1. Introduction
Rotavirus is a member of the family
Reoviridae, subfamily
Sedoreovirinae, and genus
Rotavirus. It is a nonenveloped virus with a diameter of 60–80 nm and a genome length of approximately 18.5 kb. Although rotaviruses can infect a broad spectrum of mammalian and avian species, their infectivity is generally species
-specific[
1]
.
Bovine rotavirus (BRV) group A is acknowledged as the primary cause of severe gastroenteritis in cattle, leading to significant morbidity and mortality in newborn calves and resulting in significant economic losses[
2] . The virus causes depression, acidosis, anorexia, watery diarrhea, and other clinical symptoms in calves aged 1–7 days. . When calves are secondarily infected by pathogens such as bovine coronavirus or Escherichia coli, their symptomatic manifestations become exacerbated, resulting in mortality in severe cases. The disease is prevalent in cattle-raising countries and regions worldwide, and causes serious economic losses to the cattle industry. The incidence of BRV in newborn calves is 60%–80%, with mortality as high as 30%. Although BRV has been studied for a long time, no specific therapeutic drug or effective vaccine for its prevention is yet available[
3]. BRV infection in newborn animals has a very short incubation period, and manifests as massive diarrhea and severe dehydration[
4].
The BRV genome comprises 11 segments of double-stranded RNA (dsRNA) and encodes six viral structural proteins (VP1–VP4, VP6, and VP7) and six nonstructural proteins (NSP1–NSP6). the individual segments contain 5′- and 3′-untranslated regions (UTRs). The VPs form infectious triple-layered particles that envelope the genomic dsRNA. NSPs are primarily involved in dsRNA replication and transcription, cellular pathogenesis, and the maturation of the viral particle[
5] .
Studies have demonstrated that during the invasion of host cells by rotavirus, the VP4 protein is hydrolyzed by trypsin into two peptides, VP5 and VP8[
6]. VP5 (60 kDa) is remain noncovalently associated with the infectious particle allowing the initiation of the RV entry process. VP8 (28 kDa) subunit has hemagglutinin activity, is involved in host sialic acid binding, and has a determinant role in virus tropism. VP4 contains aggregates the main antigenic sites, thus playing a crucial role in vaccine development. The activation of the VP4 structural protein through its digestion by a pancreatic enzyme significantly boosts the pathogenicity of BRV[
7] . VP6 is used to distinguish the eight primary rotaviral groups, designated A–H[
8] . Rotavirus, Classified into group A based on dual genotyping system utilizing neutralizing antibodies against capsid proteins VP7 and VP4[
9]. This system defines the genotypes G (glycoprotein) and P (protease-sensitive protein), respectively [
8]. The dependence of G and P genotypes on the host species is common, because there are host-specific barriers and restrictions to rotaviral infection. Unusual G and P genotypes of human rotaviruses have emerged as the results of interspecies transmission and the natural reassortment between human and animal viruses, especially those of cows and pigs. To prevent the transmission of new rotaviral strains to humans and animals, continuous monitoring of their emergence in animals is crucial [
10].
The use of the terms ‘serotype’ and ‘genotype’ depends upon whether antigen-based detection methods or nucleic-acid-based detection methods are used [
11]. Thus far, a minimum of 12 distinct genotypes of G (G1–G3, G5, G6, G8, G10, G11, G15, G17, G21, G24) and 11 different genotypes of P (P[1], P[3], P[5]–P[7], P[11], P[14], P[17], P[21], P[29], P[38]) have been documented in cattle. Among these, G6, G8, G10, P[1], P[5], and P[11] are the most frequently identified worldwide, according to previous studies[
12,
13]. Several G genotypes (G6, G8, and G10) and P genotypes (P[1], P[5], P[7], and P[11]) have been identified in the Chinese cattle population. Notably, the only previous relevant study reported that G6 and P[5] are the commonest G and P genotypes among dairy calves in certain regions of China, with G6P the dominant genotype combination . G10 is only frequently detected in some regions[
15].
At present, there have only been a few reports of BRV group A (BRVA) in the Yunnan province , and the prevalence rate, viral genotypes, as well as pathogenesis of the disease remain unclear. In this study, we examined fecal samples from diarrheic cows collected from a farm, a region with a strong prevalence of BRV, isolated and purified one bovine G8 rotaviral strain. The genotype of the isolated strain was further analyzed with whole-genome sequencing, to extend our understanding of the prevalence, genotypes, and pathogenic mechanisms of BRV in Yunnan.
2. Materials and Methods
2.1. Bovine Fecal Samples
In 2020–2021, 10 fecal samples were collected from a cattle farm with a severe suspected outbreak of rotavirus in southwestern Yunnan Province, China. The affected cattle showed severe fever (39.2–41.1 °C) accompanied by diarrhea. In this outbreak, unpublished data revealed that the morbidity rate was 21.5% (85/396) and the mortality rate 1% (4/396). All samples that were not analyzed immediately were stored at −80 °C.
2.2. Virus Isolation
Ten bovine diarrhea samples were detected by RT-PCR, and the positive rate was 40% (4 / 10).The fecal samples (10 ml) from diarrheic calves were collected in tubes containing 20 ml of phosphate-buffered saline (PBS). After clarification by centrifugation at 3000 rpm for 30 min, the supernatants were collected and filtered through 0.45 μm membranes (Millipore USA). Aliquots (10 ml) of supernatants were treated with 15 μg/ml trypsin (TPCK-Trypsin, Sigma-Aldrich) for 1h in a 37 °C water bath. To ensure the efficient infection of the cells, the growth medium was removed from MA-104 cell monolayers and the cells were washed with calcium- and magnesium-free PBS (Sigma-Aldrich). The confluent monolayers of MA-104 cells were inoculated with the fecal samples and incubated at 37 °C under a humidified 5% CO2 atmosphere, and monitored daily for a cytopathic effect (CPE). For serial passage, the infected cells were freeze–thawed, treated with trypsin (as described above), and added to fresh confluent cell monolayers. Cells without CPE after six passages were deemed negative for BRV isolation. The virus was collected, introduced into fresh MA-104 cells, and passaged six times, during which the CPE was monitored. The viral titer was determined in 96-well microplates after 10-fold serial dilution, and the CPE was monitored under a microscope at 4 days post-inoculation. The viral titers were calculated with the Reed–Muench method and expressed as the 50% tissue culture infectious dose per 0.1 ml (TCID50/0.1 ml).
2.3. Immunofluorescence Assay
MA-104 cells were plated and infected with the isolated strain, designated BRV-YN1-2021, in six-well plates to observe viral proliferation. The cells were then fixed in cold acetone (−20 °C) for 20 min and washed three times with PBS (pH 7.2). After reaction with BRV-positive cattle serum for 30 min at 37 °C, the cells were stained with a 100-fold-diluted fluorescein-isothiocyanate-conjugated rabbit anti-bovine antibody (BBI, Shanghai, China). The plates were washed with PBS, and the MA-104 cells were air-dried and examined under a fluorescence microscope at a magnification of 200×.
2.4. RT-PCR Amplification and Sequencing
The viral nucleic acids were extracted from the virus-containing cell-culture supernatant with the TaKaRa MiniBest Viral RNA/DNA Extraction Kit (TaKaRa, Dalian, China) and stored at −80 °C until analysis. A pair of PCR primers, BRV-F (5′-ATGGGTACGATGTGGCTCAA-3′) and BRV-R (5′-ACCGCTGGTGTCATGTTTGG-3′), was used to test the clinical samples, specifically targeting the VP6 region of BRVA [
1]. The mixture was incubated for 30 min at 50 °C according to the instructions provided in the PrimeScript™ One Step RT-PCR Kit (TaKaRa). The thermal cycling procedure included denaturation for 2 min at 94 °C, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30s, and 72 °C for 1 min. The product size was 383 base pairs (bp). Primers were designed to amplify the genes for the three structural proteins, VP4 and VP7, based on the Illumina-MiSeq-sequenced genome of strain BRV-YN1-2021 (
Table 1).
After 35 rounds of amplification (94 °C for 30 s, 55 °C for 30 s, and 72 °C for 2 min), clear signals were produced with RT–PCR, corresponding to PCR products of 2362, 1356, and 1062 bp for VP4, VP6, and VP7, respectively. The full-length PCR fragments were purified with an agarose Gel DNA Extraction Kit and cloned into the pMD-19T vector (TaKaRa), according to the manufacturer’s instructions. All nucleotide positions were confirmed with three independent sequencing reactions in both directions.
2.5. cDNA Library Construction and Illumina MiSeq Sequencing
The cDNA library was constructed and sequenced with Illumina MiSeq, as described before [
16]. Briefly ,a library of 200-bp fragments was constructed by ligating bar-coded adapters to them, using the Fast RNA-Seq Lib Prep Module (AB clonal Technology Co., Ltd, RK20304) and an RNA Adapter Module 96 Index for Illumina Set_A (Abclonal Technology Co., Ltd, RK20351), according to the manufacturer’s instructions. The library was purified with Agencourt AMPure XP magnetic beads (Beckman Coulter). The Agilent 2100 Bioanalyzer (Agilent Technologies) was used to evaluate the quality of the purified cDNA library. Nucleotide sequencing was performed on an Illumina NovaSeq 6000 sequencer (Illumina) with the NovaSeq Reagent Kit v1.5 (Illumina), generating 151 paired-end reads. The data were analyzed with SPAdes (v3.14.1;
https://github.com/ablab/spades). Using
de novo assembly, we successfully constructed contigs from the acquired sequence reads. The full-length nucleotide sequence of each gene segment was obtained by searching the non-redundant database with the assembled contigs as the query sequences for the Basic Local Alignment Search Tool (BLAST). The genetic variation in the VP4 and VP7 genes was analyzed by selecting reference sequences, and phylogenetic trees of VP4 were constructed with the MEGA software.
2.6. Full-Genome Analysis and Genotyping of Isolated Strain YN-1
The genotyping of all 11 RVA segments was determined with the online classification tool for rotavirus A genotyping, the Genotype Determination tool (
https://www.bv-brc.org/). The ORF finder tool was used to predict the putative open reading frames (ORFs) and their corresponding amino acids (
http://www.ncbi.nlm.nih.gov/gorf/gorf.html).
3. Results
3.1. Virus Isolation
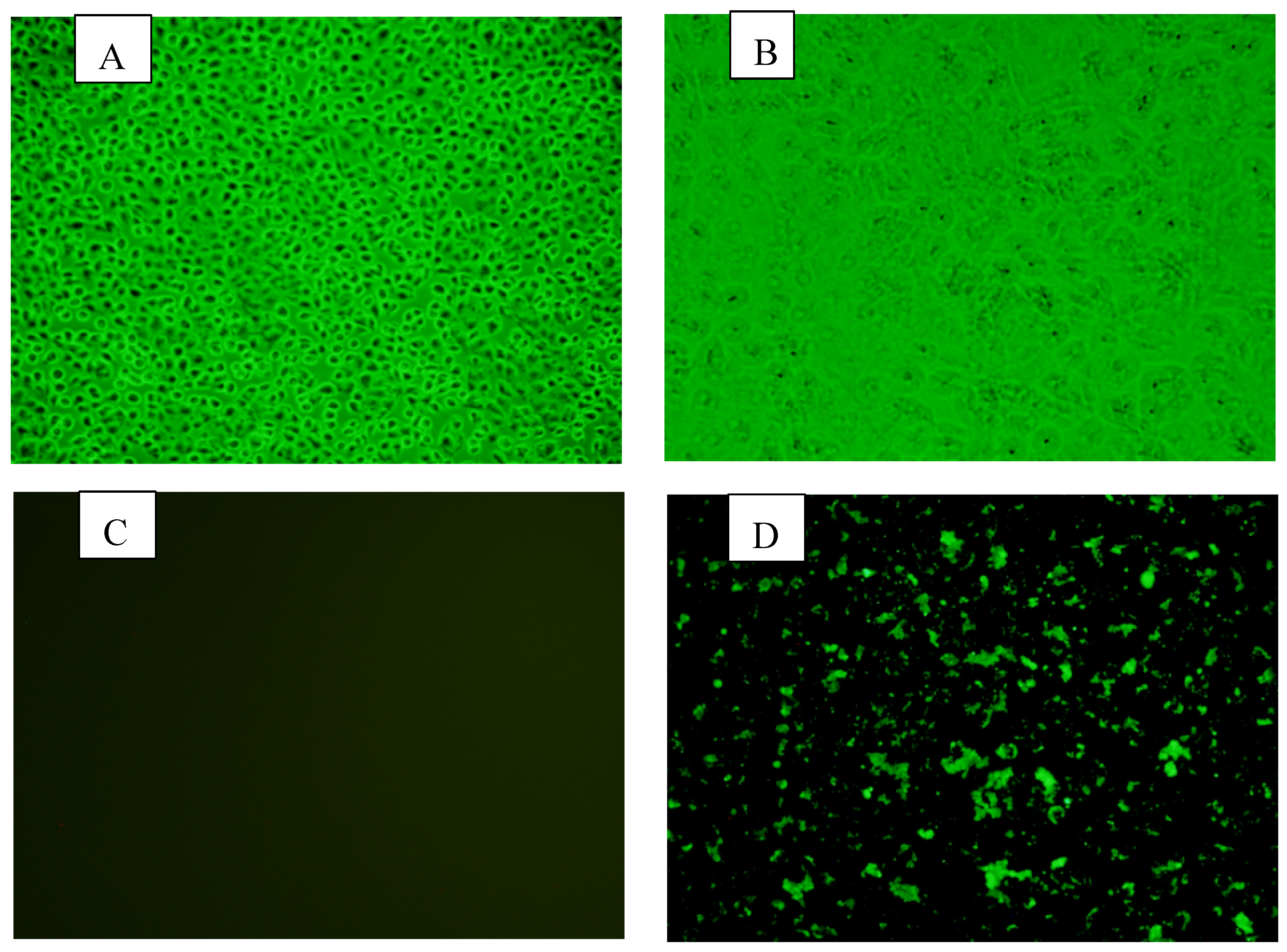
The fecal samples positive for BRV on PCR were used to isolate the virus in cell cultures.. Following inoculation, the manifestation of cytopathic effect (CPE) was evident through a series of cellular alterations. Initially, there was a notable increase in cell granularity, accompanied by the obscuring of cell boundaries. Subsequently, cells exhibited rounding, which progressed to degeneration and eventual detachment after three passages. These changes were characterized by the presence of enlarged, rounded cells with densely granular appearance, occurring in clusters. These findings indicate the successful replication of the inoculated agent within the host cells, leading to the disruption of normal cellular functions and morphology. BRV strain BRV-YN-2021 was successfully isolated from diarrheic fecal samples collected in Yunnan Province during 2020–2021. Stable CPEs were observed from passage 6 onwards. BRV-specific cytoplasmic fluorescence in the MA-104 cells could be detected by indirect immunofluorescence assay (
Figure 1) . The calculated titer of the isolated strain was 10
−5.17 TCID
50/ml.
A, No changes at the cellular level were observed on the control MA104 cells; B, Cell lysis and degeneration due to the cytopathic effect (CPE) with considerable changes in cell’s morphology; C, immunofluorescence results of the MA104 cells as a negative control after 6 passages; D, immunofluorescence results of the MA104 cells infected by RVA virus YN-1 after 6 passages.
3.2. Full-Genome Analysis of Isolated Strain BRV-YN1-2021
The complete genome of strain BRV-YN1-2021, isolated in this study, was successfully sequenced, and the sequence was submitted to the GenBank database (temporary ID as 2712923).
The results of Rotavirus A genotype determination online showed that the constellation of the strain as G8-P[
1]-I2-R2-C2-M2-A3-N2-T6-E2-H3, - which was named as RVA/Cat/CHN/YN-1/2021/G8P following the guideline [
1].
The nucleotide identities of the 11 segments of strain YN-1 with references strains are also determined during BLAST, five segments showed high nucleotide identity with a deer rotavirus strain, two segments with a human strain, and one segment with a U.S. vaccine strain. The lengths of the 11 YN-1 gene segments, putative ORFs, and promoter and terminator signals were analyzed with the National Center for Biotechnology Information ORF Finder tool (
Table 2, Table3).
3.3. Molecular Characteristics of VP4 Sequence
The sequencing analysis revealed that the complete VP4 gene of YN-1 virus comprises a total length of 2088 nucleotides and encodes 695 amino acids. The RotaC v2.0 analysis classified the sequence as the P[
1] genotype. The VP4 gene of strain YN-1 shares 75.2%–99.7% nucleotide identity and 81.6%–99.7% amino acid identity with the complete VP4 sequences of the P[
1] RVA strains available in GenBank. The strain closest to YN-1 at this locus is RVA/LabStr/USA/SA11g4O-5N/XXXX/G3P1. The VP4 gene is highly homologous to those of strains KU962131 and KJ803838 (
Figure 2). The close genetic relationship between the VP4 gene and KU962131 from the Cervus family in the United States suggests potential gene flow or species exchange between them. Due to this intimate association, there exists a potential risk of cross-border transmission between the isolate and KU962131, which could involve pathogens, genetic diseases, or ecological invasions, posing threats to the health of local animal populations.The genetic relationship between VP4 Gene and KU962131 of Cervus in the United States is very close, which indicates that there may be gene flow or species exchange between them. Due to this close connection, there may be a potential risk of cross-border transmission between the isolate and ku962131. This risk may involve pathogens, genetic diseases or ecological invasion and pose a threat to the health of local animal populations.
3.4. Molecular Characteristics of VP7 Sequence
The complete VP7 gene of YN-1 is 981 nucleotide long and encode 326 amino acids. The RotaC v2.0 analysis classified the sequence as the G8 genotype. The VP7 gene of strain YN-1 shares 88.5%–97.7% nucleotide identity and 94.2%–99.1% amino acid identity with all the complete VP7 sequences available in GenBank. The strain closest to YN-1 at this locus is RVA/Guanaco-wt/ARG/Rio_Negro/1998/G8P1,indicating its import from other countries (
Figure 3).
3.5. Molecular Characteristics of VP6 and VP1–VP3
The VP6 sequence of strain YN-1 was classified as the I2 genotype.The phylogenetic tree shows a close association between YN-1 and Chinese human RV strain PP645797, homology reached 99.5%, indicate two virus strains may have undergone similar genetic backgrounds during their evolution, potentially result from gene exchange, such as recombination events(
Figure 3). The VP1 gene of YN-1 is genotype R2, and closely related to that of Hungarian human strain BP1062, which was previously shown to be of bovine rotavirus origin; and the VP2 gene of YN-1 is genotype C2, and closely related to that of USA bovine strain NCDV. The VP3 gene of YN-1 is genotype M2, and closely related to that of a lamb rotavirus strain used as the Chinese human vaccine RVA strain LLR.
3.6 Molecular characteristics of NSP1–NSP5 genesThe genes NSP1, NSP2, NSP3, NSP4, and NSP5 of strain YN-1 belong to genotypes A3, N2, T6, E2, and H3, respectively. The NSP1 gene of YN-1 exhibits a strong genetic similarity to the Vietnamese human RVA strain NT0082. The NSP2 gene of YN-1 closely aligns with the Thai human RVA strain SKT-27. Furthermore, the NSP3 gene of YN-1 has a close relationship with the human RVA strain BP1062 from Hungary, which has been previously identified as originating from BRV. The NSP4 gene of YN-1 is closely related to the human RVA strain Hun5, also from Hungary. Lastly, the NSP5 gene of YN-1 shares a strong genetic relationship with the Argentinian guanaco RVA strain Chubut, potentially indicating a connection with a bovine strain
. [
17].
Discussion
Rotavirus is a significant zoonotic pathogen that causes severe viral intestinal diseases and is the primary cause of dehydration-related diarrhea in calves. Segmented RV is a prerequisite for the emergence of new chimeric reassortant strains. Interspecies transmission and gene reassortment drive viral evolution, enabling it to overcome host barriers and cause infections, resulting in genetically diverse BRV strains that cause severe economic losses to the cattle industry. Currently, vaccination is employed for disease prevention in clinical settings, but the evolution of the circulation process may increase the prevalence of non-vaccine strains, and RVA infections and diarrheal symptoms still remain relatively common.)
The bovine rotavirus (BRV) is a well-known pathogen associated with diarrhea in cattle. Recent research has revealed a notably high detection rate of BRV in fecal samples collected from bovine diarrhea cases. However, despite this high detection rate, the isolation of RV from these samples remains challenging, exhibiting a significantly low isolation rate. This anomaly is attributed to the unique three-layer particle structure of RV, which restricts its infectivity to specific cell types. RV must be activated by trypsin treatment before infecting cells, and serum cannot be added during infection. RV has been successfully isolated from MA104, HepG2, CaCo-2, Vero and HT-29 cells after trypsin treatment.[
18,
19] The conformational changes experienced by VP4 are the main driving force for cell membrane rupture and virus entry into host cells. The hydrolytic processing of this protein mediates the entry of rotavirus into cells. The rotavirus coat proteins VP7 and VP4 can independently stimulate the body to produce neutralizing antibodies, and VP4 can be cleaved into vp5* (60 kDa) and vp8* (28 kDa) polypeptide fragments that enhance the infectivity of the virus by trypsin action, exposing the binding site of cell receptors, eliminating the inhibitory effect of virus growth on cell culture, and enhancing the proliferation ability of virus-infected cells[
20]. In this study, 15mg of trypsin concentration activation amount and 20mg of trypsin concentration maintenance amount were used as the isolation and culture conditions for the virus solution. After six generations of blind transmission, the cells became round, shrunk, fell off, and the stable CPE with the phenomenon of net pulling appeared. Combined with the pathogenic identification results, it was proved that a g8p[
1] type BRV was successfully isolated in MA104 cells.
Bovine diarrhea is a significant disease, posing considerable harm to the cattle industry [
14]. The etiology of diarrhea is multifactorial, although rotavirus is a leading causative factor. The cattle industry in China has experienced substantial economic losses due to heightened mortality rates, increased treatment costs, and reduced weight gain in affected cattle. In this study, we isolated a BRV strain from diarrheic calves in Yunnan, designated RVA/Cattle/CHN/YN1/2021/G8P. The genotypes of the 11 gene segments of this strain were determined with the analysis of nucleotide sequence homologies. Most human RVA strains can be classified into two major and one minor genotype constellations. These include the porcine-origin Wa-like genogroup (G1/3/4/9-P[
8]-I1-R1-C1-M1-A1-N1-T1-E1-H1), the bovine-origin DS-1-like genogroup (G2-P[
4]-I2-R2-C2-M2-A2-N2-T2-E2-H2), and the feline-origin AU-1 genogroup (G3-P[
9]-I3-R3-C3-M3-A3-N3-T3-E3-H3)[
12]. The two predominant human rotaviral strains, WA-like and DS-1-like, share common origins with swine and bovine RVA strains, respectively.
Previous studies have shown that the interspecies transmission of BRVA to pigs, sheep, and other animals is a common phenomenon. Furthermore, these strains can be transmitted directly to humans or through reassortment with human RVA strains [
20]. BRVA genotype G6P[
1] and G8P[
1] strains can infect humans directly, and all 11 segments of these BRVA strains originate from bovine sources, as observed in prevalence studies conducted in China[
21,
22]. Therefore, BRVA poses a significant threat to public health.
. In conclusion, the surveillance of the rotaviral G and P genotypes undertaken in this study extends our understanding of the epidemiology of rotaviruses in Yunan Province, China.It underscores that develop an effective vaccine remains a paramount priority
Conflict of Interests Statement
The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement
This research and publication were supported by three grants from Yunnan Province Science and Technology Department: Yunnan Joint International R&D Center of Veterinary Public Health , Yunnan International Sci-Tech Special Commissioner (202203AK140015), and Yunnan Rural Revitalization & Technological Innovation Village (2024040204).
Animal Rights Statement
All animal procedures were conducted following the guidelines in the document approved by the Animal Ethics Committee of Yunnan Agricultural University.
Acknowledgements
We thank Liwen Bianji (Edanz) (
www.liwenbianji.cn) for editing the English text of a draft of this manuscript.
References
- Veletanlic V., Sartalamacchia K., Diller J.R., Ogden K.M.: Multiple rotavirus species encode fusion-associated small transmembrane (FAST) proteins with cell type-specific activity. bioRxiv 2023, doi:10.1101/2023.04.07.536061.
- Geletu U.S., Usmael M.A., Bari F.D.: Rotavirus in Calves and Its Zoonotic Importance. Vet Med Int 2021, 2021:6639701. [CrossRef]
- Kanai Y., Onishi M., Kawagishi T., Pannacha P., Nurdin J.A., Nouda R., Yamasaki M., Lusiany T., Khamrin P., Okitsu S., Hayakawa S., Ebina H., Ushijima H., Kobayashi T.: Reverse Genetics Approach for Developing Rotavirus Vaccine Candidates Carrying VP4 and VP7 Genes Cloned from Clinical Isolates of Human Rotavirus. J Virol 2020, 95. [CrossRef]
- Foster D.M., Smith G.W.: Pathophysiology of diarrhea in calves. Vet Clin North Am Food Anim Pract 2009, 25:13-36. [CrossRef]
- Nurdin J.A., Kotaki T., Kawagishi T., Sato S., Yamasaki M., Nouda R., Minami S., Kanai Y., Kobayashi T.: N-Glycosylation of Rotavirus NSP4 Protein Affects Viral Replication and Pathogenesis. J Virol 2023, 97:e186122. [CrossRef]
- Cheng X., Wu W., Teng F., Yan Y., Li G., Wang L., Wang X., Wang R., Zhou H., Jiang Y., Cui W., Tang L., Li Y., Qiao X.: Isolation and Characterization of Bovine RVA from Northeast China, 2017-2020. Life (Basel) 2021, 11. [CrossRef]
- Kaljot K.T., Shaw R.D., Rubin D.H., Greenberg H.B.: Infectious rotavirus enters cells by direct cell membrane penetration, not by endocytosis. J Virol 1988, 62:1136-1144. [CrossRef]
- Pourasgari F., Kaplon J., Karimi-Naghlani S., Fremy C., Otarod V., Ambert-Balay K., Mirjalili A., Pothier P.: The molecular epidemiology of bovine rotaviruses circulating in Iran: a two-year study. Arch Virol 2016, 161:3483-3494. [CrossRef]
- Aoki S.T., Settembre E.C., Trask S.D., Greenberg H.B., Harrison S.C., Dormitzer P.R.: Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science 2009, 324:1444-1447. [CrossRef]
- Odagiri K., Yoshizawa N., Sakihara H., Umeda K., Rahman S., Nguyen S.V., Suzuki T.: Development of Genotype-Specific Anti-Bovine Rotavirus A Immunoglobulin Yolk Based on a Current Molecular Epidemiological Analysis of Bovine Rotaviruses A Collected in Japan during 2017-2020. Viruses 2020, 12. [CrossRef]
- Esona M.D., Gautam R.: Rotavirus. Clin Lab Med 2015, 35:363-391. [CrossRef]
- Matthijnssens J., Ciarlet M., McDonald S.M., Attoui H., Banyai K., Brister J.R., Buesa J., Esona M.D., Estes M.K., Gentsch J.R., Iturriza-Gomara M., Johne R., Kirkwood C. D., Martella V., Mertens P. P., Nakagomi, O., Parreno V., Rahman M., Ruggeri F. M., Saif L. J., Santos N., Steyer A., Taniguchi K., Patton J. T., Desselberger U., Van Ranst M.: Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol 2011, 156:1397-1413. [CrossRef]
- Papp H., Matthijnssens J., Martella V., Ciarlet M., Bányai K.: Global distribution of group A rotavirus strains in horses: a systematic review. Vaccine 2013, 31:5627-5633. [CrossRef]
- Papp H., Laszlo B., Jakab F., Ganesh B., De Grazia S., Matthijnssens J., Ciarlet M., Martella V, Banyai K: Review of group A rotavirus strains reported in swine and cattle. Vet Microbiol 2013, 165:190-199. [CrossRef]
- Liu X., Yan N., Yue H., Wang Y., Zhang B., Tang C.: Detection and molecular characteristics of bovine rotavirus A in dairy calves in China. J Vet Sci 2021, 22:e69. [CrossRef]
- Ide T., Komoto S., Higo-Moriguchi K., Htun K.W., Myint Y.Y., Myat T.W., Thant K.Z., Thu H.M., Win M.M., Oo H.N., Htut T., Wakuda M., Dennis F. E., Haga K., Fujii Y., Katayama K., Rahman S., Nguyen S. V., Umeda K., Oguma K., Tsuji T., Taniguchi K.: Whole Genomic Analysis of Human G12P[6] and G12P[8] Rotavirus Strains that Have Emerged in Myanmar. Plos One 2015, 10:e124965. [CrossRef]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.: The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 2015, 10:845-858. [CrossRef]
- Kitamoto N, Ramig RF, Matson DO, Estes MK. Comparative growth of different rotavirus strains in differentiated cells (MA104, HepG2, and CaCo-2). Virology. 1991 Oct;184(2):729-37. [CrossRef]
- Superti F, Tinari A, Baldassarri L, Donelli G. HT-29 cells: a new substrate for rotavirus growth. Arch Virol. 1991;116(1-4):159-73. [CrossRef]
- Luo G, Zeng Y, Yang H, Li Y, Yang L, Li C, Song F, Zhang S, Li T, Ge S, Zhang J, Xia N. Bivalent rotavirus VP4∗ stimulates protective antibodies against common genotypes of human rotaviruses. iScience. 2022 Sep 8;25(10):105099. [CrossRef]
- Park S.I., Matthijnssens J., Saif L.J., Kim H.J., Park J.G., Alfajaro M.M., Kim D.S., Son K.Y., Yang D.K., Hyun B.H., Kang M. I., Cho K. O.: Reassortment among bovine, porcine and human rotavirus strains results in G8P[7] and G6P[7] strains isolated from cattle in South Korea. Vet Microbiol 2011, 152:55-66. [CrossRef]
- Yan N., Li R., Wang Y., Zhang B., Yue H., Tang C.: High prevalence and genomic characteristics of G6P[1] Bovine Rotavirus A in yak in China. J Gen Virol 2020, 101:701-711. [CrossRef]
- Ghosh S., Gatheru Z., Nyangao J., Adachi N., Urushibara N., Kobayashi N.: Full genomic analysis of a G8P[1] rotavirus strain isolated from an asymptomatic infant in Kenya provides evidence for an artiodactyl-to-human interspecies transmission event. J Med Virol 2011, 83:367-376. [CrossRef]
- Komoto S., Pongsuwanna Y., Ide T., Wakuda M., Guntapong R., Dennis F.E., Haga K., Fujii Y., Katayama K., Taniguchi K.: Whole genomic analysis of porcine G10P[5] rotavirus strain P343 provides evidence for bovine-to-porcine interspecies transmission. Vet Microbiol 2014, 174:577-583. [CrossRef]
- Zhu W., Dong J., Haga T., Goto Y., Sueyoshi M.: Rapid and sensitive detection of bovine coronavirus and group a bovine rotavirus from fecal samples by using one-step duplex RT-PCR assay. J Vet Med Sci 2011, 73:531-534. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).