1. Introduction
In recent years, new biomarkers have emerged that allow the monitoring of the transplant patient and that seem to be promising, especially in the case of kidney transplantation, with respect to the time of anticipation in the detection of rejection compared to renal biopsy or other classic serological/urinary biomarkers [
1].
Biopsy is currently considered the "gold standard" in the monitoring of the transplanted organ. In this way, biopsy has evolved to become an early detection method, which should meet the fundamental criteria defined for screening procedures, which include safety and acceptability for the patient, as well as the ability to detect the clinical condition to be combated at an opportune time in order to modify its eventual course [
2]. However, biopsy is far from being an ideal screening method, as it presents important limitations for patient safety and acceptability due to its invasive nature, discomfort for the patient and the complications derived from it. Comprehensive studies in the field of kidney transplantation reveal that around 1% of biopsies result in significant complications, with a risk of gross hematuria exceeding 3.5% [
3]. In addition, thanks to the new immunosuppressive therapies that are currently available, the detection of subclinical rejection is infrequent enough to justify such a risk, which has motivated many nephrology units to stop performing such routine kidney biopsies [
4].
As a result, there has been growing interest in the development of non-invasive strategies capable of detecting graft failure or rejection. The ideal non-invasive technique would be one that can be measured in plasma or urine, affordable enough for routine use, and sensitive and specific enough to detect any damage to the graft. Currently, serum creatinine and urinary indicators of renal function, such as the urinary albumin-to-creatinine ratio (UACR) are the ones that fulfill this role, as they are inexpensive, relatively reliable and easily interpreted. However, its sensitivity and specificity when it comes to detecting damage to the allograft are relatively poor [
1,
5].
There are also a number of other non-invasive methods for diagnosing and monitoring patients with rejection, such as the detection of human leukocyte antigen (HLA) and non-HLA antibodies [
6]; blood gene expression profiles such as Trugraf [
7] or kSORT [
8]; analysis of perforin, granzyme B, and inducible protein mRNA by IFN-10 [
9]; urinary chemokine levels (CXCL9 y CXCL10); proteomic and peptide signatures of rejection in urine and blood samples; enzyme-linked immunospot assay for IFN-gamma (ELISPOT) [
10]; or metabolomic changes. However, these non-invasive methods generally lack sufficient scientific evidence to support their clinical use, or they are too expensive to implement.
In this context, donor-derived circulating free DNA (dd-cfDNA), an innovative biomarker with great potential for the early identification and prevention of graft damage, is of crucial importance [
11]. This dd-cfDNA is present in the recipient's blood and comes from damaged cells in the transplanted kidney [
12]. When graft damage occurs, the number of cells affected increases, and consequently the presence of dd-cfDNA in peripheral blood. In this way, identifying an early increase in dd-cfDNA levels in the recipient would reduce the number of unnecessary biopsies, and it would also provide for the clinician with essential information in order to modify immunosuppressive therapy at the right time and prevent the progression of damage [
13].
The considerable diagnostic and prognostic potential of this innovative biomarker has prompted its inclusion in numerous and extensive multicenter trials. Among them, the following stand out: Allosure with CareDx, Prospera with Natera y Trac with Viracor-Eurofins [
14,
15,
16]. These trials have precisely defined the thresholds that discriminate the presence or absence of rejection (cfDNA >1%), as well as the associated values of sensitivity, specificity, negative predictive value (NPV) and positive predictive value (PPV) linked to this technique.
In the present prospective study, we show the results of 30 kidney transplant patients, in whom dd-cfDNA levels have been evaluated during their post-transplant follow-up, to determine if the information provided by dd-cfDNA could be of interest in the identification of possible adverse events. In addition, the aim is to verify whether the data on sensitivity, specificity, NPV and PPV obtained in our series are consistent with the values referenced in the most consolidated literature, considering that reactive renal biopsies or clinically indicated renal biopsies are performed in our center, but not prospective biopsies by protocol.
2. Materials and Methods
Our prospective study included patients undergoing kidney transplantation between 2020 and 2023 from the Hospital Clínico Universitario Virgen de la Arrixaca and Biomedical Research Institute of Murcia (IMIB) (Murcia, Spain) within the framework of the PI19/01194 National Research Project funded by the Carlos III Health Institute (Madrid, Spain) and the AP183152023 Project funded by the Madrid Mutual Foundation.
Given the high volume of patients (greater than 150) and the logistical and economic limitations, exclusion and inclusion criteria were implemented to select a sample of 30 patients representative of the population to be studied.
On the one hand, an exhaustive analysis of the medical records of the patients included in the study was carried out, with the aim of identifying and including the maximum number of patients with rejection, suspected rejection or other relevant adverse effects and who had completed monitoring in the first year after transplantation. However, only 12 patients with these characteristics could be included, and the study was completed with the first 18 patients enrolled in the trial who had not experienced any type of adverse event during their post-transplant period and who had completed follow-up.
Finally, dd-cfDNA levels were determined in 30 patients undergoing kidney transplantation with a mean age of 49.84±9.86 years and a percentage of 64% males, who were classified into groups according to their post-transplant clinical evolution, taking as a reference the Banff classification [
17,
18] for subsequent study.
Blood samples from patients were collected both at the time before transplantation and at 15 days, 3, 6 and 9 months post-transplant. A tube was removed Streck-Cell-Free DNA BCT® de 10 ml from each patient. The plasma was then isolated using a double-centrifugation protocol at 1600xg for 10 minutes at room temperature. The plasma collected was frozen at -80 °C until the time of use.
The extraction of dd-cfDNA from the plasma samples of the selected patients was performed using the commercial kit AlloSeq-cfDNA (CareDx, Brisbane, CA) following the specifications provided by the manufacturer. The analysis using next-generation sequencing (NGS) was carried out using the "MiSeq TM System" (Illumina, San Diego, CA). The analysis of NGS outputs was performed with the Alloseq cfDNA software v.1.0 (ASCFS 1.0). The data and results obtained from each patient were collected in an anonymized database with coding that prevented their identification. Data analysis was performed using the Microsoft Office Excel software and IBM SPSS Statistics v.22 software.
The results of the analysis of quantitative variables were expressed as Mean and Standard Deviation or as Median and Interquartile Range. Qualitative variables were expressed as percentages. For the contrast of means, the Student's t-test of independent and paired samples was used when the distribution was normal, or the Wilcoxon and U-Mann-Whitney test (post-hoc) for the contrast of medians in case of non-normal distributions. For multiple comparisons, the parametric ANOVA and Tukey test (post hoc), or the non-parametric Friedman test, were used. The contrast of percentages was performed using Pearson's Chi-Square test. The level of statistical significance was established for values of p<0.05.
Respect to ethical-legal aspects, this study was evaluated and approved with a favorable opinion by the Research Ethics Committee of the IMIB-Arrixaca. All participants signed an informed consent that allowed the research team to use the data and results anonymously for research purposes. Finally, none of the participating members declared any conflict of interest.
3. Results
Levels of dd-cfDNA (%) was determined in 30 kidney transplant recipients from Virgen de la Arrixaca University Hospital, with a mean age of 49.84±9.86 and 70% were men vs 30% women. The demographic and clinical characteristics of the patients included in this study are shown in
Table 1.
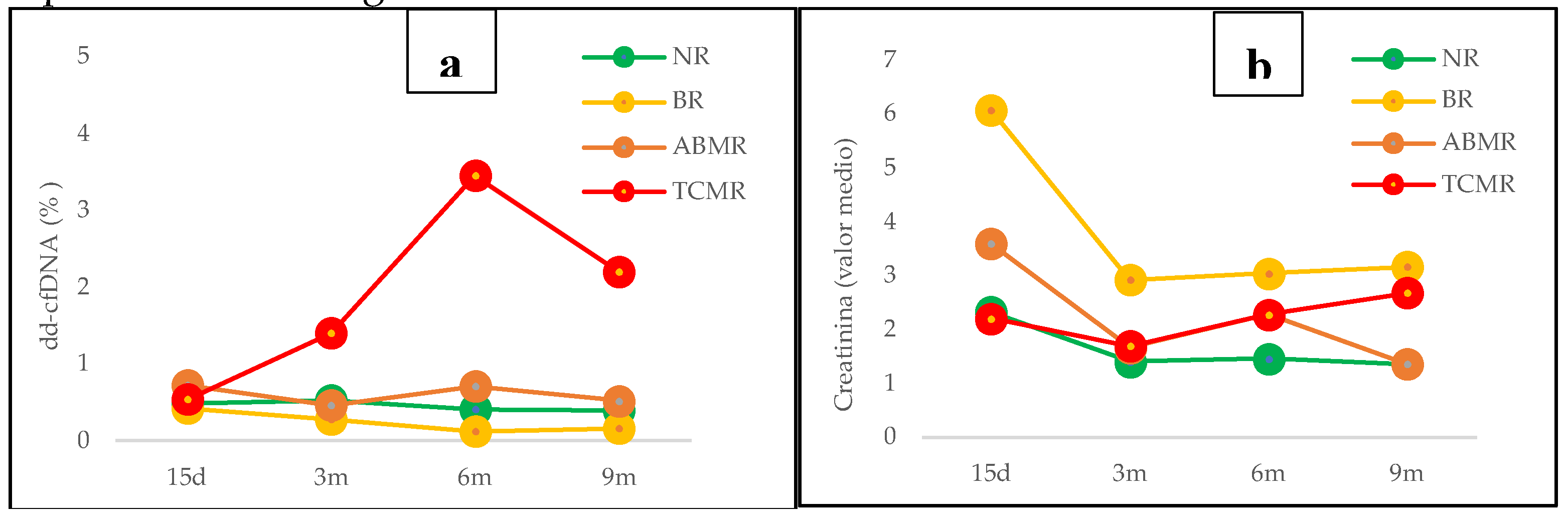
Patients were divided into groups based on their post-transplant clinical course and results for each group are shown in
Figure 1.
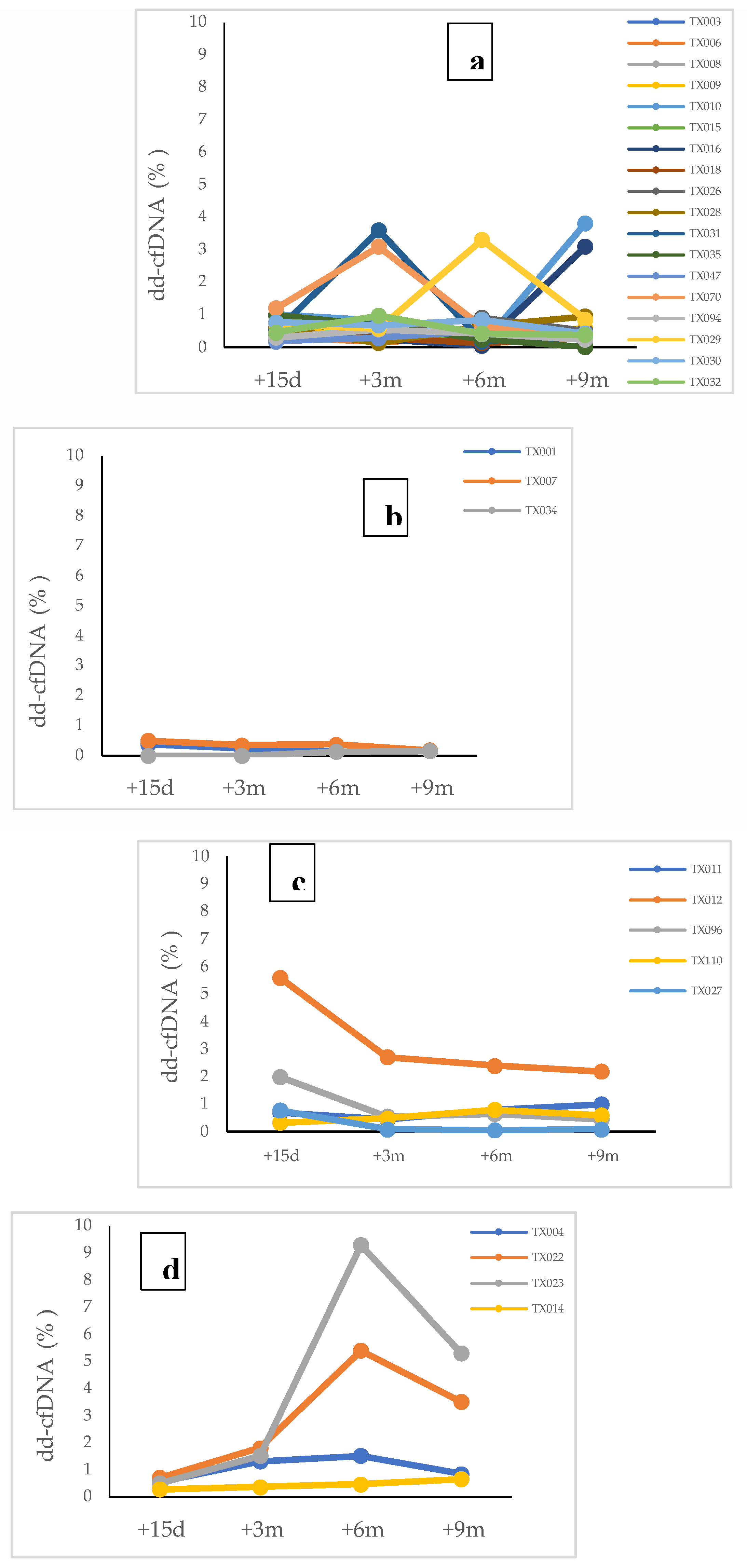
The detailed analysis of these results for each patient included in each group is shown in
Figure 2a–d.
Group 1 (N=18) included patients with no signs of graft rejection without a biopsy or with a negative biopsy, who correspond to Category 1 of Banff Classification (
Figure 2a)., showed means and medians of cfDNA percentage of dd-cfDNA≤1%, which represented a negative value of dd-cfDNA for kidney transplant, but only in 11 patients. The rest of them (n=7) exhibited dd-cfDNA levels >1%, which could be explained by other pathologies/biological processes. In this sense, the analysis of seven patients with elevated dd-cfDNA values with no apparent relation to immunological causes of rejection revealed that recurrent autoimmune pathologies in the post-transplant period and other pathologies or complications like diabetes mellitus, obesity, viral infections (CMV, EBV or HIV) and immunosuppressive drugs toxicity, may have an influence on dd-cfDNA levels, although the existence of statistically significant differences in the variables analyzed was not demonstrated due to the small sample size.
Group 2 (N=3) included patients who had borderline graft rejection (Category 3 of Banff Classification), all of them with levels of dd-cfDNA≤1% (
Figure 2b).
Group 3 (N=9) included patients with graft rejection both ABMR (Category 2 of Banff Classification) and TCRM (Category 4 of Banff Classification), and showed different profiles depending on the type of rejection.
In ABMR group (N=5), three patients showed values of dd-cfDNA>1% in post-transplants determinations corresponding with the date of rejection diagnosis by kidney biopsy or by clinical diagnosis with initiation of treatment for ABMR. In one patient, values of dd-cfDNA <1% were observed, but on the date of the rejection diagnosis showed an increased level of dd cfDNA=0,81%. The last patient in this group had an early ABMR rejection and the treatment was started before the first dd-cfDNA determination, so it was considered a false negative (
Figure 2c).
Respect to TCMR group (N=4), all patients except one showed elevated values (dd-cfDNA >0,5%) at 15 days post-transplant and positive values (dd-cfDNA>1%) for determinations made at 3, 6 and 9 moths during post-transplant period (
Figure 4d). Only one patient with a diagnosis of TCMR – IA by biopsy six months post-transplant showed negative values of dd-cfDNA. Moreover, patients with a diagnosis of rejection (ABMR or TCRM) after 3 months (66%) showed dd-cfDNA levels >1%, anticipating at least 1 month the graft rejection diagnosis.
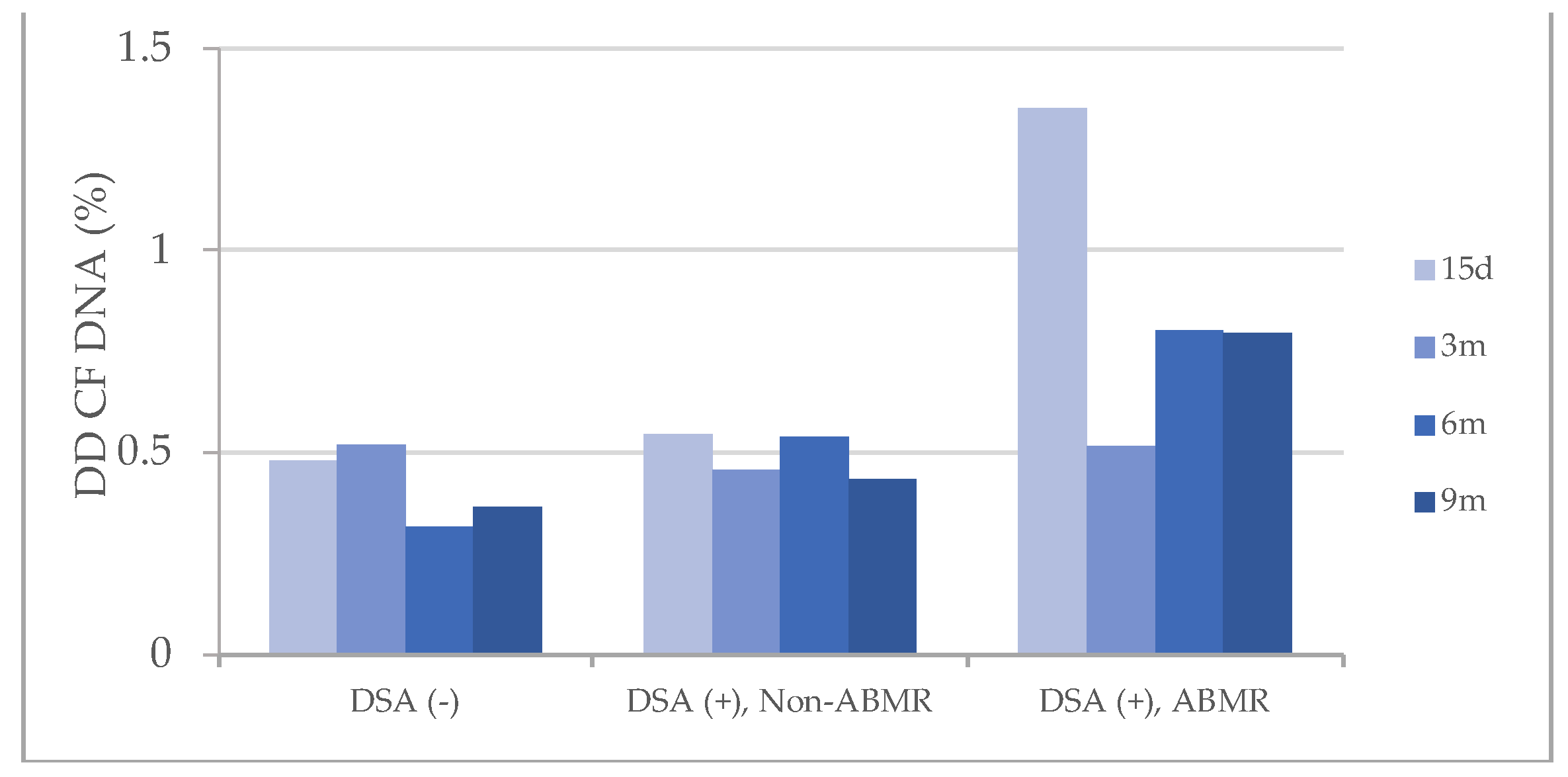
These results of dd-cfDNA were also compared with the presence of donor specific antibodies (DSAs) in transplant patient against HLA molecules from kidney graft (
Figure 3), and similar profiles were observed with dd-cfDNA levels that barely reached 0.5% in the groups of patients who did not present DSAs and in those who had DSAs but without ABMR. However, patients with ABMR (DSAs+) showed elevated dd-cfDNA levels>0.5% in all post-transplant determinations.
Finally, the results of the diagnostic validity test are shown in
Table 2 and
Figure 4. Regarding rejection in general, sensitivity for the technique of 50% was observed in our study, in accordance with the data that has been reported in the studies carried out to date. However, a specificity value of 61% was lower than those reported in the literature reviews. Accordingly, PPV and NPV were also low. However, when these parameters were analyzed for each type of rejection, an improvement in NPV was observed, especially in TCMR.
Figure 4.
Frecuencies of negative and positive pacientes for dd-cfDNA leves in each group. NR: Non Rejection, BR: Borderline Rejection, ABMR: Antibody Mediated Rejection, TCMR: T Cell Mediated Rejection.
Figure 4.
Frecuencies of negative and positive pacientes for dd-cfDNA leves in each group. NR: Non Rejection, BR: Borderline Rejection, ABMR: Antibody Mediated Rejection, TCMR: T Cell Mediated Rejection.
4. Discussion
In the present study, we have observed in 30 patients with a kidney transplantation that shows levels of dd-cfDNA>1% in 6 of 9 of them with active rejection (ABMR or TCMR) and elevated values (>0,5%) in other 2 patients into this group. Taking into account the main studies and clinical trials published to date [
5,
11,
12,
13,
14,
15,
16], our results are consistent with regard to patients with ABMR, where 60% of patients presented positive values coinciding with the diagnosis by biopsy and with the detection of DSA.
However, a finding that differentiates our study from other describing dd-cfDNA as a better biomarker to predict ABMR than TCMR, was the behavior of dd-cfDNA levels observed in 3 of the 4 patients diagnosed with TCMR, since they showed values above 1% from the third post-transplant month and were maintained throughout the follow-up period, regardless of the date of diagnosis of cell rejection (+4 months, +12 months and +8 months, respectively), anticipating diagnosis by 1 to 3 months. In fact, these patients are the ones who show the highest levels of dd-cfDNA in our series. In addition, in these 3 patients, the dd-cfDNA determination performed at 15 post-transplant days was greater than 0.5%, which was considered as elevation. The only patient in this group who had levels below 1% was diagnosed with TCMR-IA, and this result is consistent with other studies, where they obtain values similar and even more similar to those obtained for patients with borderline rejection, and in this sense, it appears to have no positive predictive value for borderline rejection or TCMR-IA.
Respect to low plasma levels in patient with no rejection signs, most important studies about the predictive value of dd-cfDNA of kidney graft rejection like DART (Diagnosis Acute Rejection in Kidney Transplant) [
15] report that the high NPV of dd-cfDNA is the main factor to consider as a biomarker with an essential role in avoiding unnecessary biopsies. However, our results show a low NPV of the technique because other pathologies or biological post-transplant complications can also affect dd-cfDNA levels and must necessarily be considered in the interpretation of the obtained result. Specifically, we found 7 patients with no signs of rejection with dd-cfDNA values>1%, among which the following were observed: 2 with obesity, 3 with recurrent autoimmune pathologies and 2 suffering infections.
On the other hand, some studies [
5,
11,
12,
13,
14,
15,
16,
19], in addition to relating dd-cfDNA levels with biopsy results and verifying that this biomarker turned out to be more effective than creatinine in defining clinical and subclinical rejection used in routine clinical routine, have reported an association of dd-cfDNA values>0.5% with an increase in developing DSA antibodies.
Nevertheless, dd-cfDNA, could allow together with other biomarkers in the context of a future score, avoid invasive biopsies in most cases, mainly in the case of suspected TCMR or ABMR.
This present article has several interesting limitations that should be indicated and appointed. Further addittional research, including a more extensive cohort and more thorough follow-up, will be required to validate our preliminary results and evaluate the connection between dd-cfDNA, allograft rejection and kidney transplant outcome in future studies. Another eventual limitation of our study is the low number of rejection episodes in our recipients, although it should be taken into account the shorter follow-up period of this study. Finally and in conclusion, increasing the number of analyzed kidney transplants is strictly necessary to corroborate and validate our obtained results and also more extended follow-up periods are also required.
5. Conclusions
Plasma levels of dd-cfDNA could be considered a novel biomarker of graft rejection after first term post-transplant up to several months before its clinical presentation, mainly in TCMR or ABMR. In the present study, it appears to have no positive predictive value for borderline rejection or TCMR IA. Our results show a low negative predictive value of the technique because other pathologies or biological post-transplant complications can also affect dd-cfDNA levels and they must necessarily be considered in the interpretation of the obtained results. However, dd-cfDNA, could allow together with other biomarkers in the context of a future score, avoid invasive biopsies in most cases, mainly in the case of suspected TCMR or ABMR.
Author Contributions
Conceptualization, Botella, C. and Muro, M.; methodology, Jiménez-Coll, V. and Fernández, M.; software, Botella, C.; Jiménez-Coll, V; validation, Fernández, M., Moya-Quiles, MR.; Minguela, A.; Botella, C and Muro, M; formal analysis, Botella, C; Fernández, M.; investigation, Botella, C., Jiménez-Coll, V., Fernández, M.; Galian, J.A.; Morales, F.; Martinez-Gomez, G.; Moya-Quiles, MR.; Gonzalez, R; Alegría, M.J.; resources, Jiménez-Coll, V., Fernandez, M.; Minguela, A.; Alegría, M.J ; data curation, Botella, C., Jimenez-Coll, V., Fernández, M. ; writing—original draft preparation, Botella, C.; writing—review and editing, Muro, M.; visualization, Legaz, I.; Llorente, S.; supervision, Muro, M.; project administration, Muro, M.; funding acquisition, Muro, M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Virgen de la Arrixaca Universitary Clinic Hospital and Biomedical Research Institute of Murcia (IMIB) (Murcia, Spain) within the framework of the PI19/01194 National Research Project funded by the Carlos III Health Institute (Madrid, Spain) and the AP183152023 Project funded by the Madrid Mutual Foundation.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board ( Ethics Committee) of Biomedical Research Institute of Murcia (IMIB) (Murcia, Spain) for studies involving humans.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study and written informed consent has been obtained from the patient(s) to publish this paper.
Data Availability Statement
Data is unavailable due to privacy or ethical restrictions, a statement is still required.
Acknowledgments
We want to acknowledge the support given for all technical and administrative personal of the Immunology Service at HCUVA.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Jimenez-Coll V, Llorente S, Boix F, Alfaro R, Galián JA, Martinez-Banaclocha H, et al. Monitoring of Serological, Cellular and Genomic Biomarkers in Transplantation, Computational Prediction Models and Role of Cell-Free DNA in Transplant Outcome. Int J Mol Sci. 2023;24(4):3908. [CrossRef]
- O’Callaghan JM, Knight SR. Noninvasive biomarkers in monitoring kidney allograft health. Curr Opin Organ Transplant. 2019;24(4):411–5. [CrossRef]
- Schwarz A, Gwinner W, Hiss M, Radermacher J, Mengel M, Haller H. Safety and Adequacy of Renal Transplant Protocol Biopsies. American Journal of Transplantation. 2005;5(8):1992–6. [CrossRef]
- Rush DN, Gibson IW. Subclinical Inflammation in Renal Transplantation. Transplantation. 2019;103(6):e139–45. [CrossRef]
- Jiménez-Coll V, El kaaoui El band J, Llorente S, González-López R, Fernández-González M, Martínez-Banaclocha H, et al. All That Glitters in cfDNA Analysis Is Not Gold or Its Utility Is Completely Established Due to Graft Damage: A Critical Review in the Field of Transplantation. Diagnostics. 2023;13(12):1982. [CrossRef]
- Haas M, Loupy A, Lefaucheur C, Roufosse C, Glotz D, Seron D, et al. The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell–mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. American Journal of Transplantation. 2018;18(2):293–307. [CrossRef]
- Marsh CL, Kurian SM, Rice JC, Whisenant TC, David J, Rose S, et al. Application of TruGraf v1: A Novel Molecular Biomarker for Managing Kidney Transplant Recipients With Stable Renal Function. Transplant Proc. 2019;51(3):722–8. [CrossRef]
- Van Loon E, Giral M, Anglicheau D, Lerut E, Dubois V, Rabeyrin M, et al. Diagnostic performance of kSORT, a blood-based mRNA assay for noninvasive detection of rejection after kidney transplantation: A retrospective multicenter cohort study. American Journal of Transplantation. 2021;21(2):740–50. [CrossRef]
- Bontha SV, Maluf DG, Mueller TF, Mas VR. Systems Biology in Kidney Transplantation: The Application of Multi-Omics to a Complex Model. American Journal of Transplantation. 2017;17(1):11–21. [CrossRef]
- Udomkarnjananun S, Kerr SJ, Townamchai N, van Besouw NM, Hesselink DA, Baan CC. Donor-specific ELISPOT assay for predicting acute rejection and allograft function after kidney transplantation: A systematic review and meta-analysis. Clin Biochem. 2021;94:1–11. [CrossRef]
- Abdulhadi T, Alrata L, Dubrawka C, Amurao G, Kalipatnapu SM, Isaac C, et al. Donor-derived cell free DNA as a biomarker in kidney transplantation. Pharmacogenomics. 2023; [CrossRef]
- Kataria A, Kumar D, Gupta G. Donor-derived Cell-free DNA in Solid-organ Transplant Diagnostics: Indications, Limitations, and Future Directions. Transplantation. 2021;105(6):1203–11. [CrossRef]
- Martuszewski A, Paluszkiewicz P, Król M, Banasik M, Kepinska M. Donor-Derived Cell-Free DNA in Kidney Transplantation as a Potential Rejection Biomarker: A Systematic Literature Review. J Clin Med. 2021;10(2):193. [CrossRef]
- Kueht ML, Dongur LP, Cusick M, Stevenson HL, Mujtaba M. The Current State of Donor-Derived Cell-Free DNA Use in Allograft Monitoring in Kidney Transplantation. J Pers Med. 2022;12(10):1700. [CrossRef]
- Bloom RD, Bromberg JS, Poggio ED, Bunnapradist S, Langone AJ, Sood P, et al. Cell-Free DNA and Active Rejection in Kidney Allografts. Journal of the American Society of Nephrology. 2017;28(7):2221–32. [CrossRef]
- Huang E, Jordan SC. Donor-derived cell-free DNA in kidney transplantation: evolving concepts and potential limitations. Kidney Int. 2022;101(4):676–7. [CrossRef]
- Loupy A, Mengel M, Haas M. Thirty years of the International Banff Classification for Allograft Pathology: the past, present, and future of kidney transplant diagnostics. Vol. 101, Kidney International Elsevier B.V.; 2022. p. 678–91.
- Callemeyn J, Lamarthée B, Koenig A, Koshy P, Thaunat O, Naesens M. Allorecognition and the spectrum of kidney transplant rejection. Vol. 101, Kidney International Elsevier B.V.; 2022. p. 692–710.
- Bu, L.; Gupta, G.; Pai, A.; Anand, S.; Stites, E.; Moinuddin, I.; Bowers, V.; Jain, P.; Axelrod, D.A.; Weir, M.R.; et al. Clinical outcomes from the Assessing Donor-derived cell-free DNA Monitoring Insights of kidney Allografts with Longitudinal surveillance (ADMIRAL) study. Kidney Int. 2022, 101, 793–803. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).