Submitted:
04 July 2024
Posted:
09 July 2024
You are already at the latest version
Abstract
Keywords:
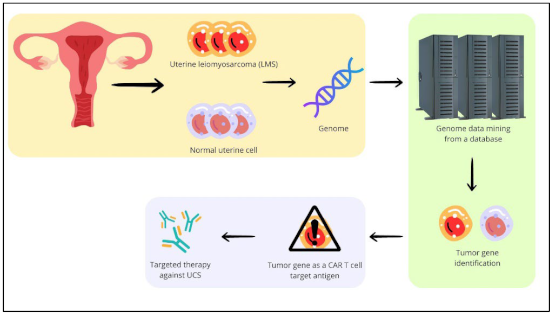
1. Introduction
2. Results
2.1. Topology of the GSE Samples Used
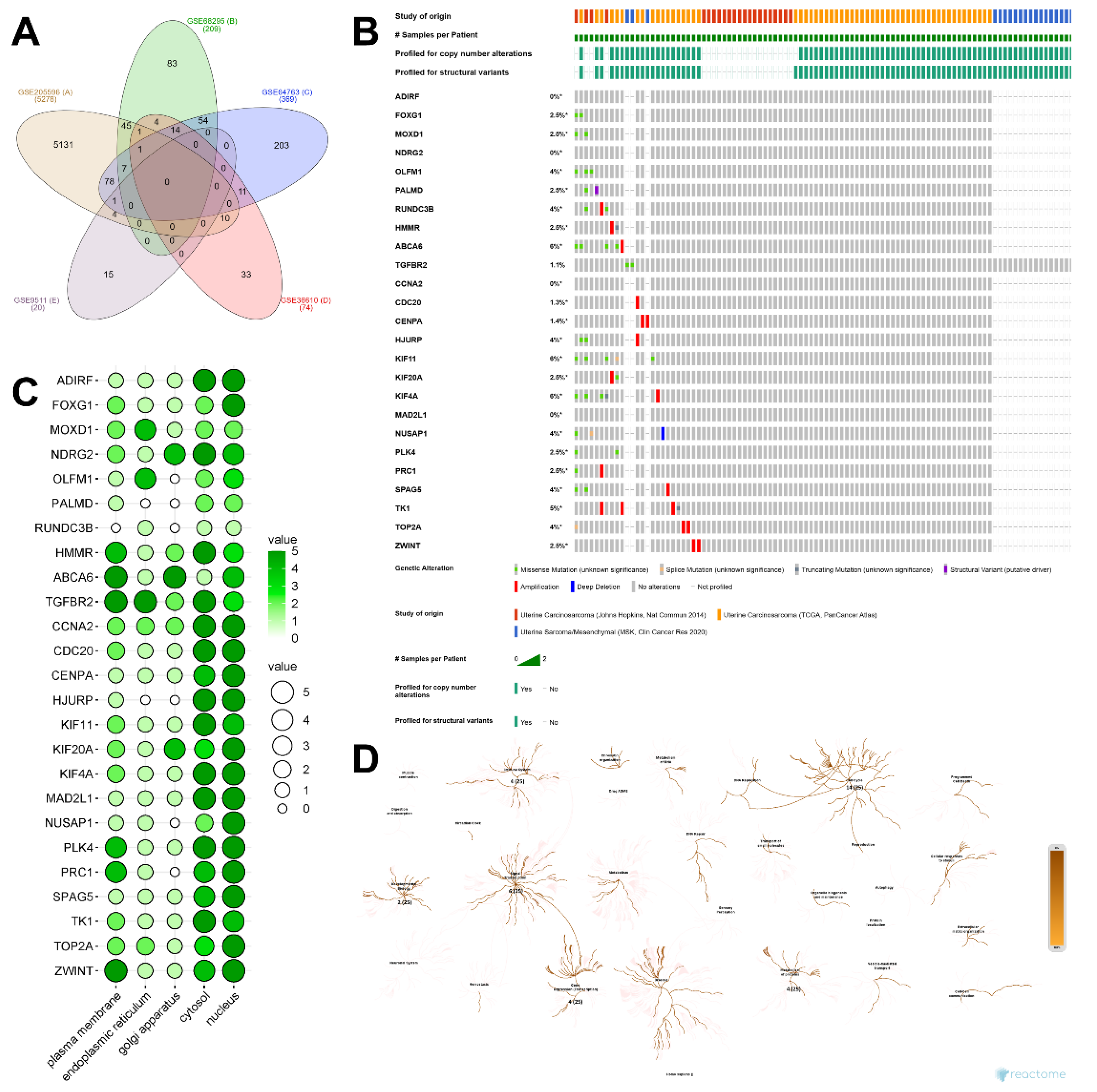
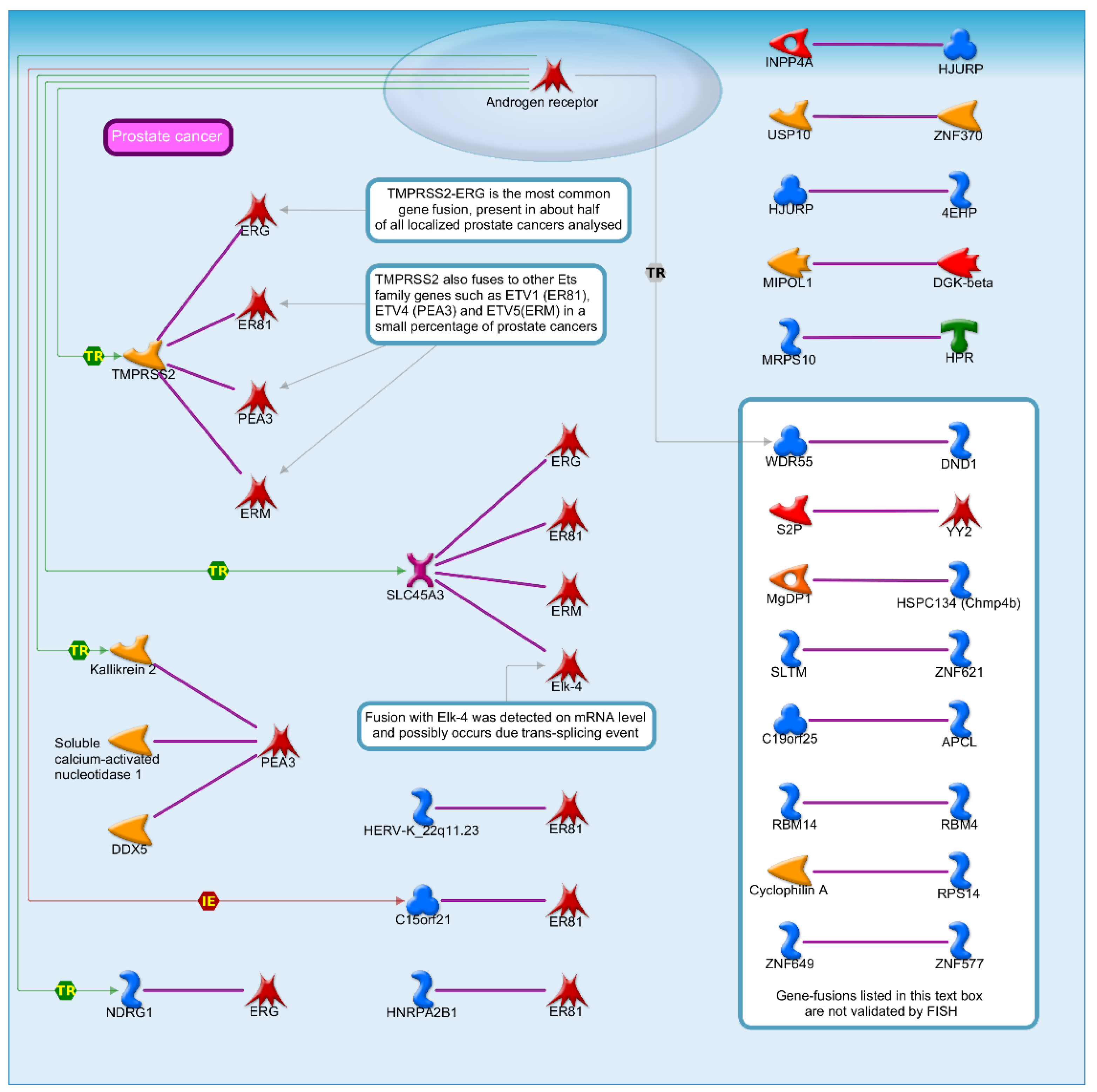
2.2. Identification of DEGs and Their Discovery in Cancer Cases
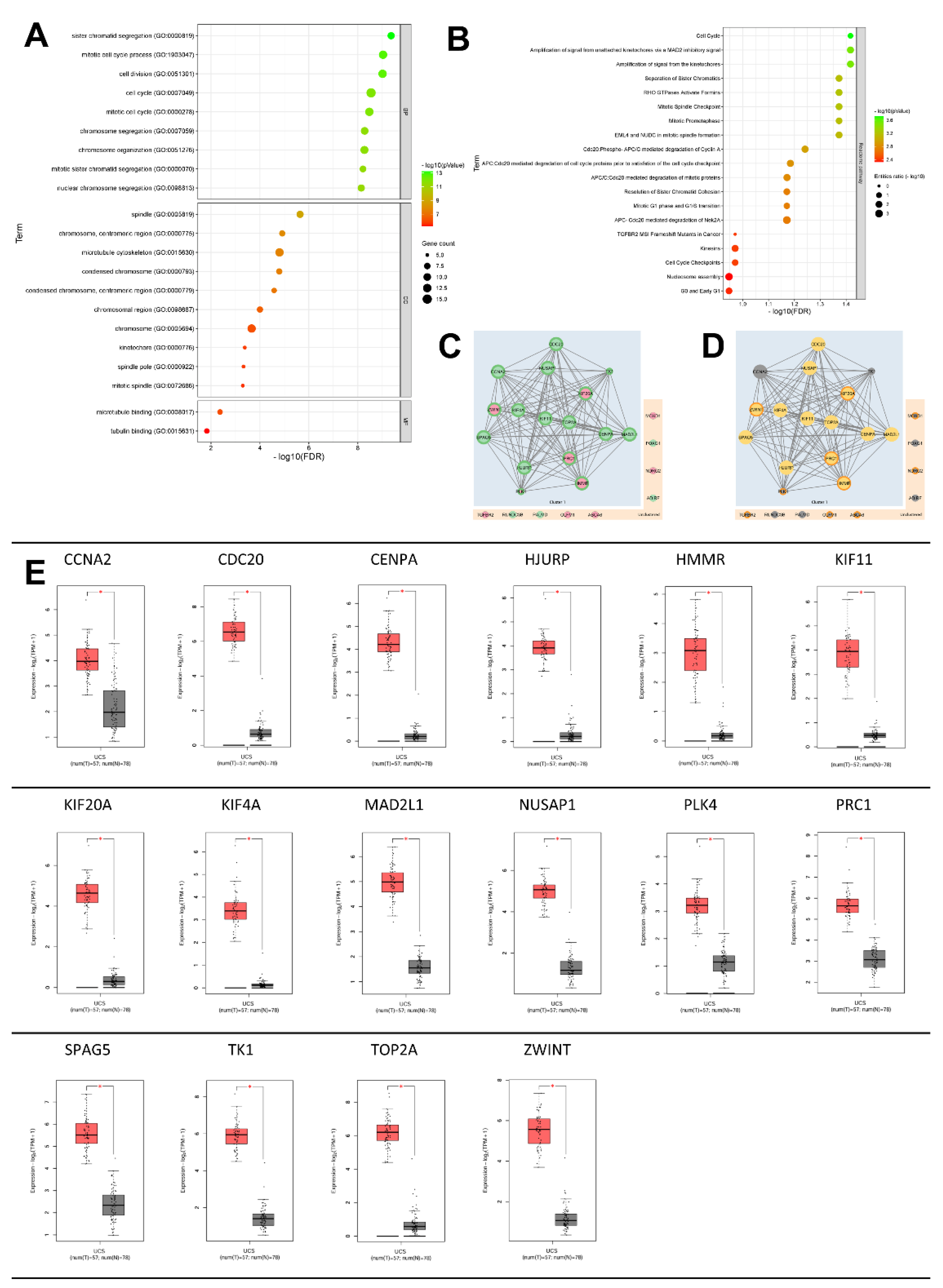
2.3. GeneOntology of DEGs and their Related Biological Process
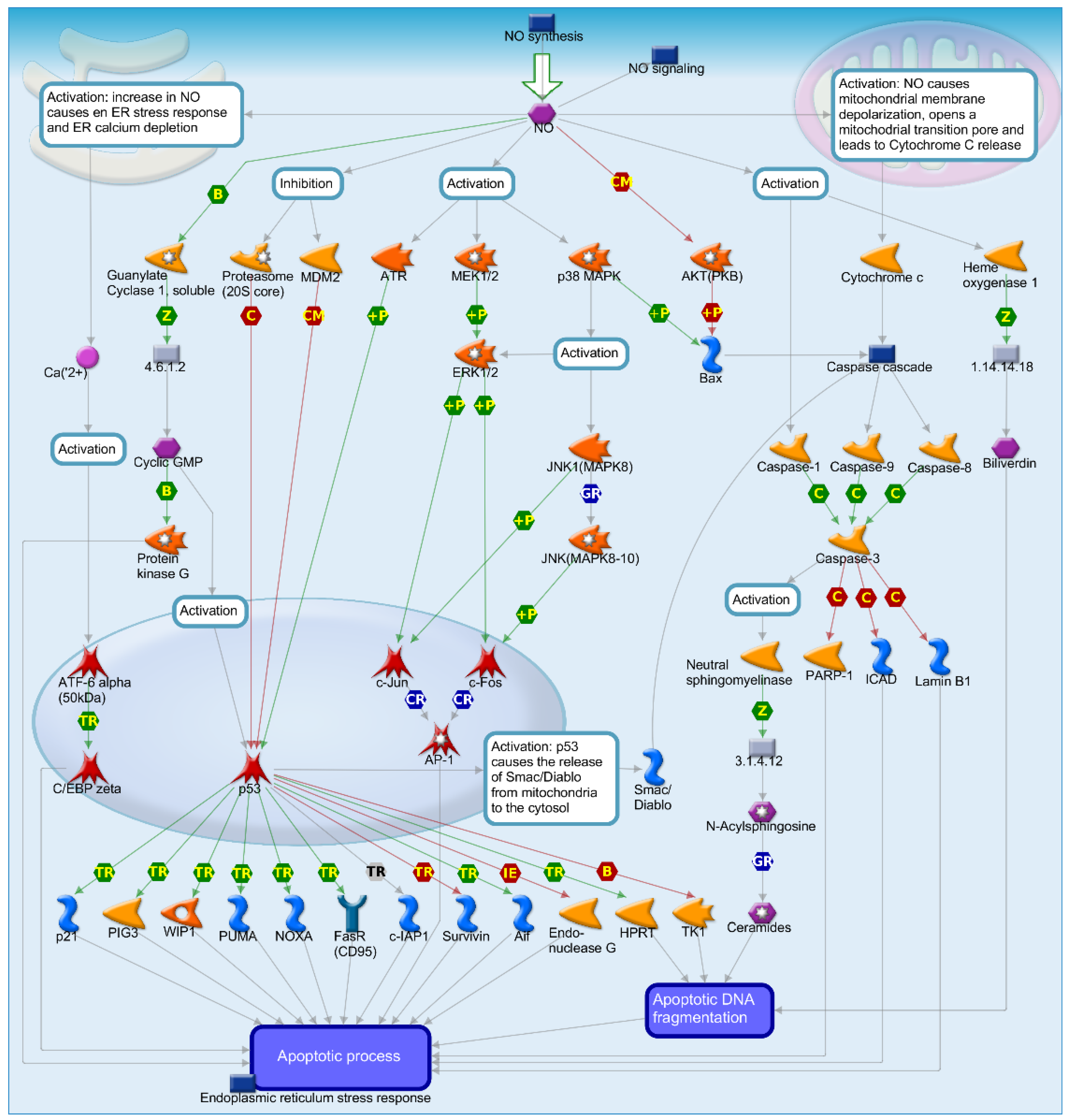
2.4. Mechanisms of Several DEGs Related to Cell Cycle Based on the MetaCore Database
3. Discussion
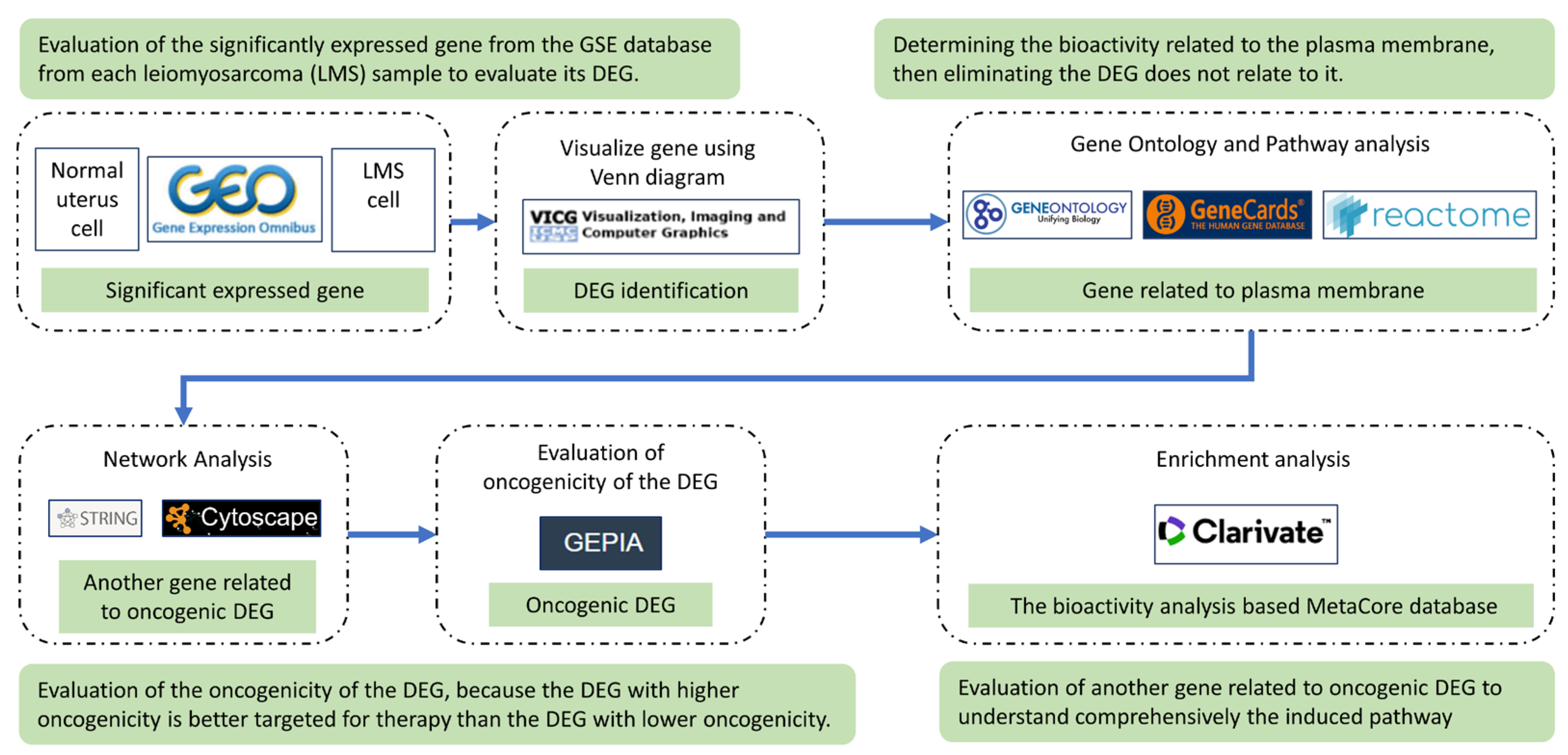
4. Material and Method
4.1. Data Sources
4.2. Identification of Significant Expressed Genes and Their Protein Localization
4.3. Construction of Protein Network from the Significant Expressed Gene
4.4. Oncogenicity Prediction Analysis
4.6. Evaluation of the DEGs Pathway and Biological Process by Metacore
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Byar, K.L.; Fredericks, T. Uterine Leiomyosarcoma. J Adv Pract Oncol 2022, 13, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Delisle, M.; Alshamsan, B.; Nagaratnam, K.; Smith, D.; Wang, Y.; Srikanthan, A. Metastasectomy in Leiomyosarcoma: A Systematic Review and Pooled Survival Analysis. Cancers 2022, 14, 3055. [Google Scholar] [CrossRef] [PubMed]
- Lacuna, K.; Bose, S.; Ingham, M.; Schwartz, G. Therapeutic Advances in Leiomyosarcoma. Front. Oncol. 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Asano, H.; Isoe, T.; Ito, Y.M.; Nishimoto, N.; Watanabe, Y.; Yokoshiki, S.; Watari, H. Status of the Current Treatment Options and Potential Future Targets in Uterine Leiomyosarcoma: A Review. Cancers 2022, 14, 1180. [Google Scholar] [CrossRef] [PubMed]
- Kyriazoglou, A.; Liontos, M.; Ntanasis-Stathopoulos, I.; Gavriatopoulou, M. The Systemic Treatment of Uterine Leiomyosarcomas: A Systematic Review. No News Is Good News? Medicine 2021, 100, e25309. [Google Scholar] [CrossRef] [PubMed]
- Kostine, M.; Briaire-de Bruijn, I.H.; Cleven, A.H.G.; Vervat, C.; Corver, W.E.; Schilham, M.W.; Van Beelen, E.; van Boven, H.; Haas, R.L.; Italiano, A.; et al. Increased Infiltration of M2-Macrophages, T-Cells and PD-L1 Expression in High Grade Leiomyosarcomas Supports Immunotherapeutic Strategies. OncoImmunology 2018, 7, e1386828. [Google Scholar] [CrossRef] [PubMed]
- Feins, S.; Kong, W.; Williams, E.F.; Milone, M.C.; Fraietta, J.A. An Introduction to Chimeric Antigen Receptor (CAR) T-Cell Immunotherapy for Human Cancer. American Journal of Hematology 2019, 94, S3–S9. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, J.; Ruella, M.; Houot, R. Overcoming Intrinsic Resistance of Cancer Cells to CAR T-Cell Killing. Clinical Cancer Research 2021, 27, 6298–6306. [Google Scholar] [CrossRef] [PubMed]
- Marofi, F.; Motavalli, R.; Safonov, V.A.; Thangavelu, L.; Yumashev, A.V.; Alexander, M.; Shomali, N.; Chartrand, M.S.; Pathak, Y.; Jarahian, M.; et al. CAR T Cells in Solid Tumors: Challenges and Opportunities. Stem Cell Research & Therapy 2021, 12, 81. [Google Scholar] [CrossRef]
- Iuliano, L.; Dalla, E.; Picco, R.; Mallarapu, S.; Brancolini, C. Proteotoxic Stress-Induced Apoptosis in Cancer Cells: Understanding the Susceptibility and Enhancing the Potency 2022.
- Miyata, T.; Nakabayashi, K. Genomic, Epigenomic, and Transcriptomic Profiling towards Identifying Omics-Features and Specific Biomarkers That Distinguish Uterine Leiomyosarcoma and Leiomyoma at Molecular Levels (Expression) 2017.
- Levine, D.; Boyd, J. Expression Data from Normal Myometrium, Leiomyomata, and Leiomyosarcomas 2015.
- Matthew, A.; Creighton, C.; Weiwei, S. Gene Expression Profiles of Uterine Leiomyosarcoma (ULMS) 2012.
- Quade, B.; Morton, C.; Hodge, J. Characterization of Plexiform Leiomyomata Provide Further Evidence for Genetic Heterogeneity Underlying Uterine Fibroids 2008.
- Genta, S.; Bruce, J.; Li, X.; Felicen, S.; Abdul Razak, A.R.; Saibil, S.; Butler, M.O.; Bedard, P.L.; Lam, B.; Tsao, M.S.; et al. Deciphering Primary and Acquired Immunotherapy Resistance with Whole Genome and Transcriptome Analysis (WGTA). JCO 2023, 41, 2602–2602. [Google Scholar] [CrossRef]
- Bruno, L.P.; Doddato, G.; Valentino, F.; Baldassarri, M.; Tita, R.; Fallerini, C.; Bruttini, M.; Lo Rizzo, C.; Mencarelli, M.A.; Mari, F.; et al. New Candidates for Autism/Intellectual Disability Identified by Whole-Exome Sequencing. International Journal of Molecular Sciences 2021, 22, 13439. [Google Scholar] [CrossRef]
- Khan, A.; Bruno, L.P.; Alomar, F.; Umair, M.; Pinto, A.M.; Khan, A.A.; Khan, A.; Saima; Fabbiani, A. ; Zguro, K.; et al. SPTBN5, Encoding the βV-Spectrin Protein, Leads to a Syndrome of Intellectual Disability, Developmental Delay, and Seizures. Front. Mol. Neurosci. 2022, 15. [Google Scholar] [CrossRef]
- Nogrady, B. How Cancer Genomics Is Transforming Diagnosis and Treatment. Nature 2020, 579, S10–S11. [Google Scholar] [CrossRef] [PubMed]
- Paudel, P.; Dhungana, B.; Shrestha, E.; Verma, D.; Paudel, P.; Dhungana, B.; Shrestha, E.; Verma, D. Leiomyosarcoma of the Uterus: A Rare Diagnosis. Cureus 2021, 13. [Google Scholar] [CrossRef]
- Juhasz-Böss, I.; Gabriel, L.; Bohle, R.M.; Horn, L.C.; Solomayer, E.-F.; Breitbach, G.-P. Uterine Leiomyosarcoma. Oncology Research and Treatment 2018, 41, 680–686. [Google Scholar] [CrossRef]
- Kanayama, S.; Oi, H.; Kawaguchi, R.; Furukawa, N.; Kobayashi, H. Immunohistochemical Analysis of P16 Expression in Uterine Smooth Muscle Tumors. Open Journal of Obstetrics and Gynecology 2015, 5, 688–697. [Google Scholar] [CrossRef]
- Sterner, R.C.; Sterner, R.M. CAR-T Cell Therapy: Current Limitations and Potential Strategies. Blood Cancer J. 2021, 11, 1–11. [Google Scholar] [CrossRef]
- Sadelain, M.; Brentjens, R.; Riviere, I. The Basic Principles of Chimeric Antigen Receptor (CAR) Design. Cancer Discov 2013, 3, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Han, X.; Bo, J.; Han, W. Target Selection for CAR-T Therapy. Journal of Hematology & Oncology 2019, 12, 62. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discovery 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Ravi, S.; Alencar, A.M.; Arakelyan, J.; Xu, W.; Stauber, R.; Wang, C.-C.I.; Papyan, R.; Ghazaryan, N.; Pereira, R.M. An Update to Hallmarks of Cancer. Cureus 2022, 14, e24803. [Google Scholar] [CrossRef]
- He, Z.; Mei, L.; Connell, M.; Maxwell, C.A. Hyaluronan Mediated Motility Receptor (HMMR) Encodes an Evolutionarily Conserved Homeostasis, Mitosis, and Meiosis Regulator Rather than a Hyaluronan Receptor. Cells 2020, 9, 819. [Google Scholar] [CrossRef]
- Zhang, H.; Ren, L.; Ding, Y.; Li, F.; Chen, X.; Ouyang, Y.; Zhang, Y.; Zhang, D. Hyaluronan-Mediated Motility Receptor Confers Resistance to Chemotherapy via TGFβ/Smad2-Induced Epithelial-Mesenchymal Transition in Gastric Cancer. The FASEB Journal 2019, 33, 6365–6377. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, Z.; Song, K. AR-mTOR-SRF Axis Regulates HMMR Expression in Human Prostate Cancer Cells. Biomol Ther (Seoul) 2021, 29, 667–677. [Google Scholar] [CrossRef]
- Li, W.; Wang, R.; Wang, W. Exploring the Causality and Pathogenesis of Systemic Lupus Erythematosus in Breast Cancer Based on Mendelian Randomization and Transcriptome Data Analyses. Front. Immunol. 2023, 13. [Google Scholar] [CrossRef]
- Tang, Y.; Yin, Y.; Xie, M.; Liang, X.; Li, J.; Li, K.; Hu, B. Systematic Analysis of the Clinical Significance of Hyaluronan-Mediated Motility Receptor in Colorectal Cancer. Front. Mol. Biosci. 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Chen, B.; Kong, L.; Chen, X.; Ouyang, Y.; Bai, J.; Yu, D.; Zhang, H.; Li, X.; Zhang, D. HMMR Promotes Peritoneal Implantation of Gastric Cancer by Increasing Cell–Cell Interactions. Discov Onc 2022, 13, 81. [Google Scholar] [CrossRef]
- Zhao, X.; Zhou, L.; Li, X.; Ni, J.; Chen, P.; Ma, R.; Wu, J.; Feng, J. Overexpression of KIF20A Confers Malignant Phenotype of Lung Adenocarcinoma by Promoting Cell Proliferation and Inhibiting Apoptosis. Cancer Medicine 2018, 7, 4678–4689. [Google Scholar] [CrossRef] [PubMed]
- Zhan, P.; Zhang, B.; Xi, G.; Wu, Y.; Liu, H.; Liu, Y.; Xu, W.; Zhu, Q.; Cai, F.; Zhou, Z.; et al. PRC1 Contributes to Tumorigenesis of Lung Adenocarcinoma in Association with the Wnt/β-Catenin Signaling Pathway. Molecular Cancer 2017, 16, 108. [Google Scholar] [CrossRef]
- Qiao, Y.; Pei, Y.; Luo, M.; Rajasekaran, M.; Hui, K.M.; Chen, J. Cytokinesis Regulators as Potential Diagnostic and Therapeutic Biomarkers for Human Hepatocellular Carcinoma. Exp Biol Med (Maywood) 2021, 246, 1343–1354. [Google Scholar] [CrossRef]
- Gluszek-Kustusz, A.; Craske, B.; Legal, T.; McHugh, T.; Welburn, J.P. Phosphorylation Controls Spatial and Temporal Activities of motor-PRC1 Complexes to Complete Mitosis. The EMBO Journal 2023, 42, e113647. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, J.; Xu, L.; Wei, W.; Cheng, A.; Zhang, L.; Zhang, M.; Wu, G.; Cai, C. CDK16 Promotes the Progression and Metastasis of Triple-Negative Breast Cancer by Phosphorylating PRC1. Journal of Experimental & Clinical Cancer Research 2022, 41, 149. [Google Scholar] [CrossRef]
- Bu, H.; Li, Y.; Jin, C.; Yu, H.; Wang, X.; Chen, J.; Wang, Y.; Ma, Y.; Zhang, Y.; Kong, B. Overexpression of PRC1 Indicates a Poor Prognosis in Ovarian Cancer. International Journal of Oncology 2020, 56, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Y.; Meng, L.; Liu, X.-Y.; Peng, A.; Chen, Y.; Liu, C.; Chen, H.; Sun, S.; Miao, X.; et al. Reducing Protein Regulator of Cytokinesis 1 as a Prospective Therapy for Hepatocellular Carcinoma. Cell Death Dis 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Xu, T.; Wang, X.; Jia, X.; Gao, W.; Li, J.; Gao, F.; Zhan, P.; Ji, W. Overexpression of Protein Regulator of Cytokinesis 1 Facilitates Tumor Growth and Indicates Unfavorable Prognosis of Patients with Colon Cancer. Cancer Cell International 2020, 20, 528. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Shi, X.; Zhang, R.; Tian, Y.; Wang, X.; Wei, C.; Li, D.; Li, X.; Kong, X.; Liu, Y.; et al. Regulation of Proliferation and Cell Cycle by Protein Regulator of Cytokinesis 1 in Oral Squamous Cell Carcinoma. Cell Death Dis 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Liu, Z.; Zhang, A.; Li, N. Identification of Key Genes and Pathways in Hepatocellular Carcinoma: A Preliminary Bioinformatics Analysis. Medicine 2019, 98, e14287. [Google Scholar] [CrossRef] [PubMed]
- Chau, C.H.; Steeg, P.S.; Figg, W.D. Antibody-Drug Conjugates for Cancer. Lancet 2019, 394, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Criscitiello, C.; Morganti, S.; Curigliano, G. Antibody–Drug Conjugates in Solid Tumors: A Look into Novel Targets. Journal of Hematology & Oncology 2021, 14, 20. [Google Scholar] [CrossRef]
- Tarantino, P.; Pestana, R.C.; Corti, C.; Modi, S.; Bardia, A.; Tolaney, S.M.; Cortes, J. Antibody–Drug Conjugates: Smart Chemotherapy Delivery across Tumor Histologies. CA CANCER J CLIN 2022, 72. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets—Update. Nucleic Acids Research 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wang, X.; Huang, G.; Li, R.; Liu, X.; Cao, L.; Ye, J.; Zhang, P. Bioinformatic Identification of Differentially Expressed Genes Associated with Hepatocellular Carcinoma Prognosis. Medicine 2022, 101, e30678. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, D.; Zhu, F.; Chen, F.; Zhu, Y.; Yu, R.; Fan, L. Disordered APC/C-Mediated Cell Cycle Progression and IGF1/PI3K/AKT Signalling Are the Potential Basis of Sertoli Cell-Only Syndrome. Andrologia 2019, 51, e13288. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Athar, M.; Manne, U.; Varambally, S. Comparative Transcriptome Analyses Reveal Genes Associated with SARS-CoV-2 Infection of Human Lung Epithelial Cells. Sci Rep 2021, 11, 16212. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Current Protocols in Bioinformatics 2016, 54, 1–30. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.-P.; Mi, H. PANTHER: Making Genome-Scale Phylogenetics Accessible to All. Protein Sci 2022, 31, 8–22. [Google Scholar] [CrossRef]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The Reactome Pathway Knowledgebase 2022. Nucleic Acids Research 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Forner, O.; Marin-Garcia, P.; Arnau, V.; D’Eustachio, P.; Stein, L.; Hermjakob, H. Reactome Pathway Analysis: A High-Performance in-Memory Approach. BMC Bioinformatics 2017, 18, 142. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.-E.; Lengauer, T.; Albrecht, M. Computing Topological Parameters of Biological Networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhong, J.; Chen, G.; Li, M.; Wu, F.; Pan, Y. ClusterViz: A Cytoscape APP for Cluster Analysis of Biological Network. IEEE/ACM Transactions on Computational Biology and Bioinformatics 2015, 12, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Research 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Cirillo, E.; Parnell, L.D.; Evelo, C.T. A Review of Pathway-Based Analysis Tools That Visualize Genetic Variants. Front Genet 2017, 8, 174. [Google Scholar] [CrossRef] [PubMed]
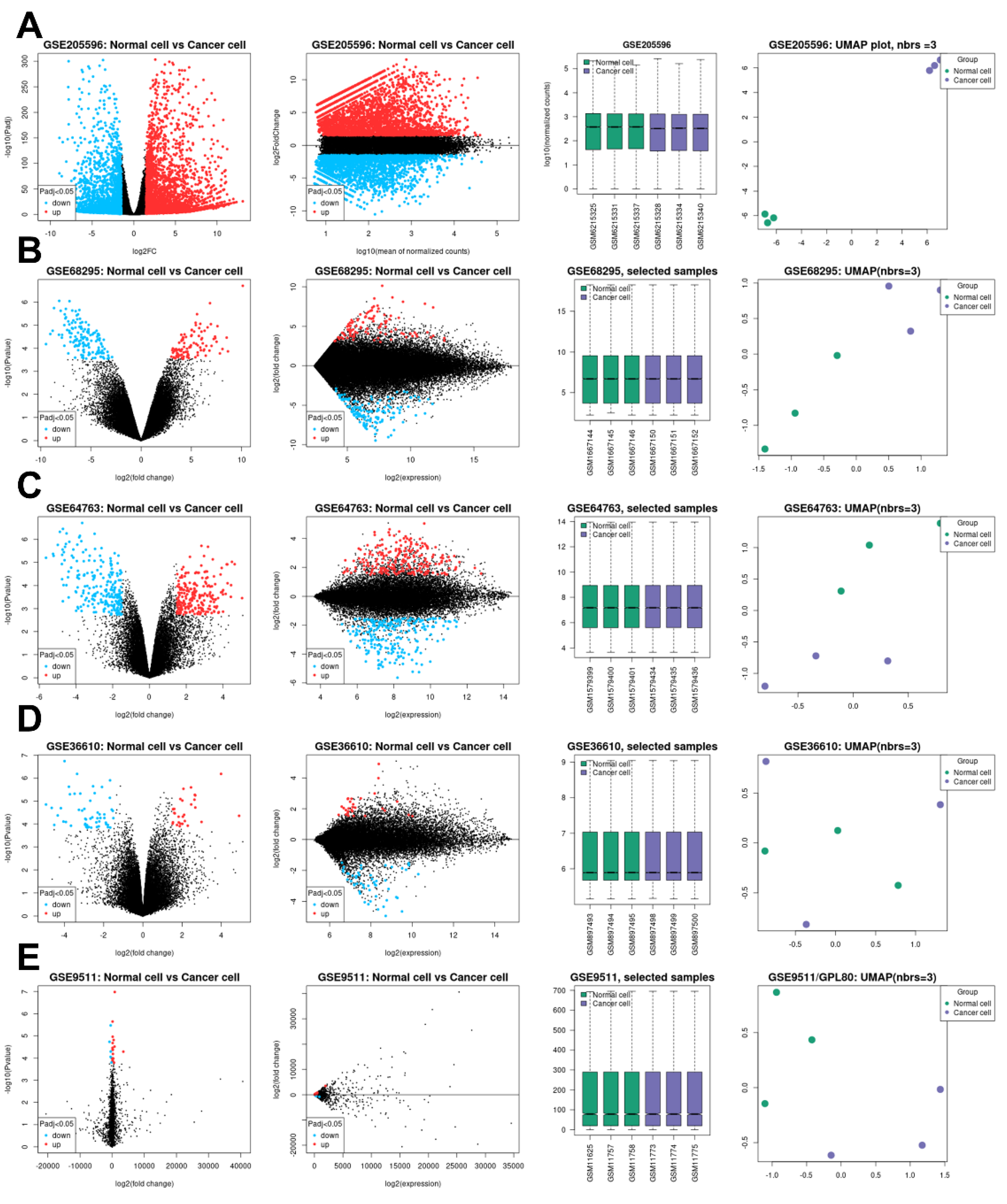
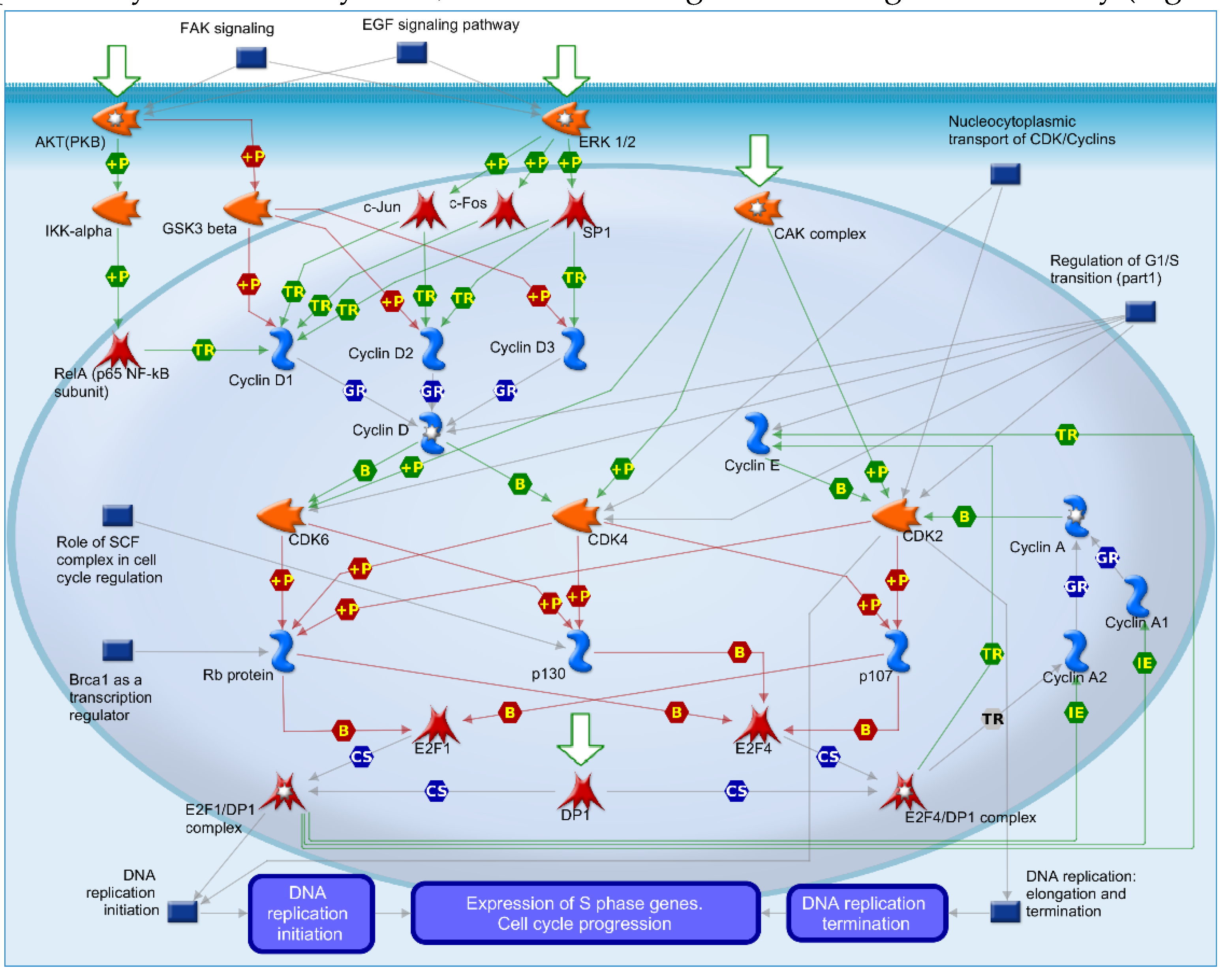
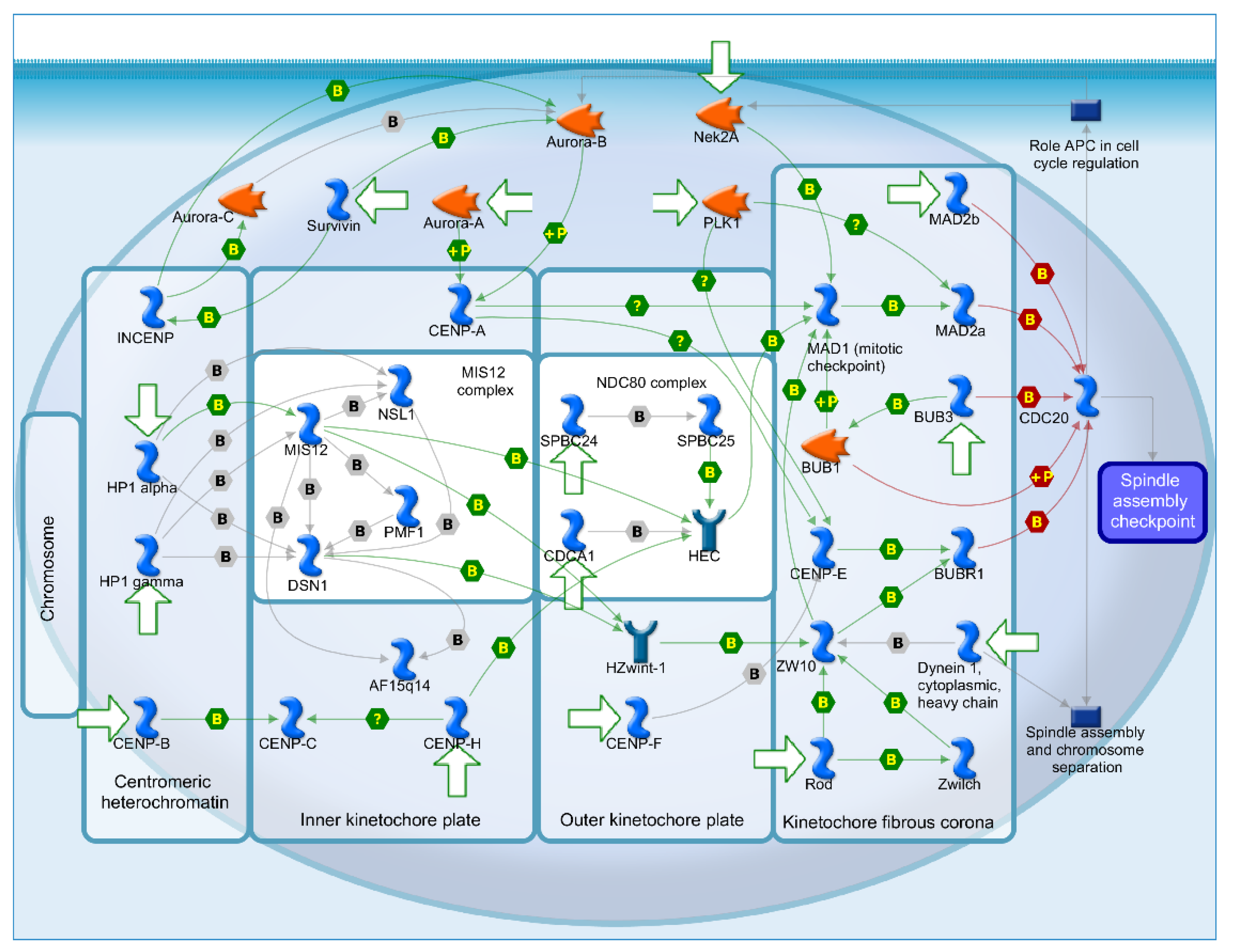
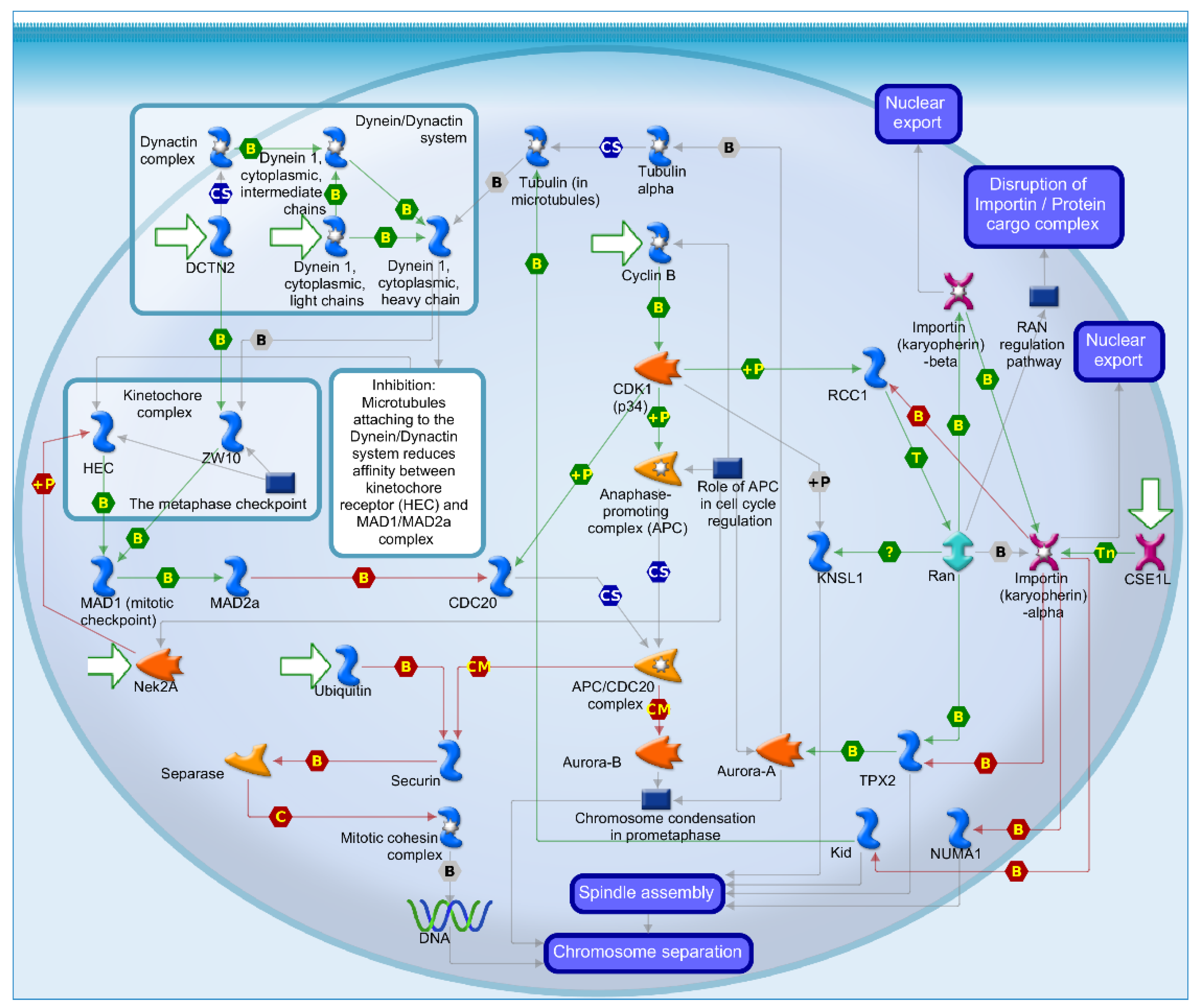
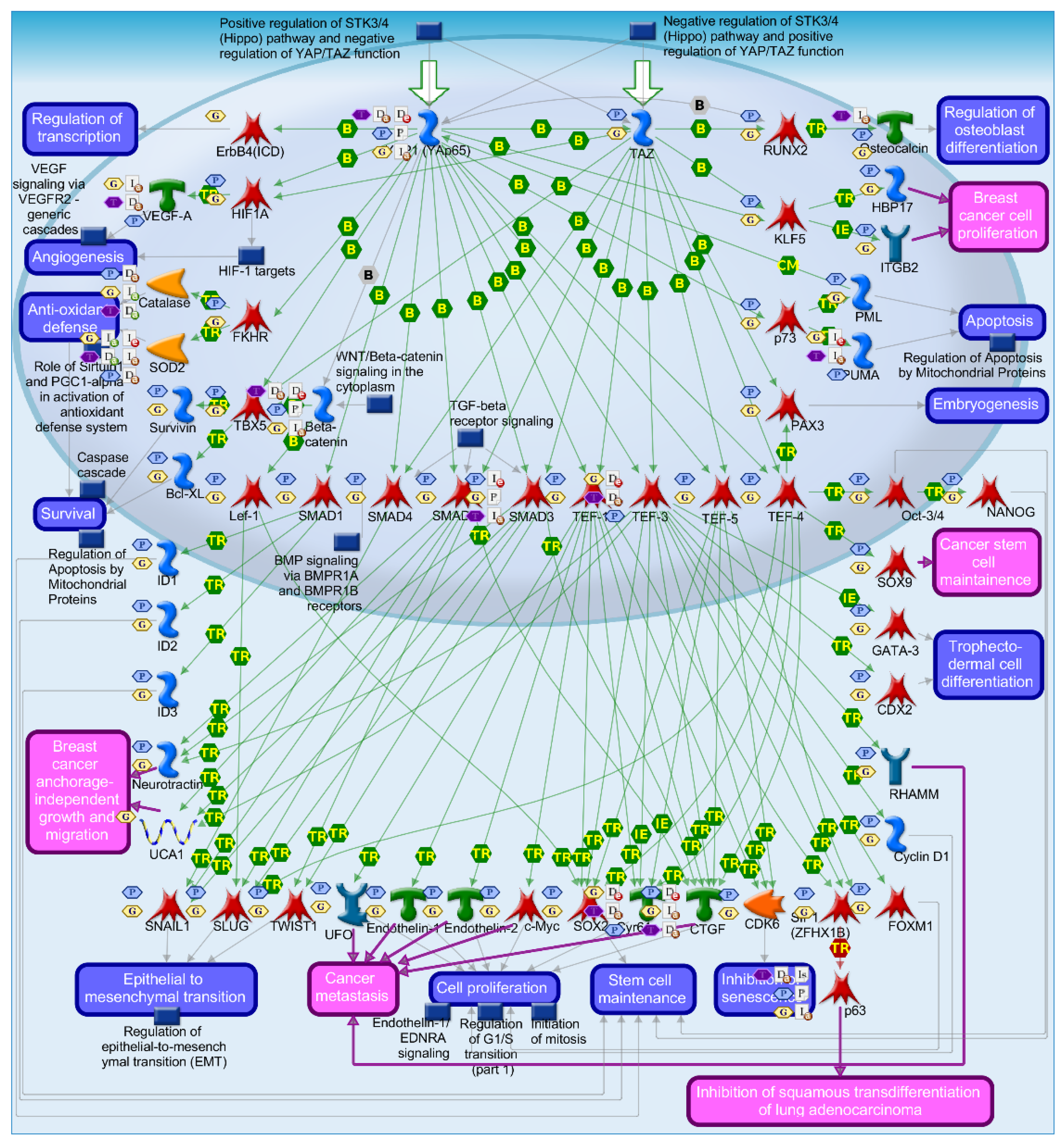
| GEO profile | Year | Cancer sample | Normal sample | Platform | Annotation platform |
|---|---|---|---|---|---|
| GSE205596 [10] | 2023 | Leiomyosarcoma (9) | Uterine smooth muscle (9) | GPL24676 | Illumina NovaSeq 6000 (Homo sapiens) |
| GSE68295 [11] | 2017 | Leiomyosarcoma (3) | Uterine myometrium (3) | GPL6480 | Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Probe Name version) |
| GSE64763 [12] | 2015 | Leiomyosarcoma (25) | Uterine myometrium (29) | GPL571 | [HG-U133A_2] Affymetrix Human Genome U133A 2.0 Array |
| GSE36610 [13] | 2012 | Leiomyosarcoma (12) | Uterine myometrium (10) | GPL7363 | Illumina HumanWG-6_V2_0_R2 |
| GSE9511 [14] | 2008 | Leiomyosarcoma (8) | Uterine myometrium (4) | GPL80 | [Hu6800] Affymetrix Human Full Length HuGeneFL Array |
| Protein name | Betweenness Centrality | Degree | Cluster |
|---|---|---|---|
| ABCA6 | 0.00E+00 | 0 | Unclustered |
| ADIRF | 0.00E+00 | 0 | Unclustered |
| CCNA2 | 6.80E-04 | 15 | Cluster 1 |
| CDC20 | 6.80E-04 | 15 | Cluster 1 |
| CENPA | 6.80E-04 | 15 | Cluster 1 |
| FOXG1 | 0.00E+00 | 0 | Unclustered |
| HJURP | 6.80E-04 | 15 | Cluster 1 |
| HMMR | 6.80E-04 | 15 | Cluster 1 |
| KIF11 | 6.80E-04 | 15 | Cluster 1 |
| KIF20A | 6.80E-04 | 15 | Cluster 1 |
| KIF4A | 6.80E-04 | 15 | Cluster 1 |
| MAD2L1 | 6.80E-04 | 15 | Cluster 1 |
| MOXD1 | 0.00E+00 | 0 | Unclustered |
| NDRG2 | 0.00E+00 | 0 | Unclustered |
| NUSAP1 | 6.80E-04 | 15 | Cluster 1 |
| OLFM1 | 0.00E+00 | 0 | Unclustered |
| PALMD | 0.00E+00 | 0 | Unclustered |
| PLK4 | 0.00E+00 | 14 | Cluster 1 |
| PRC1 | 6.80E-04 | 15 | Cluster 1 |
| RUNDC3B | 0.00E+00 | 0 | Unclustered |
| SPAG5 | 6.80E-04 | 15 | Cluster 1 |
| TGFBR2 | 0.00E+00 | 0 | Unclustered |
| TK1 | 0.00E+00 | 14 | Cluster 1 |
| TOP2A | 6.80E-04 | 15 | Cluster 1 |
| ZWINT | 6.80E-04 | 15 | Cluster 1 |
| Term name | Description | FDR value | Genes ratio | Gene input name |
|---|---|---|---|---|
| CL:6604 | Mixed, incl. Amplification of signal from the kinetochores, and Condensin complex | 6.08E-18 | 12/93 | KIF11, SPAG5, CENPA, CDC20, ZWINT, KIF4A, HMMR, PRC1, KIF20A, TOP2A, HJURP, NUSAP1. |
| CL:6596 | Mixed, incl. Mitotic Spindle Checkpoint, and Mitotic sister chromatid segregation | 6.11E-18 | 13/151 | KIF11, MAD2L1, SPAG5, CENPA, CDC20, ZWINT, KIF4A, HMMR, PRC1, KIF20A, TOP2A, HJURP, NUSAP1. |
| CL:6608 | Mixed, incl. Regulation of mitotic sister chromatid segregation, and Kinesin motor domain, conserved site | 1.79E-14 | 9/51 | KIF11, SPAG5, CDC20, KIF4A, HMMR, PRC1, KIF20A, TOP2A, NUSAP1. |
| CL:6610 | Mixed, incl. Spindle elongation, and Polo-like kinase mediated events | 4.76E-13 | 8/41 | KIF11, SPAG5, KIF4A, HMMR, PRC1, KIF20A, TOP2A, NUSAP1. |
| CL:6619 | Mixed, incl. Spindle elongation, and Outer kinetochore | 2.74E-12 | 7/24 | KIF11, KIF4A, HMMR, PRC1, KIF20A, TOP2A, NUSAP1. |
| CL:6620 | Mixed, incl. Spindle elongation, and Axon hillock | 1.66E-11 | 6/12 | KIF11, KIF4A, HMMR, PRC1, TOP2A, NUSAP1. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





