Submitted:
09 July 2024
Posted:
10 July 2024
You are already at the latest version
Abstract
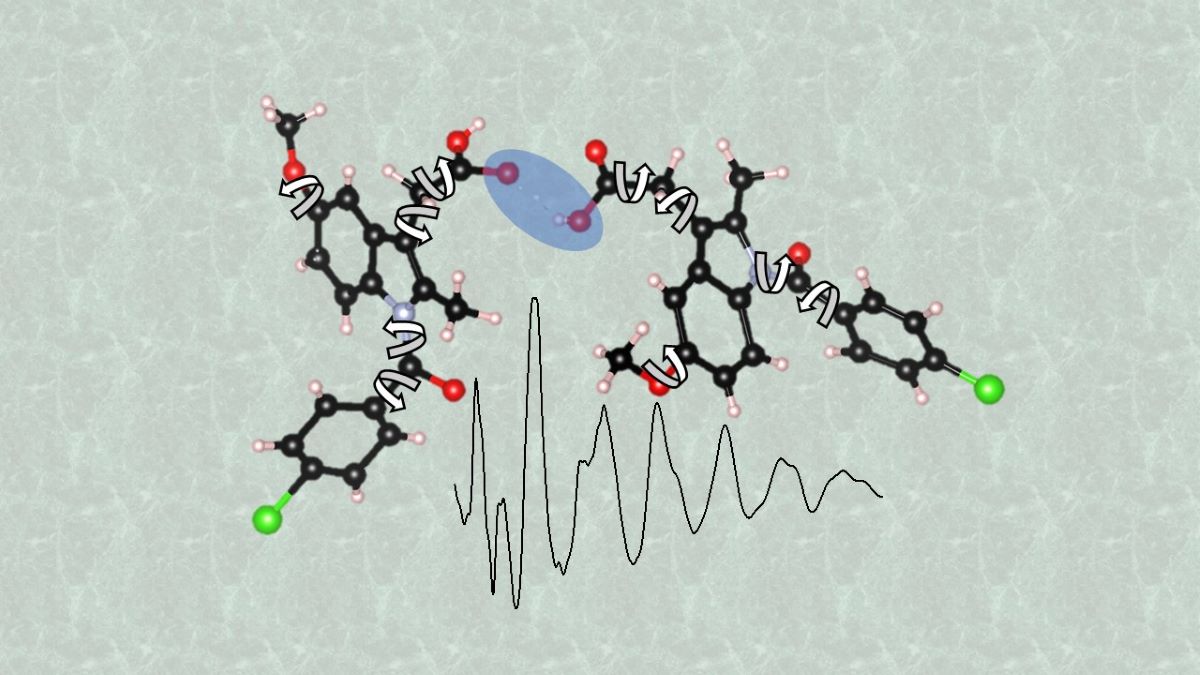
Keywords:
1. Introduction
2. Materials and Methods
3. Results
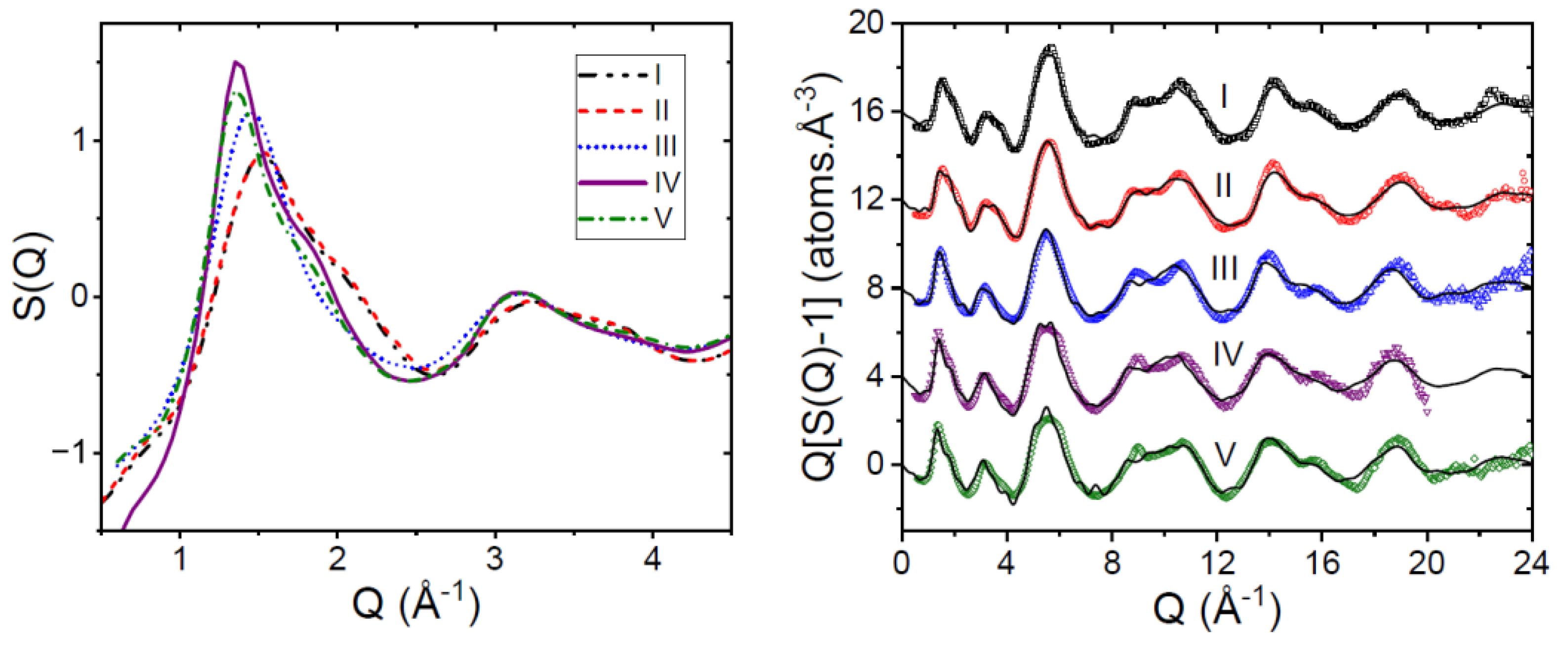
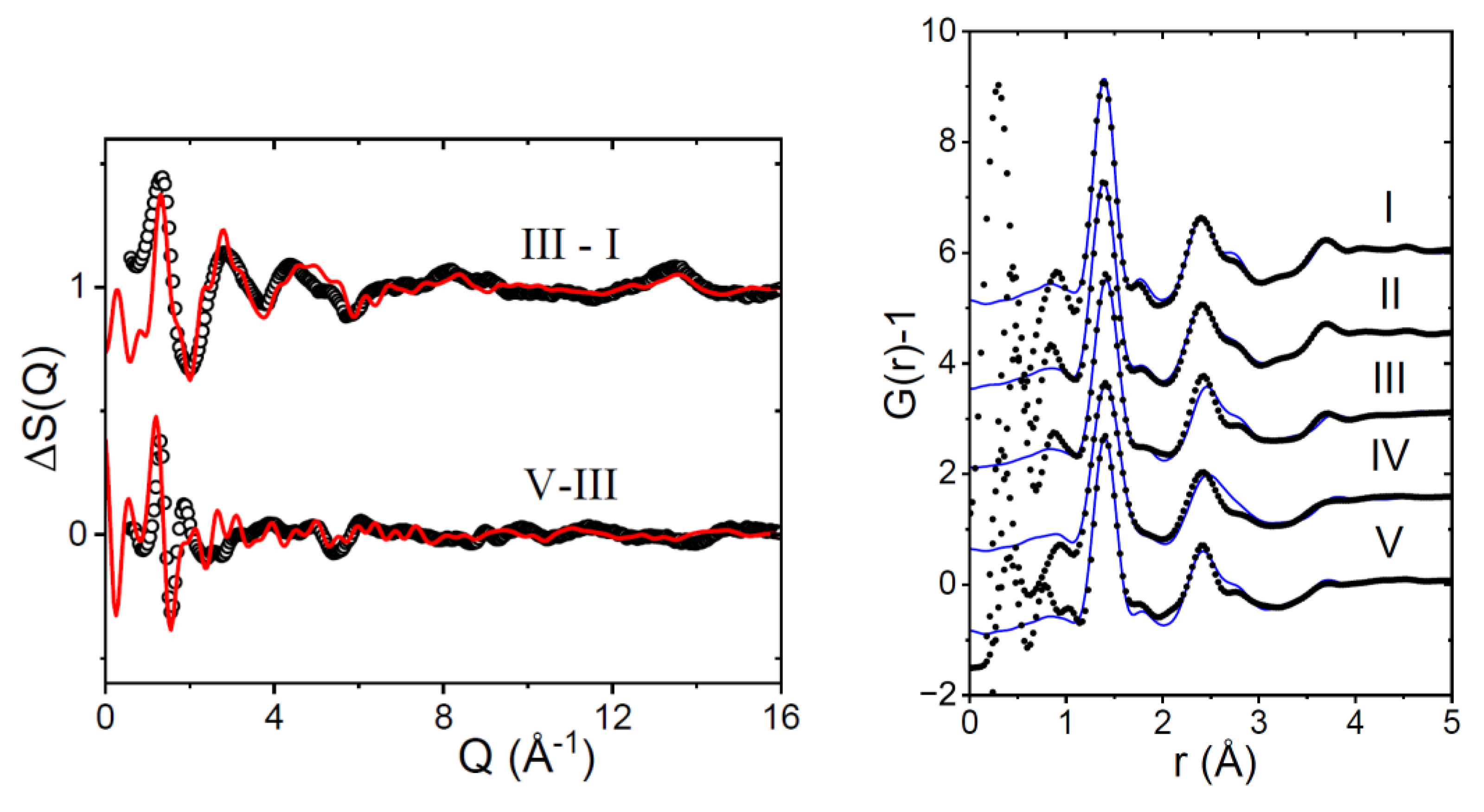
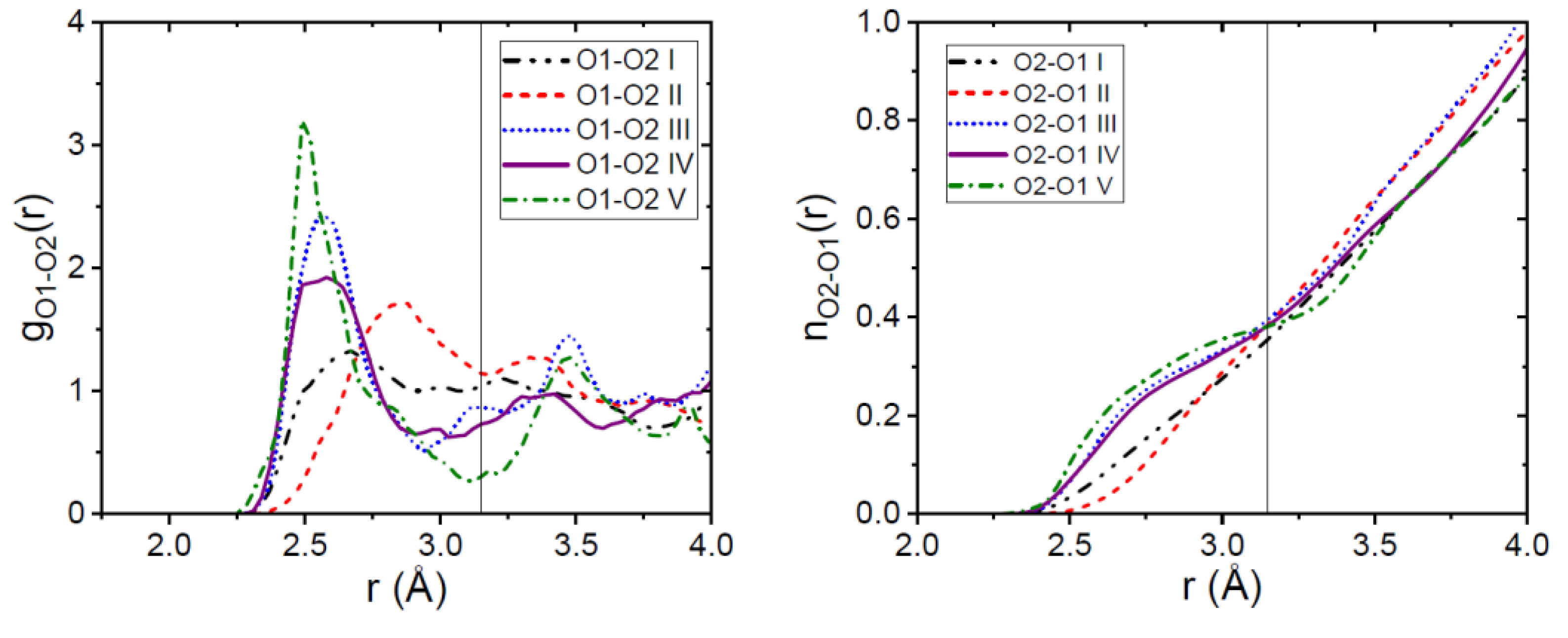
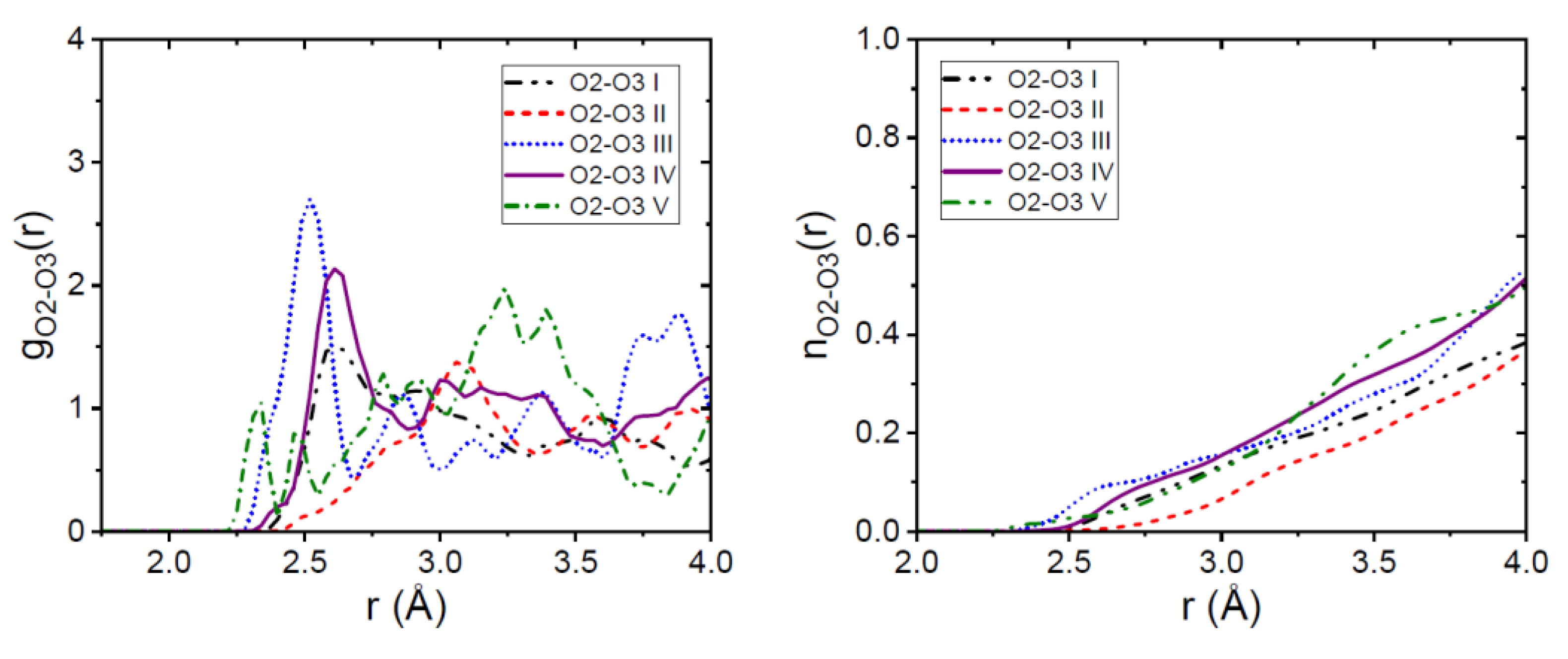
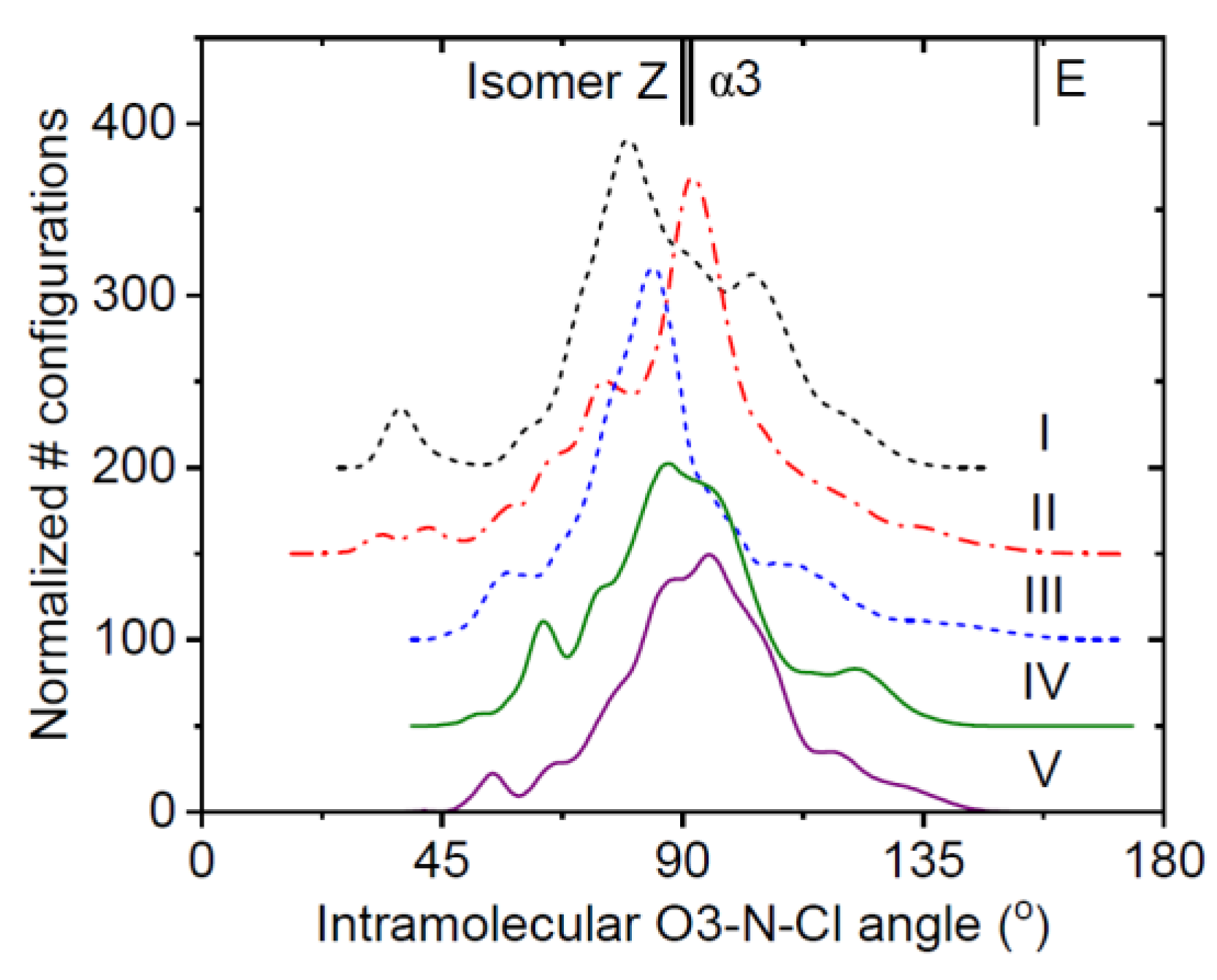
3.1. EPSR Models
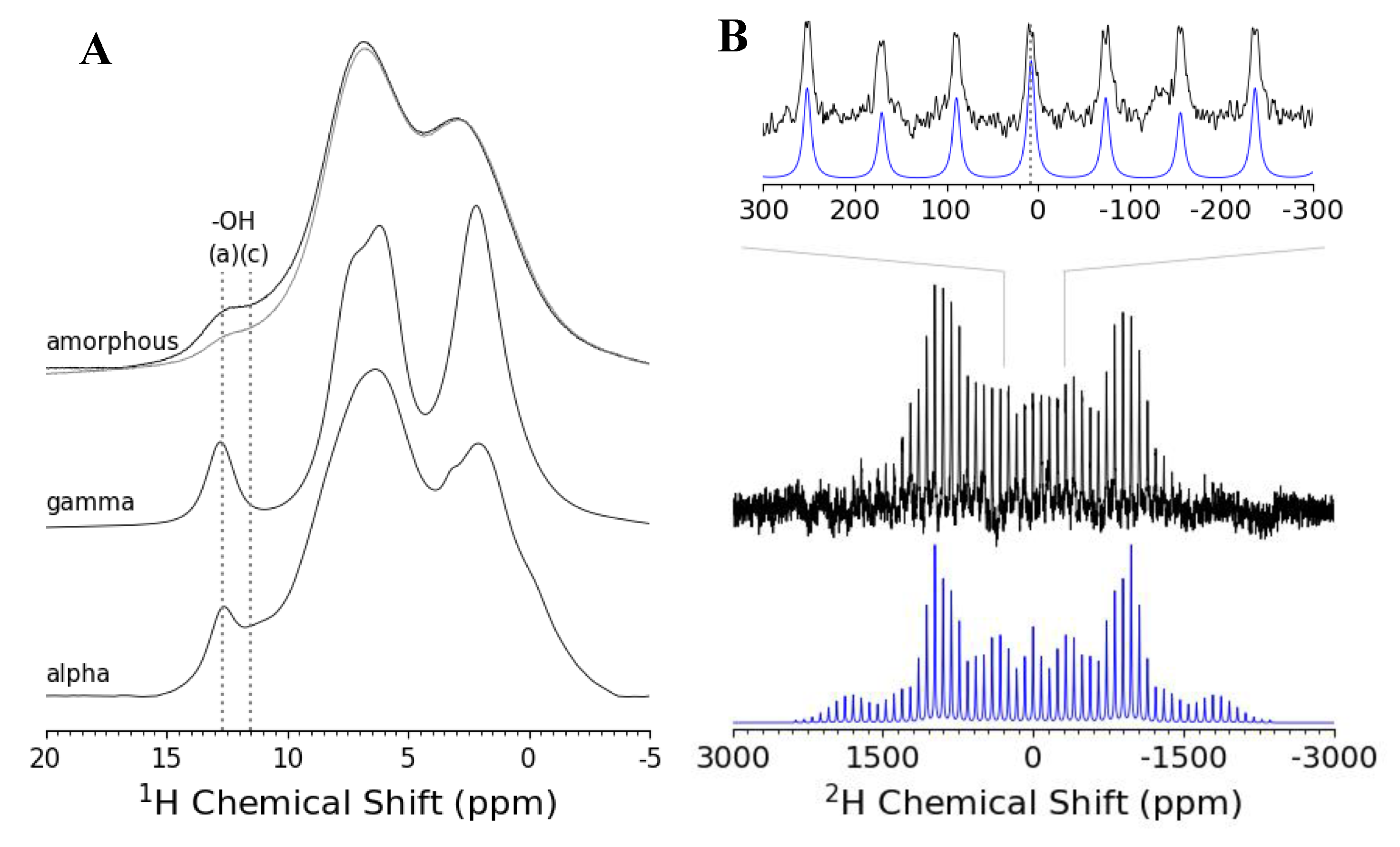
3.2. ssNMR Experiments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Byrn S.R., Z.G., Chen X.S., Solid State Properties of Pharmaceutical Material. 2017, Hoboken: John Wiley & Sons, Inc.
- Healy, A.M., et al., Pharmaceutical solvates, hydrates and amorphous forms: A special emphasis on cocrystals. Adv Drug Deliv Rev, 2017. 117: p. 25-46. [CrossRef]
- Hancock, B.C. and G. Zografi, Characteristics and significance of the amorphous state in pharmaceutical systems. J Pharm Sci, 1997. 86(1): p. 1-12. [CrossRef]
- Andronis, V., M. Yoshioka, and G. Zografi, Effects of sorbed water on the crystallization of indomethacin from the amorphous state. J Pharm Sci, 1997. 86(3): p. 346-51. [CrossRef]
- Bates, S., et al., Analysis of amorphous and nanocrystalline solids from their X-ray diffraction patterns. Pharm Res, 2006. 23(10): p. 2333-49. [CrossRef]
- Fukuoka, E., M. Makita, and S. Yamamura, Glassy state of pharmaceuticals. II. Bioinequivalence of glassy and crystalline indomethacin. Chem Pharm Bull (Tokyo), 1987. 35(7): p. 2943-8. [CrossRef]
- Crowley, K.J. and G. Zografi, Cryogenic grinding of indomethacin polymorphs and solvates: assessment of amorphous phase formation and amorphous phase physical stability. J Pharm Sci, 2002. 91(2): p. 492-507. [CrossRef]
- Yoshioka, M., B.C. Hancock, and G. Zografi, Crystallization of indomethacin from the amorphous state below and above its glass transition temperature. J Pharm Sci, 1994. 83(12): p. 1700-5. [CrossRef]
- Andronis, V. and G. Zografi, Crystal nucleation and growth of indomethacin polymorphs from the amorphous state. Journal of Non-crystalline Solids, 2000. 271: p. 236-248. [CrossRef]
- Greco, K. and R. Bogner, Crystallization of amorphous indomethacin during dissolution: effect of processing and annealing. Mol Pharm, 2010. 7(5): p. 1406-18. [CrossRef]
- Karmwar, P., et al., Investigation of properties and recrystallisation behaviour of amorphous indomethacin samples prepared by different methods. Int J Pharm, 2011. 417(1-2): p. 94-100.
- Xiang, T.-X. and B.D. Anderson, Molecular Dynamics Simulation of Amorphous Indomethacin. Molecular Pharmaceutics, 2013. 10(1): p. 102-114. [CrossRef]
- Kistenmacher, T.J. and R.E. Marsh, Crystal and molecular structure of an antiinflammatory agent, indomethacin, 1-(p-chlorobenzoyl)-5-methoxy-2-methylindole-3-acetic acid. Journal of the American Chemical Society, 1972. 94(4): p. 1340-1345. [CrossRef]
- Chen, X., et al., Reactivity differences of indomethacin solid forms with ammonia gas. J Am Chem Soc, 2002. 124(50): p. 15012-9. [CrossRef]
- Andrusenko, I., et al., Structure determination, thermal stability and dissolution rate of δ-indomethacin. International Journal of Pharmaceutics, 2021. 608: p. 121067. [CrossRef]
- Surwase, S.A., et al., Indomethacin: new polymorphs of an old drug. Molecular pharmaceutics, 2013. 10(12): p. 4472-4480. [CrossRef]
- Benmore, C.J., et al., A High Energy X-ray Diffraction Study of Amorphous Indomethacin. Journal of Pharmaceutical Sciences, 2022. 111(3): p. 818-824. [CrossRef]
- Benmore, C.J., et al. The Structure of Liquid and Glassy Carbamazepine. Quantum Beam Science, 2022. 6. [CrossRef]
- Benmore, C.J., et al., X-ray Diffraction of Water in Polyvinylpyrrolidone. Molecular Pharmaceutics, 2023. 20(7): p. 3645-3652. [CrossRef]
- Åman, K., P. Håkansson, and P.-O. Westlund, A general approach to the calculation of 2H2O NMR lineshapes in microheterogeneous systems: a distorted bicontinuous cubic phase. Physical Chemistry Chemical Physics, 2005. 7(7): p. 1394-1401. [CrossRef]
- Soper, A.K., Partial structure factors from disordered materials diffraction data: An approach using empirical potential structure refinement. Physical Review B, 2005. 72(10): p. 104204. [CrossRef]
- Gallington, L.C., et al. Review of Current Software for Analyzing Total X-ray Scattering Data from Liquids. Quantum Beam Science, 2023. 7. [CrossRef]
- Soper, A.K., Empirical potential Monte Carlo simulation of fluid structure. Chemical Physics, 1996. 202(2): p. 295-306. [CrossRef]
- Soper, A.K., Joint structure refinement of x-ray and neutron diffraction data on disordered materials: application to liquid water. Journal of Physics: Condensed Matter, 2007. 19(33): p. 335206.
- Benmore, C., L. Gallington, and E. Soignard, Intermediate range order in supercooled water. Molecular Physics, 2019. 117(18): p. 2470, 2476.
- Berglund, B. and R.W. Vaughan, Correlations between proton chemical shift tensors, deuterium quadrupole couplings, and bond distances for hydrogen bonds in solids. The Journal of Chemical Physics, 1980. 73(5): p. 2037-2043. [CrossRef]
- Di Martino, R.M.C., B.D. Maxwell, and T. Pirali, Deuterium in drug discovery: progress, opportunities and challenges. Nature Reviews Drug Discovery, 2023. 22(7): p. 562-584. [CrossRef]
- Lu, X., et al., Solid-state NMR analysis of crystalline and amorphous Indomethacin: An experimental protocol for full resonance assignments. J Pharm Biomed Anal, 2019. 165: p. 47-55. [CrossRef]
- Heinz, A., et al., Quantifying ternary mixtures of different solid-state forms of indomethacin by Raman and near-infrared spectroscopy. Eur J Pharm Sci, 2007. 32(3): p. 182-92. [CrossRef]
- Gerges, J. and F. Affouard, Insight From Molecular Dynamics Simulations on the Crystallization Tendency of Indomethacin Polymorphs in the Undercooled Liquid State. J Pharm Sci, 2020. 109(2): p. 1086-1095. [CrossRef]
- Terban, M.W. and S.J.L. Billinge, Structural Analysis of Molecular Materials Using the Pair Distribution Function. Chemical Reviews, 2022. 122(1): p. 1208-1272. [CrossRef]
- Benmore, C.J., 10.14 - X-ray and neutron diffraction from glasses and liquids, in Comprehensive Inorganic Chemistry III (Third Edition), J. Reedijk and K.R. Poeppelmeier, Editors. 2023, Elsevier: Oxford. p. 384-424.
- Chen, S., A.Y. Sheikh, and R. Ho, Evaluation of effects of pharmaceutical processing on structural disorders of active pharmaceutical ingredient crystals using nanoindentation and high-resolution total scattering pair distribution function analysis. J Pharm Sci, 2014. 103(12): p. 3879-3890. [CrossRef]
- Fronza, G., et al., 1H NMR and Molecular Modeling Study on the Inclusion Complex β-Cyclodextrin−Indomethacin. The Journal of Organic Chemistry, 1996. 61(3): p. 909-914. [CrossRef]
- Mantsch, H.H., H. Saitô, and I.C.P. Smith, Deuterium magnetic resonance, applications in chemistry, physics and biology. Progress in Nuclear Magnetic Resonance Spectroscopy, 1977. 11(4): p. 211-272. [CrossRef]
- Massiot, D., et al., Modelling one- and two-dimensional solid-state NMR spectra. Magnetic Resonance in Chemistry, 2002. 40(1): p. 70-76. [CrossRef]
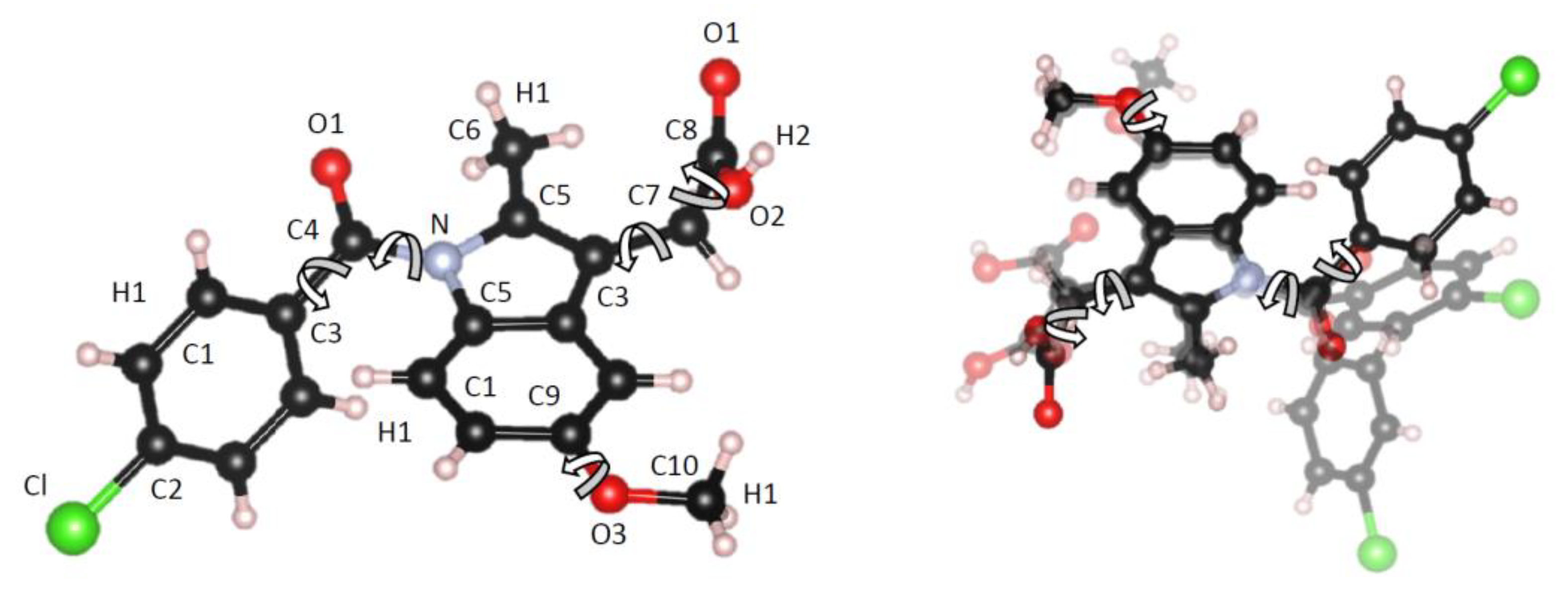
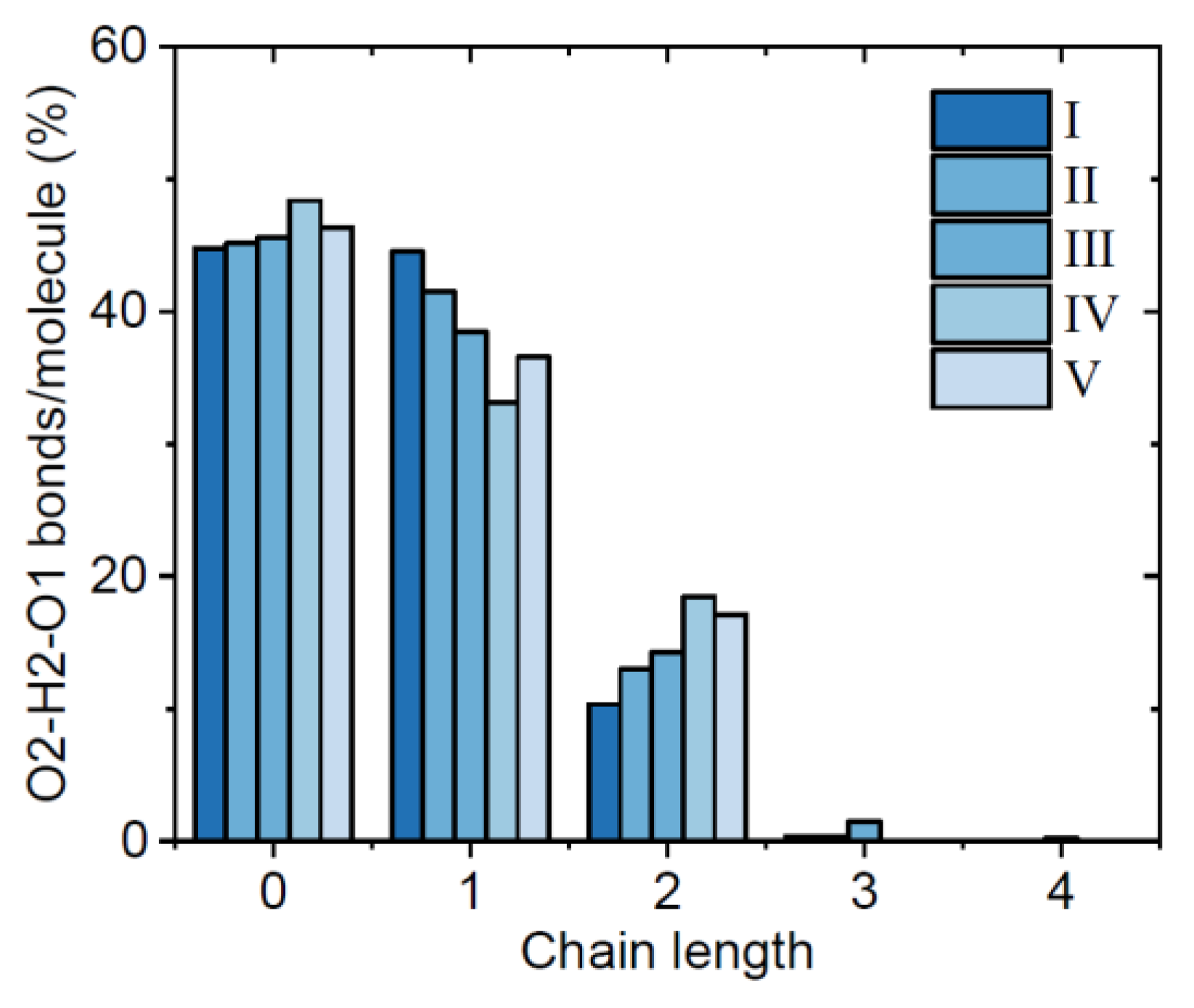
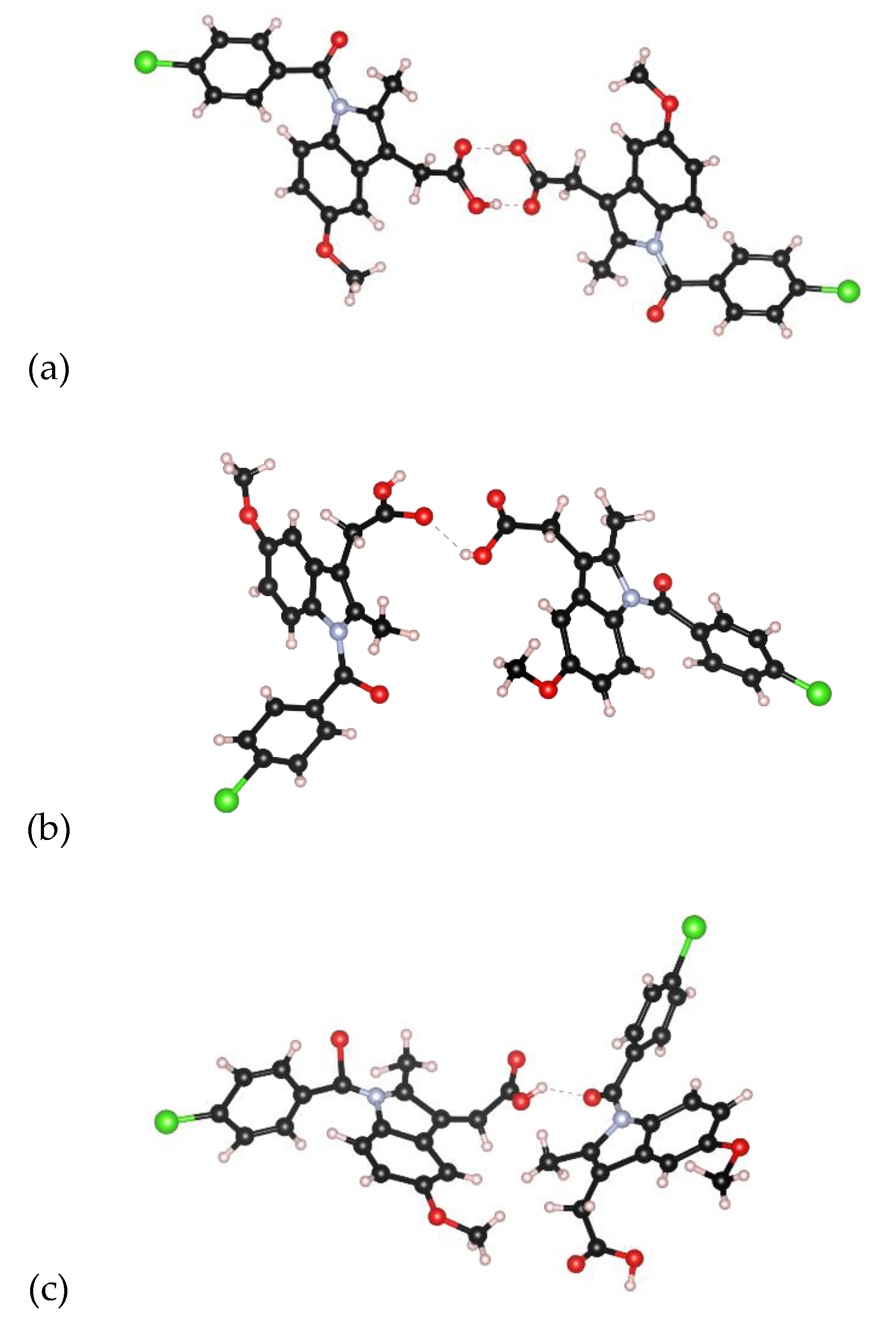
| Atom | ε (KJ/mol) | σ (Å) | Partial charge, Q |
|---|---|---|---|
| O1 (acceptor), O2 (donor) | 0.65 | 3.1 | -0.6 |
| O3 (acceptor) | 0.65 | 3.1 | -0.4 |
| C4, C8 | 0.8 | 3.7 | +0.7 |
| H2 | 0 | 0 | +0.8 |
| H1 | 0 | 0 | 0.0 |
| C1,C2,C3,C5,C6,C7,C9,C10 | 0.8 | 3.7 | 0.0 |
| Cl | 0.8 | 3.2 | 0.0 |
| Sample | Atomic number density (atomsÅ-3) | Number of rotations within molecule | Intra-molecular C-X bond length or expansion (Å) |
| γ-form (crystal) | 0.0952 | None (Z isomer fixed geometry) | 1.380 (initial bond) |
| I | 0.0900 | 5 | +0.015 |
| II | 0.0900 | 5 | +0.015 |
| III | 0.0925 | 5 | +0.036 |
| IV | 0.0950 | None (slight variation) | +0.041 |
| V | 0.0950 | None (slight variation) | +0.036 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





