melanoma Cells by Cooperating with Integrins. Int J Mol Sci 2021, 23. [Google Scholar] [CrossRef]
- Noujarède, J.; Carrié, L.; Garcia, V.; Grimont, M.; Eberhardt, A.; Mucher, E.; Genais, M.; Schreuder, A.; Carpentier, S.; Ségui, B.; et al. Sphingolipid Paracrine Signaling Impairs Keratinocyte Adhesion to Promote Melanoma Invasion. Cell Rep 2023, 42. [Google Scholar] [CrossRef] [PubMed]
- Portoukalian, J.; Zwingelstein, G.; Doré, J. -F Lipid Composition of Human Malignant Melanoma Tumors at Various Levels of Malignant Growth. Eur J Biochem 1979, 94, 19–23. [Google Scholar] [CrossRef]
- Realini, N.; Palese, F.; Pizzirani, D.; Pontis, S.; Basit, A.; Bach, A.; Ganesan, A.; Piomelli, D. Acid Ceramidase in Melanoma: EXPRESSION, LOCALIZATION, AND EFFECTS OF PHARMACOLOGICAL INHIBITION. J Biol Chem 2016, 291, 2422–2434. [Google Scholar] [CrossRef] [PubMed]
- Perez-Valle, A.; Abad-García, B.; Fresnedo, O.; Barreda-Gómez, G.; Aspichueta, P.; Asumendi, A.; Astigarraga, E.; Fernández, J.A.; Boyano, M.D.; Ochoa, B. A UHPLC-Mass Spectrometry View of Human Melanocytic Cells Uncovers Potential Lipid Biomarkers of Melanoma. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Huergo-Baños, C.; Velasco, V.; Garate, J.; Fernández, R.; Martín-Allende, J.; Zabalza, I.; Artola, J.L.; Martí, R.M.; Asumendi, A.; Astigarraga, E.; et al. Lipid Fingerprint-Based Histology Accurately Classifies Nevus, Primary Melanoma, and Metastatic Melanoma Samples. Int J Cancer 2024, 154, 712–722. [Google Scholar] [CrossRef]
- Prouteau, A.; André, C. Canine Melanomas as Models for Human Melanomas: Clinical, Histological, and Genetic Comparison. Genes (Basel) 2019, 10. [Google Scholar] [CrossRef]
- Schiffman, J.D.; Breen, M. Comparative Oncology: What Dogs and Other Species Can Teach Us about Humans with Cancer. Philos Trans R Soc Lond B Biol Sci 2015, 370. [Google Scholar] [CrossRef]
- Abdelmegeed, S.M.; Mohammed, S. Canine Mammary Tumors as a Model for Human Disease. Oncol Lett 2018, 15, 8195–8205. [Google Scholar] [CrossRef]
- Pinho, S.S.; Carvalho, S.; Cabral, J.; Reis, C.A.; Gärtner, F. Canine Tumors: A Spontaneous Animal Model of Human Carcinogenesis. Transl Res 2012, 159, 165–172. [Google Scholar] [CrossRef]
- Stevenson, V.B.; Klahn, S.; LeRoith, T.; Huckle, W.R. Canine Melanoma: A Review of Diagnostics and Comparative Mechanisms of Disease and Immunotolerance in the Era of the Immunotherapies. Front Vet Sci 2023, 9. [Google Scholar] [CrossRef]
- Gillard, M.; Cadieu, E.; De Brito, C.; Abadie, J.; Vergier, B.; Devauchelle, P.; Degorce, F.; Dréano, S.; Primot, A.; Dorso, L.; et al. Naturally Occurring Melanomas in Dogs as Models for Non-UV Pathways of Human Melanomas. Pigment Cell Melanoma Res 2014, 27, 90–102. [Google Scholar] [CrossRef]
- Gardner, H.L.; Fenger, J.M.; London, C.A. Dogs as a Model for Cancer. Annu Rev Anim Biosci 2016, 4, 199–222. [Google Scholar] [CrossRef] [PubMed]
- Nishiya, A.T.; Massoco, C.O.; Felizzola, C.R.; Perlmann, E.; Batschinski, K.; Tedardi, M.V.; Garcia, J.S.; Mendonça, P.P.; Teixeira, T.F.; Dagli, M.L.Z. Comparative Aspects of Canine Melanoma. Vet Sci 2016, 3. [Google Scholar] [CrossRef]
- Palma, S. Di; McConnell, A.; Verganti, S.; Starkey, M. Review on Canine Oral Melanoma: An Undervalued Authentic Genetic Model of Human Oral Melanoma? Vet Pathol 2021, 58, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, T.F.; Gentile, L.B.; Da Silva, T.C.; Mennecier, G.; Chaible, L.M.; Cogliati, B.; Roman, M.A.L.; Gioso, M.A.; Dagli, M.L.Z. Cell Proliferation and Expression of Connexins Differ in Melanotic and Amelanotic Canine Oral Melanomas. Vet Res Commun 2014, 38, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Alexander, A.N.; Huelsmeyer, M.K.; Mitzey, A.; Dubielzig, R.R.; Kurzman, I.D.; MacEwen, E.G.; Vail, D.M. Development of an Allogeneic Whole-Cell Tumor Vaccine Expressing Xenogeneic Gp100 and Its Implementation in a Phase II Clinical Trial in Canine Patients with Malignant Melanoma. Cancer Immunol Immunother 2006, 55, 433–442. [Google Scholar] [CrossRef] [PubMed]
- van der Weyden, L.; Brenn, T.; Patton, E.E.; Wood, G.A.; Adams, D.J. Spontaneously Occurring Melanoma in Animals and Their Relevance to Human Melanoma. J Pathol 2020, 252, 4–21. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.M.; Bastian, B.C.; Michael, H.T.; Webster, J.D.; Prasad, M.L.; Conway, C.M.; Prieto, V.M.; Gary, J.M.; Goldschmidt, M.H.; Esplin, D.G.; et al. Sporadic Naturally Occurring Melanoma in Dogs as a Preclinical Model for Human Melanoma. Pigment Cell Melanoma Res 2014, 27, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Spangler, W.L.; Kass, P.H. The Histologic and Epidemiologic Bases for Prognostic Considerations in Canine Melanocytic Neoplasia. Vet Pathol 2006, 43, 136–149. [Google Scholar] [CrossRef]
- Shuman, A.G.; Light, E.; Olsen, S.H.; Pynnonen, M.A.; Taylor, J.M.G.; Johnson, T.M.; Bradford, C.R. Mucosal Melanoma of the Head and Neck: Predictors of Prognosis. Arch Otolaryngol Head Neck Surg 2011, 137, 331–337. [Google Scholar] [CrossRef]
- Millanta, F.; Fratini, F.; Corazza, M.; Castagnaro, M.; Zappulli, V.; Poli, A. Proliferation Activity in Oral and Cutaneous Canine Melanocytic Tumours: Correlation with Histological Parameters, Location, and Clinical Behaviour. Res Vet Sci 2002, 73, 45–51. [Google Scholar] [CrossRef]
- Oh, J.H.; Cho, J.Y. Comparative Oncology: Overcoming Human Cancer through Companion Animal Studies. Exp Mol Med 2023, 55, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Ranieri, G.; Gadaleta, C.D.; Patruno, R.; Zizzo, N.; Daidone, M.G.; Hansson, M.G.; Paradiso, A.; Ribatti, D. A Model of Study for Human Cancer: Spontaneous Occurring Tumors in Dogs. Biological Features and Translation for New Anticancer Therapies. Crit Rev Oncol Hematol 2013, 88, 187–197. [Google Scholar] [CrossRef]
- K, L.-T.; CM, W.; TS, M.; EK, K.; DB, J.; M, K.; M, C.; JL, C.; EJ, K.; MC, Z.; et al. Genome Sequence, Comparative Analysis and Haplotype Structure of the Domestic Dog. Nature 2005, 438, 803–819. [Google Scholar] [CrossRef]
- Broniec, A.; Goto, M.; Matsuki, H. A Peculiar Phase Transition of Plasmalogen Bilayer Membrane under High Pressure. Langmuir 2009, 25, 11265–11268. [Google Scholar] [CrossRef]
- Aramaki, S.; Tsuge, S.; Islam, A.; Eto, F.; Sakamoto, T.; Oyama, S.; Li, W.; Zhang, C.; Yamaguchi, S.; Takatsuka, D.; et al. Lipidomics-Based Tissue Heterogeneity in Specimens of Luminal Breast Cancer Revealed by Clustering Analysis of Mass Spectrometry Imaging: A Preliminary Study. PLoS One 2023, 18. [Google Scholar] [CrossRef]
- Wu, Q.; Yu, J.; Zhang, M.; Xiong, Y.; Zhu, L.; Wei, B.; Wu, T.; Du, Y. Serum Lipidomic Profiling for Liver Cancer Screening Using Surface-Assisted Laser Desorption Ionization MS and Machine Learning. Talanta 2024, 268. [Google Scholar] [CrossRef] [PubMed]
- Bifarin, O.O.; Sah, S.; Gaul, D.A.; Moore, S.G.; Chen, R.; Palaniappan, M.; Kim, J.; Matzuk, M.M.; Fernández, F.M. Machine Learning Reveals Lipidome Remodeling Dynamics in a Mouse Model of Ovarian Cancer. J Proteome Res 2023, 22, 2092–2108. [Google Scholar] [CrossRef]
- Xiao, Y.; Ma, D.; Yang, Y.S.; Yang, F.; Ding, J.H.; Gong, Y.; Jiang, L.; Ge, L.P.; Wu, S.Y.; Yu, Q.; et al. Comprehensive Metabolomics Expands Precision Medicine for Triple-Negative Breast Cancer. Cell Res 2022, 32, 477. [Google Scholar] [CrossRef]
- Li, R.; Li, L.; Xu, Y.; Yang, J. Machine Learning Meets Omics: Applications and Perspectives. Brief Bioinform 2022, 23. [Google Scholar] [CrossRef]
- Cai, Z.; Poulos, R.C.; Liu, J.; Zhong, Q. IScience Machine Learning for Multi-Omics Data Integration in Cancer. iScience 2022, 25, 103798. [Google Scholar] [CrossRef] [PubMed]
- Arjmand, B.; Hamidpour, S.K.; Tayanloo-Beik, A.; Goodarzi, P.; Aghayan, H.R.; Adibi, H.; Larijani, B. Machine Learning: A New Prospect in Multi-Omics Data Analysis of Cancer. Front Genet 2022, 13, 824451. [Google Scholar] [CrossRef] [PubMed]
- Csala, A.; Zwinderman, A.H. Multivariate Statistical Methods for High-Dimensional Multiset Omics Data Analysis. Computational Biology 2019, 71–83. [Google Scholar] [CrossRef]
- Caponigro, V.; Tornesello, A.L.; Merciai, F.; La Gioia, D.; Salviati, E.; Basilicata, M.G.; Musella, S.; Izzo, F.; Megna, A.S.; Buonaguro, L.; et al. Integrated Plasma Metabolomics and Lipidomics Profiling Highlights Distinctive Signature of Hepatocellular Carcinoma in HCV Patients. J Transl Med 2023, 21, 1–15. [Google Scholar] [CrossRef]
- Wolrab, D.; Jirásko, R.; Cífková, E.; Höring, M.; Mei, D.; Chocholoušková, M.; Peterka, O.; Idkowiak, J.; Hrnčiarová, T.; Kuchař, L.; et al. Lipidomic Profiling of Human Serum Enables Detection of Pancreatic Cancer. Nat Commun 2022, 13. [Google Scholar] [CrossRef]
- Jiang, N.; Zhang, Z.; Chen, X.; Zhang, G.; Wang, Y.; Pan, L.; Yan, C.; Yang, G.; Zhao, L.; Han, J.; et al. Plasma Lipidomics Profiling Reveals Biomarkers for Papillary Thyroid Cancer Diagnosis. Front Cell Dev Biol 2021, 9. [Google Scholar] [CrossRef]
- Kim, H.Y.; Lee, H.; Kim, S.H.; Jin, H.; Bae, J.; Choi, H.K. Discovery of Potential Biomarkers in Human Melanoma Cells with Different Metastatic Potential by Metabolic and Lipidomic Profiling. Sci Rep 2017, 7. [Google Scholar] [CrossRef]
- Kurokawa, G.A.; Hamamoto Filho, P.T.; Delafiori, J.; Galvani, A.F.; de Oliveira, A.N.; Dias-Audibert, F.L.; Catharino, R.R.; Pardini, M.I.M.C.; Zanini, M.A.; Lima, E. de O.; et al. Differential Plasma Metabolites between High- and Low-Grade Meningioma Cases. Int J Mol Sci 2023, 24, 394. [Google Scholar] [CrossRef]
- Gallart-Ayala, H.; Courant, F.; Severe, S.; Antignac, J.P.; Morio, F.; Abadie, J.; Le Bizec, B. Versatile Lipid Profiling by Liquid Chromatography-High Resolution Mass Spectrometry Using All Ion Fragmentation and Polarity Switching. Preliminary Application for Serum Samples Phenotyping Related to Canine Mammary Cancer. Anal Chim Acta 2013, 796, 75–83. [Google Scholar] [CrossRef]
- Safari Yazd, H.; Bazargani, S.F.; Fitzpatrick, G.; Yost, R.A.; Kresak, J.; Garrett, T.J. Metabolomic and Lipidomic Characterization of Meningioma Grades Using LC-HRMS and Machine Learning. J Am Soc Mass Spectrom 2023, 34, 2187–2198. [Google Scholar] [CrossRef]
- Shang, X.; Zhang, C.; Kong, R.; Zhao, C.; Wang, H. Construction of a Diagnostic Model for Small Cell Lung Cancer Combining Metabolomics and Integrated Machine Learning. Oncologist 2024, 29, e392–e401. [Google Scholar] [CrossRef] [PubMed]
- Starodubtseva, N.L.; Tokareva, A.O.; Rodionov, V. V.; Brzhozovskiy, A.G.; Bugrova, A.E.; Chagovets, V. V.; Kometova, V. V.; Kukaev, E.N.; Soares, N.C.; Kovalev, G.I.; et al. Integrating Proteomics and Lipidomics for Evaluating the Risk of Breast Cancer Progression: A Pilot Study. Biomedicines 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Manzi, M.; Palazzo, M.; Knott, M.E.; Beauseroy, P.; Yankilevich, P.; Giménez, M.I.; Monge, M.E. Coupled Mass-Spectrometry-Based Lipidomics Machine Learning Approach for Early Detection of Clear Cell Renal Cell Carcinoma. J Proteome Res 2021, 20, 841–857. [Google Scholar] [CrossRef]
- Wang, G.; Yao, H.; Gong, Y.; Lu, Z.; Pang, R.; Li, Y.; Yuan, Y.; Song, H.; Liu, J.; Jin, Y.; et al. Metabolic Detection and Systems Analyses of Pancreatic Ductal Adenocarcinoma through Machine Learning, Lipidomics, and Multi-Omics. Sci Adv 2021, 7, 2724. [Google Scholar] [CrossRef]
- Yang, C.; Zhou, S.; Zhu, J.; Sheng, H.; Mao, W.; Fu, Z.; Chen, Z. Plasma Lipid-Based Machine Learning Models Provides a Potential Diagnostic Tool for Colorectal Cancer Patients. Clinica Chimica Acta 2022, 536, 191–199. [Google Scholar] [CrossRef]
- Krishnan, S.T.; Winkler, D.; Creek, D.; Anderson, D.; Kirana, C.; Maddern, G.J.; Fenix, K.; Hauben, E.; Rudd, D.; Voelcker, N.H. Staging of Colorectal Cancer Using Lipid Biomarkers and Machine Learning. Metabolomics 2023, 19. [Google Scholar] [CrossRef]
- Zhou, J.; Ji, N.; Wang, G.; Zhang, Y.; Song, H.; Yuan, Y.; Yang, C.; Jin, Y.; Zhang, Z.; Zhang, L.; et al. Metabolic Detection of Malignant Brain Gliomas through Plasma Lipidomic Analysis and Support Vector Machine-Based Machine Learning. EBioMedicine 2022, 81. [Google Scholar] [CrossRef] [PubMed]
- Saito, R.; Yoshimura, K.; Shoda, K.; Furuya, S.; Akaike, H.; Kawaguchi, Y.; Murata, T.; Ogata, K.; Iwano, T.; Takeda, S.; et al. Diagnostic Significance of Plasma Lipid Markers and Machine Learning-Based Algorithm for Gastric Cancer. Oncol Lett 2021, 21. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.C.; Wu, W.H.; Huang, S.; Li, Z.W.; Li, X.; Shui, G.H.; Lam, S.M.; Li, B.W.; Li, Z.X.; Zhang, Y.; et al. Plasma Lipids Signify the Progression of Precancerous Gastric Lesions to Gastric Cancer: A Prospective Targeted Lipidomics Study. Theranostics 2022, 12, 4671. [Google Scholar] [CrossRef]
- Kujala, M.; Nevalainen, J. A Case Study of Normalization, Missing Data and Variable Selection Methods in Lipidomics. Stat Med 2015, 34, 59–73. [Google Scholar] [CrossRef]
- Del Prete, E.; Campos, A.M.; Della Rocca, F.; Gallo, C.; Fontana, A.; Nuzzo, G.; Angelini, C. ADViSELipidomics: A Workflow for Analyzing Lipidomics Data. Bioinformatics 2022, 38, 5460–5462. [Google Scholar] [CrossRef]
- Ding, X.; Yang, F.; Chen, Y.; Xu, J.; He, J.; Zhang, R.; Abliz, Z. Norm ISWSVR: A Data Integration and Normalization Approach for Large-Scale Metabolomics. Anal Chem 2022, 94, 7500–7509. [Google Scholar] [CrossRef]
- Ulmer, C.Z.; Ragland, J.M.; Koelmel, J.P.; Heckert, A.; Jones, C.M.; Garrett, T.J.; Yost, R.A.; Bowden, J.A. LipidQC: Method Validation Tool for Visual Comparison to SRM 1950 Using NIST Interlaboratory Comparison Exercise Lipid Consensus Mean Estimate Values. Anal Chem 2017, 89, 13069–13073. [Google Scholar] [CrossRef]
- Köfeler, H.C.; Ahrends, R.; Baker, E.S.; Ekroos, K.; Han, X.; Hoffmann, N.; Holcapek, M.; Wenk, M.R.; Liebisch, G. Recommendations for Good Practice in Ms-Based Lipidomics. J Lipid Res 2021, 62. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, V.B.; Fitzgerald, G.A.; Murphy, R.C.; Liebisch, G.; Dennis, E.A.; Quehenberger, O.; Subramaniam, S.; Wakelam, M.J.O. Steps Toward Minimal Reporting Standards for Lipidomics Mass Spectrometry in Biomedical Research Publications. Circ Genom Precis Med 2020, 13, E003019. [Google Scholar] [CrossRef]
- Triebl, A.; Burla, B.; Selvalatchmanan, J.; Oh, J.; Tan, S.H.; Chan, M.Y.; Mellet, N.A.; Meikle, P.J.; Torta, F.; Wenk, M.R. Shared Reference Materials Harmonize Lipidomics across MS-Based Detection Platforms and Laboratories. J Lipid Res 2020, 61, 105–115. [Google Scholar] [CrossRef]
- Tsuchida, S.; Nakayama, T. MALDI-Based Mass Spectrometry in Clinical Testing: Focus on Bacterial Identification. Applied Sciences 2022, Vol. 12, Page 2814 2022, 12, 2814. [Google Scholar] [CrossRef]
- Poirion, O.B.; Jing, Z.; Chaudhary, K.; Huang, S.; Garmire, L.X. DeepProg: An Ensemble of Deep-Learning and Machine-Learning Models for Prognosis Prediction Using Multi-Omics Data. Genome Med 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Song, M.; Shen, H.; Hong, H.; Gong, P.; Deng, H.-W.; Zhang, C. Deep Learning Methods for Omics Data Imputation. Biology (Basel) 2023, 12. [Google Scholar] [CrossRef]
- Kang, M.; Ko, E.; Mersha, T.B. A Roadmap for Multi-Omics Data Integration Using Deep Learning. Brief Bioinform 2022, 23. [Google Scholar] [CrossRef]
- Albaradei, S.; Thafar, M.; Alsaedi, A.; Van Neste, C.; Gojobori, T.; Essack, M.; Gao, X. Machine Learning and Deep Learning Methods That Use Omics Data for Metastasis Prediction. Comput Struct Biotechnol J 2021, 19, 5008–5018. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.A.; Kondrashova, O.; Bradley, A.; Williams, E.D.; Pearson, J. V.; Waddell, N. Deep Learning in Cancer Diagnosis, Prognosis and Treatment Selection. Genome Med 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.; Snyder, M. Multi-Omics Profiling for Health. Mol Cell Proteomics 2023, 22, 100561. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.R.; Carvalho, M.; Aveiro, S.S.; Melo, T.; Domingues, M.R.; Macedo-Silva, C.; Coimbra, N.; Jerónimo, C.; Henrique, R.; Bastos, M. de L.; et al. Comprehensive Metabolomics and Lipidomics Profiling of Prostate Cancer Tissue Reveals Metabolic Dysregulations Associated with Disease Development. J Proteome Res 2022, 21, 727–739. [Google Scholar] [CrossRef]
- Jung, J.H.; Yang, D.Q.; Song, H.; Wang, X.; Wu, X.; Kim, K.P.; Pandey, A.; Byeon, S.K. Characterization of Lipid Alterations by Oncogenic PIK3CA Mutations Using Untargeted Lipidomics in Breast Cancer. OMICS 2023, 27, 327–335. [Google Scholar] [CrossRef]
| Reference | Cancer Origin | Analyzed Tissue | Acquisition | ML Algorithms | Notes |
|---|---|---|---|---|---|
| [65] | Melanoma | Solid biopsy | MALDI-MS | LR, NB, SVM | Lipid imaging |
| [89] | Mammary | Solid biopsy | UHPLC-MS | LASSO, SVM | SRAA |
| [86] | Mammary | Solid biopsy | DESI | KNN | Lipid imaging |
| [100] | Meningioma | Solid biopsy | LC-HRMS | DT, KNN, LR, NB, RF, SVM | |
| [88] | Mouse ovarian | Solid biopsy | UHPLC-MS | LR, RF, KNN, SVM, VC | SRAA |
| [101] | Lung | Serum | LC-MS/MS | LR, RF, SVM | Panel of 8 metabolites, SRAA |
| [102] | Mammary | Serum | LC-MS | LR | Tumor metastatic potential |
| [87] | Liver | Serum | MALDI-MS | LDA, LR, MLP, RF, SVM | |
| [103] | Renal | Serum | UPLC-MS | LASSO-SVM | Coupled ML algorithms |
| [104] | Pancreas | Serum | MALDI-MS | SVM | |
| [105] | Colorectal | Plasma | LC-MS | KNN, PLS, RF, SVM | |
| [106] | Colorectal | Plasma | LC-MS | MLR-EM, BRANN | Tumor Stage Classification |
| [107] | Glioma | Plasma | HPLC-MS | SVM | |
| [108] | Gastric | Plasma | LC/ESI-MS | LR | |
| [109] | Gastric | Plasma | UHPLC-MS | LR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





