1. Introduction
Vibrio parahaemolyticus poses a significant threat to public health through the consumption of contaminated seafood [
1]. According to the U.S. Centers for Disease Control and Prevention (CDC), approximately 84,000 people suffer from Vibrio-related illnesses annually [
2,
3]. This Gram-negative halophilic bacterium inhabits estuarine and marine environments and can naturally infiltrate oysters [
2,
3,
4]. However, the number of bacteria significantly increases in oysters during the warm-water season, and improper distribution or handling can further accelerate bacterial contamination, leading to public health concerns [
5,
6]. For instance, the Pacific
V. parahaemolyticus strain (O4 serotype/sequence type 36) severely impacted Oyster Bay, NY, causing a significant increase in reported illnesses and extended closures in Long Island Sound, including major areas in Connecticut [
7]. By 2013, the outbreak had spread from Virginia to Massachusetts, causing over 100 reported illnesses and resulting in unprecedented closures and recalls [
8]. This crisis made the industry view the situation as an existential threat and become more open to implementing controls to restore their critical summer operations [
9].
A multiplex polymerase chain reaction (PCR) protocol is listed as one of the standard methods in the U.S. Food and Drug Administration (FDA)’s Bacteriological Analytical Manual (BAM) to identify
V. parahaemolyticus and to determine its pathogenicity simultaneously, while information for the real-time PCR assay is still not available [
10]. This assay is designed to yield three bands showing the thermolabile hemolysin (
tlh) gene as a unique identification marker and the thermostable-related hemolysin (
trh) and thermostable-direct hemolysin (
tdh) genes as pathogenic markers of
V. parahaemolyticus. Studies have demonstrated that all examined
V. parahaemolyticus showed the amplification of
tlh gene, with no positive results in closely related
Vibrio spp. and other foodborne pathogens [
11,
12,
13]. Moreover, clinical investigations have indicated that
V. parahaemolyticus isolated from human patients carry the
trh and
tdh genes, which are potentially responsible for seafood-related illnesses and deaths [
14,
15,
16]. Therefore, identifying
V. parahaemolyticus and determining its pathogenicity in oysters are crucial steps to prevent foodborne illnesses and protect the domestic seafood industry.
Recently, we conducted the multiplex PCR recommended by BAM to identify
V. parahaemolyticus and assessed its pathogenicity in oysters from the U.S. Gulf Coast. After 25 PCR cycles using a positive control strain (F11-3A), the amplicons were loaded onto a 1.5 % tris-borate-EDTA (TBE) agarose gel [
2,
11]. However, the three bands did not appear even after 90 minutes of electrophoresis. The bands for
trh (486 bp) and
tlh (450 bp) were not separated well, and the
tdh band (270 bp) was weaker compared to others. This method was originally developed by Bej et al, and the major difference between the multiplex PCR method of BAM and Bej et al. was the number of PCR cycle (25 cycles of BAM and 30 cycles of Bej et al) [
11]. We found that the original method produced thicker
tdh band than BAM, but the bands for
trh and
tdh were not well-separated even after 90 minutes of electrophoresis.
In the present study, we aimed to improve the multiplex PCR method recommended by BAM by modifying the primers to achieve efficient separation of the three target genes and enhance the amplification of the tdh gene. Additionally, we examined the relative concentrations of primers, PCR cycling conditions, amounts of template DNA, the percentage of agarose, and electrophoresis time to produce three even amplicons, thereby confirming the specificity of the three sets of primers. The detection limit of the assay was evaluated using various concentrations of V. parahaemolyticus DNA, and the specificity of the assay was tested using nine Vibrio strains known to be human pathogens and 18 well-documented foodborne pathogens.
2. Materials and Methods
2.1. Bacteria, Genomic DNA, and Primers
Vibrio parahaemolyticus F11-3A was used as the reference strain to amply
tlh,
trh and
tdh genes [
2,
11]. Bacterial DNA was extracted using the Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research, Irvine, CA, USA) following the manufacturer’s instructions. The concentration of bacterial DNA was measured using the NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) by measuring the absorbance at 260 nm. Primers listed in table 1 were used for multiplex PCR. The
tlh gene primers were designed using SnapGene software (version 5.2, San Diego, CA, USA) targeting a specific region within the gene (gene ID: GU971655.1, MH047289.1, OP270227.1, accessed on 15 January 2024). Primers for the amplification of
trh and
tdh genes were adopted from both Bej et al. and BAM protocols [
2,
11] (
Table 1).
2.2. Multiplex PCR Condition
Three multiplex PCR methods were employed. The PCR mixture (50 µl) of both BAM and Bej et al consisted of the bacterial DNA (10 ng to 1 pg) from V. parahaemolyticus F11-3A, 1 µM of each of the primers (5 µl of each primer from 10 µM stock), 5 µl of a 10 X PCR buffer, 320 µM of each of the dNTPs (8 µl of a 8 mM stock dNTPs), 2.5 units of Dream Taq Green DNA polymerase (0.5 µl of 5 units/µL, Thermo Scientific, Vilnius, Lithuania), and 5.5 µl of water. The amplification condition of BAM was 1 cycle at 94 °C for 3 min, followed by 25 cycles of 94 °C for 1 min, 60 °C for 1 min, and 72 °C for 2 min, with a final extension at 72 °C for 3 min, while the amplification condition of Bej et al (ref) was 1 cycle at 94 °C for 3 min, followed by 30 cycles of 94 °C for 1 min, 58 °C for 1 min, and 72 °C for 1 min, with a final extension at 72 °C for 5 min.
Our PCR mixture was composed of the bacterial DNA (10 ng to 1 pg), 0.25 µM of each of the primers (1.25 µl of each primer from 10 µM stock), 5 µl of a 10 X PCR buffer, 320 µM of each of the dNTPs (8 µl of a 8 mM stock dNTPs), 1.5 units of Dream Taq Green DNA polymerase (0.3 µl of 5 units/µL, Thermo Scientific, Vilnius, Lithuania), and 28.2 µl of water. The amplification condition for this study was 1 cycle at 94 °C for 3 min, followed by 35 cycles of 94 °C for 1 min, 58 °C for 1 min, and 72 °C for 1 min, with a final extension at 72 °C for 5 min.
Electrophoresis was conducted using 1.5% TBE (TBE, Alfa Aesar, Ward Hill, MA, USA) agarose gels containing the SYBR Safe DNA gel stain (Invitrogen, Waltham, MA, USA) for various durations (30, 60, and 90 min) to optimize band separation. Similarly, electrophoresis through 2% TBE gel was run for 15 min, 30min, and 45min. The gel was visualized using the Gel Doc XR+ system (Bio-Rad, Hercules, CA, USA).
2.3. Optimization, Sensitivity, and Specificity of Multiplex PCR Assay
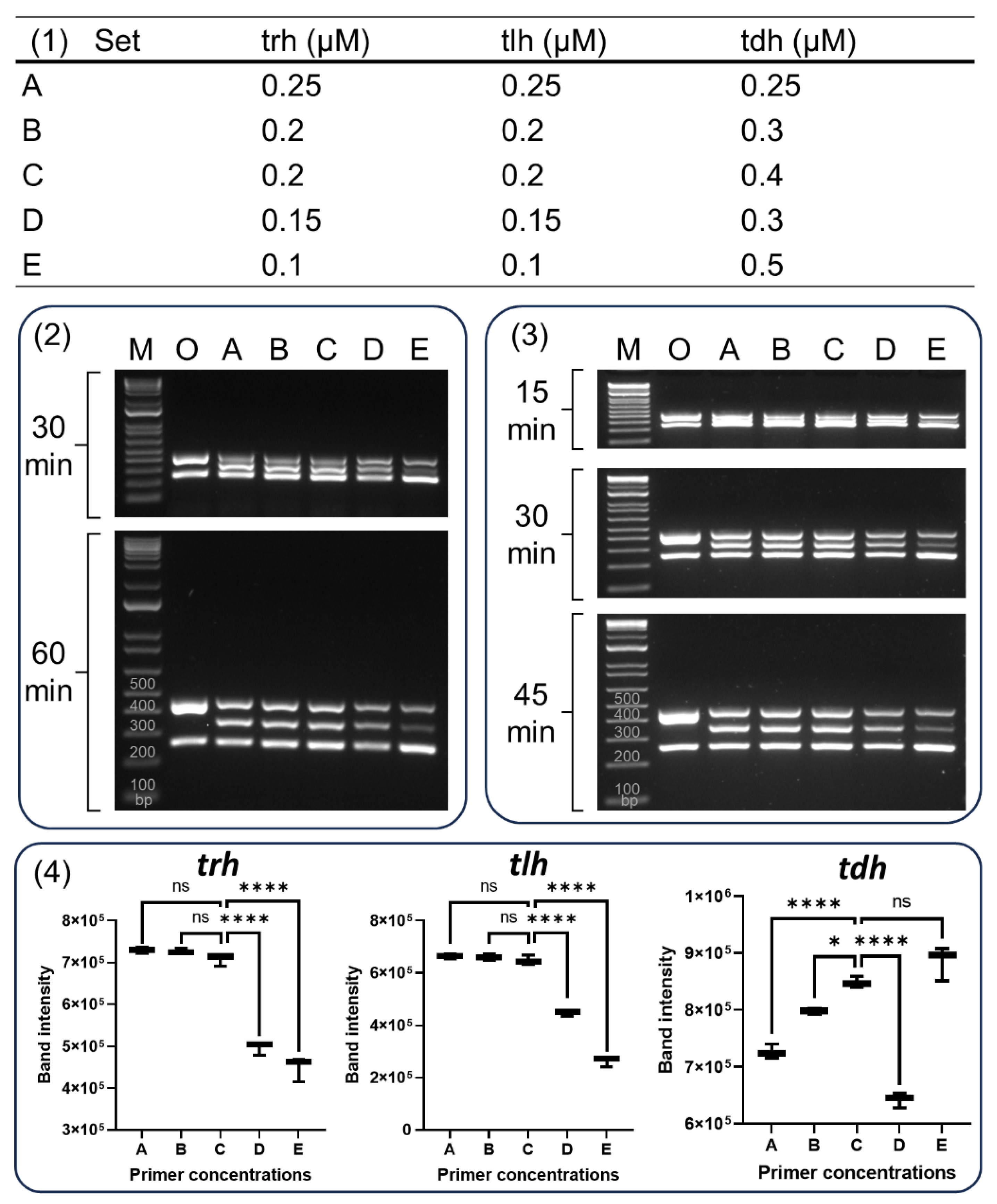
For the optimization of primer concentrations, 5 different PCR reaction mixtures were examined (
Figure 2-1). Set A contained 0.25 µM of each of the primers. Set B contained 0.2 µM of primers for
tlh and
trh, and 0.3 µM of
tdh. Set C contained 0.2 µM of primers for
tlh and
trh, and 0.4 µM of
tdh. Set 4 contained 0.15 µM of primers for
tlh and
trh, and 0.3 µM of
tdh. Finally, Set E contained 0.1 µM of primers for
tlh and
trh, and 0.5 µM of
tdh. To determine the optimal PCR cycles for our multiplex PCR, the amplification condition was examined 1 cycle at 94 °C for 3 min, followed by 25, 30, or 35 cycles of 94 °C for 1 min, 58 °C for 1 min, and 72 °C for 1 min, with a final extension at 72 °C for 5 min. Various concentrations (10 ng, 1 ng, 100 pg, 10 pg, 1 pg, and 100 fg) of
V. parahaemolyticus F11-3A DNA were used to determine the sensitivity of the multiplex PCR. After genomic DNAs were extracted from other
Vibrio strains and foodborne pathogenic bacteria listed in figure 4, one ng of each of bacterial DNA samples was employed to determine the specificity of the multiplex PCR.
2.4. Statistical Analysis
The band intensity of amplicons was quantified using the Image Lab Software (Bio-Rad, Hercules, CA, USA). The statistical analysis was conducted using the Prism software (Version 9, GraphPad, Boston, MA, USA) and significant differences were determined by Ordinary one-way ANOVA analysis (Dunnett’s multiple comparisons test, P < 0.05). The data were presented as mean ± standard deviation (SD, n=3).
3. Results and Discussion
3.1. Multiplex PCR Assay of BAM and Bej et al.
Multiplex PCR assays were conducted based on the method described by Bej et al and BAM to evaluate the simultaneous detection of
tlh,
trh and
tdh genes of
V. parahaemolyticus [
10,
11]. Since the BAM protocol originated from Bej et al., both protocols shared identical reaction components, including the PCR mixture and primer sequences except for modifications to the cycling conditions in the BAM protocol. To evaluate the performance of target gene amplifications, the assay was conducted with various primer combinations using 1 ng of F11-3A genomic DNA (
Figure 1, panels A-G). Additionally, the sensitivity of the assay was assessed using different concentrations of F11-3A DNA with all three primer sets (
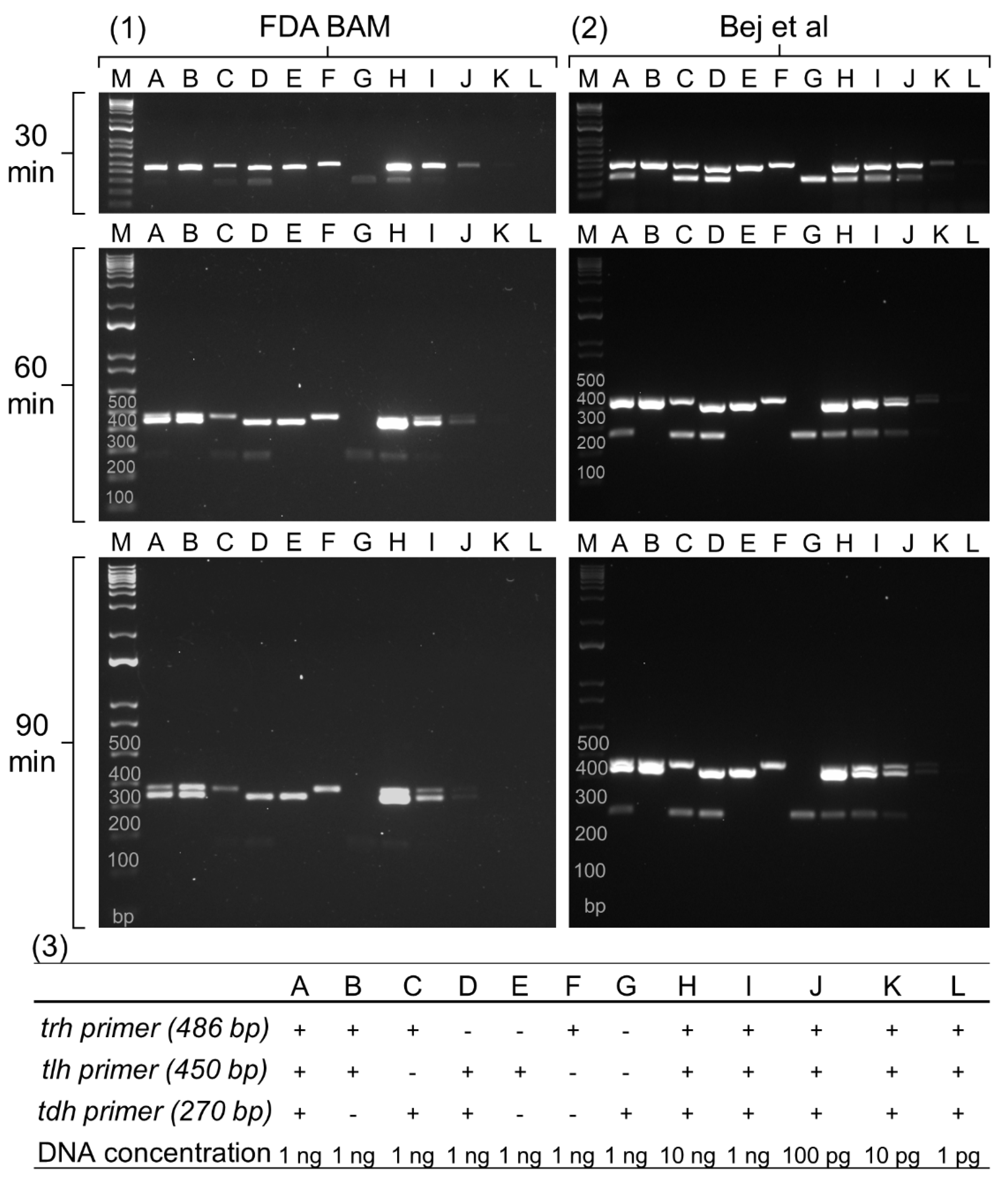
Figure 1, panels H-L). Finally, the electrophoresis run time was optimized by running gels at different durations (30, 60, and 90 minutes) to achieve optimal band separation on the 1.5% TBE gel.
The multiplex PCR assay of BAM (
Figure 1-1) exhibited insufficient separation of
trh and
tlh gene amplicons at 30 and 60 minutes of electrophoresis. While separation of both genes was achieved at 90 minutes, bands remained unresolved with 10 ng of bacterial DNA (
Figure 1-1H). The
tdh gene displayed a faint band in the sample containing 1 ng of bacterial DNA (
Figure 1-1A, C, D, G, and I) and became visible at 10 ng of DNA (
Figure 1-1H). Therefore, the limit of detection (LOD) of the BAM multiplex PCR was determined to be 10 ng of genomic DNA (
Figure 1-1H, I, J, K, and L).
The multiplex PCR assay of Bej et al. (
Figure 1-2) showed stronger band intensity for all three genes compared to BAM. This difference was probably due to the increased number of PCR cycles employed by Bej et al. (30 cycles) compared to BAM (25 cycles). Although a band for the
tdh gene appeared with 1 ng of bacterial DNA, its intensity diminished with longer electrophoresis times (
Figure 1-2A, C, D, G, and I). Additionally, both
trh and
tlh genes remained inseparable even at 90 minutes (
Figure 1-2A, B, D, G, and I). The LOD of this method was determined to be 100 pg of bacterial DNA (
Figure 1-2H, I, J, K, and L).
Both multiplex PCR assays exhibited limitations in separating the amplicons of the trh (486 bp) and tlh (450 bp) genes due to their similar sizes. This small size difference makes it difficult to distinguish the two amplicons on a gel. Additionally, the tdh gene amplicon (270 bp) often displays weaker band intensity compared to trh and tlh, potentially hindering its detection. To address these limitations, our multiplex PCR was designed to effectively separate the amplicons of trh and tlh genes, allowing for clear identification of both targets. Furthermore, the assay would be optimized to generate amplicons for all three genes (trh, tlh, and tdh) with similar band intensities, facilitating easier detection and analysis. This improved design aimed to overcome the limitations of the existing assays and provide a more reliable method for detecting these genes.
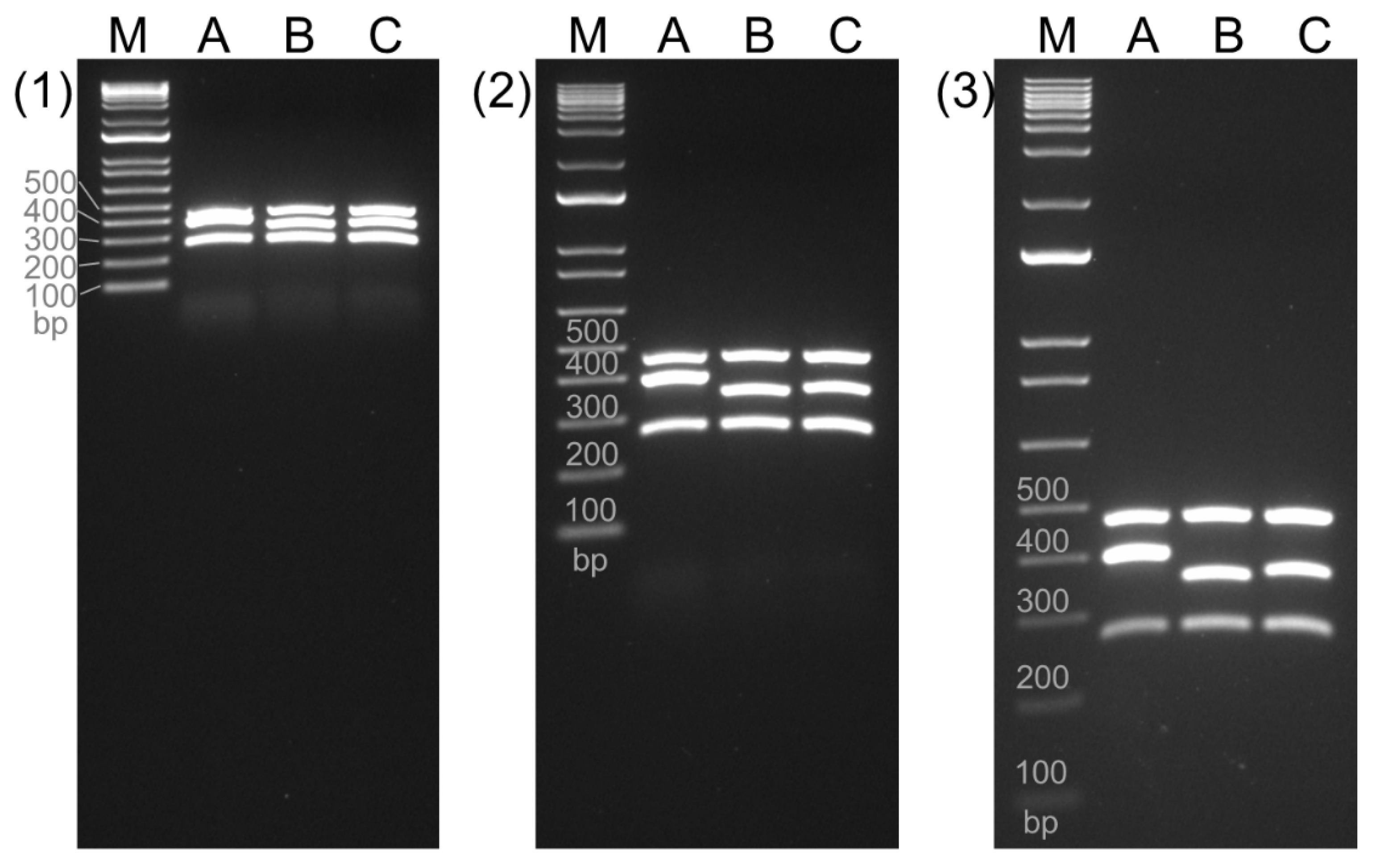
3.2. Optimization of the Current Multiplex PCR
To optimize a multiplex PCR assay for efficient separation and detection of three genes (
tlh,
trh, and
tdh) in
Vibrio parahaemolyticus, three primer sets for
tlh gene were examined for our enhanced multiplex PCR (
Table 1). The middle band of lane A, B, and C in
Figure 2 were amplified using the combination of VP_TLH_L and VP_TLH_R2 (403 bp), VP_TLH_F2 and VP_TLH_R (359 bp), and VP_TLH_F2 and VP_TLH_R2 (369 bp), respectively. All three candidates for
tlh gene displayed specific amplicons along with the expected amplicons for
trh (486 bp) and
tdh (252 bp) genes on the gel, with no non-specific products observed. Notably, Panel C using the VP_TLH_F2/VP_TLH_R2 primers displayed the most consistent separation between all three target bands even after shorter electrophoresis times. Therefore, this primer set was selected for the optimized multiplex PCR.
Our results, similar to those of previously reported methods (BAM and Bej et al.), showed a weaker band intensity for the
tdh gene (
Figure 2-3) compared to the
trh and
tlh genes at longer electrophoresis times (90 minutes). This was likely due to amplification bias, which was influenced by primer selection to a greater extent than by the template used [
17]. To address this and improve band separation, five different primer concentrations (set A to E) were tested (
Figure 3-1). The resulting amplicons were loaded onto 1.5% (
Figure 3-2) and 2% (
Figure 3-3) TBE agarose gels to evaluate the optimal electrophoresis conditions. While the Bej et al. multiplex PCR (Lane O in
Figure 3-2 and 3-3) failed to separate
trh and
tlh genes, our optimized multiplex PCR (Lane A to E) achieved separation at shorter electrophoresis times: 60 min on a 1.5% gel and 30 min or 45 min on a 2% gel. Additionally, the bands displayed sharper and clearer intensity after 45 min on a 2% gel compared to 60 min on a 1.5% gel. Therefore, electrophoresis on 2% TBE gel for 45 min was used in further band intensity analysis for optimization of primer concentrations (
Figure 3-4).
A previous study has indicated that the final concentration of the primers (0.04–0.5 µM) may vary considerably among the loci and was often established empirically [
18]. To achieve even amplification in multiplex PCR, it was recommended to adjust primer concentrations. This involves increasing the concentration for "weak" loci and decreasing it for "strong" loci [
19]. In this study, it was aimed to produce three amplicons evenly by decreasing the concentration of
trh and
tdh primers and by increasing the concentration of
tdh primers. Both
trh and
tlh genes displayed the significant band intensity at a concentration of 0.2 µM within the set C combination (
Figure 3-4). The
tdh gene also displayed the significant band intensity in set C, which contained 0.4 µM concentration of the
tdh primer. Interestingly, the same concentration of
tdh primer (0.3 µM) resulted in different band intensities between set B and D. This observation highlights the importance of multiple attempts to optimize reaction conditions for efficient multiplex PCR, as noted by Markoulatos et al. [
19].
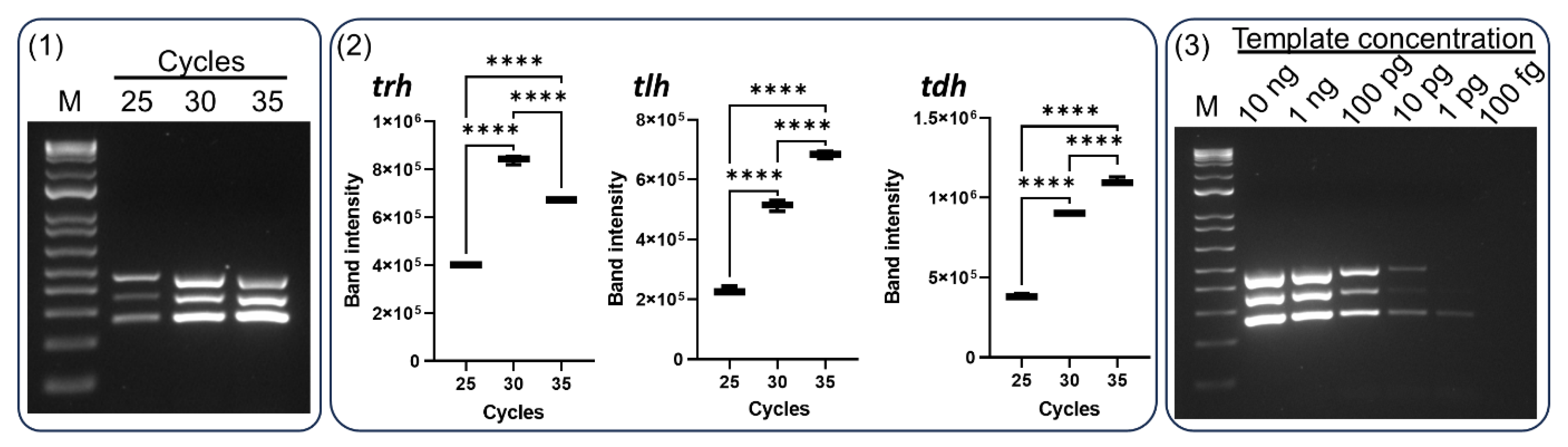
3.3. Sensitivity and Specificity of our Multiplex PCR
A previous study reported a multiplex PCR for detection of
groEL,
trh, and
tdh genes with a limit of detection (LOD) of 200 pg of
V. parahaemolyticus DNA using 30 PCR cycles [
20]. Another study demonstrated that the LOD of food-borne bacterial DNA in their multiplex PCR assay was 6.4 pg for
Staphylococcus aureus, 32 pg for
Escherichia coli O157:H7, 800 pg for
Listeria monocytogenes, 160 pg for
Shigella flexneri, and 32 pg for
Salmonella enterica serovar Enteritidis using 35 cycles [
21]. In this study, the sensitivity of our multiplex PCR assay was determined using various concentrations of F11-3A DNA (
Figure 4-3). The LOD for the
trh,
tlh, and
tdh genes were 10 pg, 10 pg, and 1 pg, respectively. The overall LOD for all three genes was 10 pg of bacterial DNA. This sensitivity was 1,000 times and 10 times higher than the results obtained using the BAM and Bej et al. methods, respectively (
Figure 1).
Figure 4 The specificity of the multiplex PCR assay was evaluated using three
V. parahaemolyticus (Lane 1-3 in
Figure 5), nine
Vibrio strains known to be human pathogens (Lane 4-12), and 18 well-documented food-borne pathogens (Lane 13-30) ([
10,
22,
23,
24]. All three target genes were successfully amplified using F11-3A (Lane 1). In contrast,
V. parahaemolyticus ATCC 178802 showed only
tlh-positive result, while
V. parahaemolyticus ATCC 35118 displayed both
tlh and
tdh-positive, consistent with previous studies [
11,
13]. Notably, our multiplex PCR did not produce any amplified products from other
Vibrio species and food-borne pathogenic bacteria. Therefore, this assay could be considered as an improved version of the multiplex PCR protocol recommended by BAM.
4. Conclusions
This study presents an efficient multiplex PCR assay for the detection of the species-specific tlh gene and two pathogenic trh and tdh genes. To enhance the multiplex PCR recommended by BAM, we redesigned primer set, optimized the concentration of primers, and adjusted the conditions of PCR cycles and gel electrophoresis. Our assay effectively separated the amplicons of three genes with similarly clear band intensities, facilitating their detection. Given its high sensitivity and specificity, this improved multiplex PCR method could significantly enhance the BAM recommended multiplex PCR protocol.
Author Contributions
Conceptualization, S.P. and Y.Z.; methodology, S.P; software, S.P.; formal analysis, S.P.; investigation, S.P.; resources, Y.Z.; data curation, S.P.; writing—original draft preparation, S.P.; writing—review and editing, Y.Z.; visualization, S.P.; supervision, Y.Z.; project administration, Y.Z.; funding acquisition, Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by USDA-ARS-SCA agreement number 58-6066-7081 and state CRIS project number MIS 081710 for MS Center for Food Safety and Postharvest Technology.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Su, Y.-C.; Liu, C. Vibrio parahaemolyticus: a concern of seafood safety. Food microbiology 2007, 24, 549–558. [Google Scholar] [CrossRef] [PubMed]
- DePaola, A.; Nordstrom, J.L.; Bowers, J.C.; Wells, J.G.; Cook, D.W. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Applied and environmental microbiology 2003, 69, 1521–1526. [Google Scholar] [CrossRef] [PubMed]
- Froelich, B.A.; Noble, R.T. Vibrio bacteria in raw oysters: managing risks to human health. Philosophical Transactions of the Royal Society B: Biological Sciences 2016, 371, 20150209. [Google Scholar] [CrossRef] [PubMed]
- Broberg, C.A.; Calder, T.J.; Orth, K. Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes and infection 2011, 13, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Andrews, L. Strategies to control Vibrios in molluscan shellfish. 2004.
- Nordstrom, J.; Kaysner, C.; Blackstone, G.; Vickery, M.; Bowers, J.; DePaola, A. Effect of intertidal exposure on Vibrio parahaemolyticus levels in Pacific Northwest oysters. Journal of food protection 2004, 67, 2178–2182. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Urtaza, J.; Baker-Austin, C.; Jones, J.L.; Newton, A.E.; Gonzalez-Aviles, G.D.; DePaola, A. Spread of Pacific northwest Vibrio parahaemolyticus strain. New England Journal of Medicine 2013, 369, 1573–1574. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.E.; Garrett, N.; Stroika, S.G.; Halpin, J.L.; Turnsek, M.; Mody, R.K. Notes from the field: increase in Vibrio parahaemolyticus infections associated with consumption of atlantic coast shellfish-2013. MMWR Morb Mortal Wkly Rep 2014, 63, 335–336. [Google Scholar] [PubMed]
- DePaola, A. Managing Vibrio Risk in Oysters. Food Protection Trends 2019, 39, 338–347. [Google Scholar]
- Kaysner, C.A.; DePaola, A.; Jones, J. Bacteriological analytical manual chapter 9: Vibrio. Food and Drug Administration, Maryland. [https://www. fda. gov/food/laboratory-methods-food/bam-chapter-9-vibrio]. Reviewed: December 2004, 19, 2019.
- Bej, A.K.; Patterson, D.P.; Brasher, C.W.; Vickery, M.C.; Jones, D.D.; Kaysner, C.A. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. Journal of microbiological methods 1999, 36, 215–225. [Google Scholar] [CrossRef]
- Nordstrom, J.L.; Vickery, M.C.; Blackstone, G.M.; Murray, S.L.; DePaola, A. Development of a multiplex real-time PCR assay with an internal amplification control for the detection of total and pathogenic Vibrio parahaemolyticus bacteria in oysters. Applied and Environmental Microbiology 2007, 73, 5840–5847. [Google Scholar] [CrossRef]
- Park, S.B.; Zhang, Y. Development of Multienzyme Isothermal Rapid Amplification (MIRA) Combined with Lateral-Flow Dipstick (LFD) Assay to Detect Species-Specific tlh and Pathogenic trh and tdh Genes of Vibrio parahaemolyticus. Pathogens 2024, 13, 57. [Google Scholar] [CrossRef]
- Hongping, W.; Jilun, Z.; Ting, J.; Yixi, B.; Xiaoming, Z. Insufficiency of the Kanagawa hemolytic test for detecting pathogenic Vibrio parahaemolyticus in Shanghai, China. Diagnostic microbiology and infectious disease 2011, 69, 7–11. [Google Scholar] [CrossRef]
- Shirai, H.; Ito, H.; Hirayama, T.; Nakamoto, Y.; Nakabayashi, N.; Kumagai, K.; Takeda, Y.; Nishibuchi, M. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infection and immunity 1990, 58, 3568–3573. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, X.; Hu, Z.; Xue, X.; Zhang, M.; Wu, Q.; Zhang, W.; Zhang, Y.; Lu, R. Characterization of Vibrio parahaemolyticus isolated from stool specimens of diarrhea patients in Nantong, Jiangsu, China during 2018–2020. Plos one 2022, 17, e0273700. [Google Scholar] [CrossRef]
- Suzuki, M.T.; Giovannoni, S.J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Applied and environmental microbiology 1996, 62, 625–630. [Google Scholar] [CrossRef]
- Markoulatos, P.; Samara, V.; Siafakas, N.; Plakokefalos, E.; Spyrou, N.; Moncany, M.L. Development of a quadriplex polymerase chain reaction for human cytomegalovirus detection. Journal of clinical laboratory analysis 1999, 13, 99–105. [Google Scholar] [CrossRef]
- Markoulatos, P.; Siafakas, N.; Moncany, M. Multiplex polymerase chain reaction: a practical approach. Journal of clinical laboratory analysis 2002, 16, 47–51. [Google Scholar] [CrossRef]
- Hossain, M.T.; Kim, Y.-O.; Kong, I.-S. Multiplex PCR for the detection and differentiation of Vibrio parahaemolyticus strains using the groEL, tdh and trh genes. Molecular and cellular probes 2013, 27, 171–175. [Google Scholar] [CrossRef]
- Chen, J.; Tang, J.; Liu, J.; Cai, Z.; Bai, X. Development and evaluation of a multiplex PCR for simultaneous detection of five foodborne pathogens. Journal of applied microbiology 2012, 112, 823–830. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nature Reviews Disease Primers 2018, 4, 1–19. [Google Scholar] [CrossRef]
- Bintsis, T. Foodborne pathogens. AIMS microbiology 2017, 3, 529. [Google Scholar] [CrossRef] [PubMed]
- Priyanka, B.; Patil, R.K.; Dwarakanath, S. A review on detection methods used for foodborne pathogens. Indian Journal of Medical Research 2016, 144, 327–338. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Multiplex PCR analysis of Vibrio parahaemolyticus tlh, trh and tdh genes. The protocols were based on the Bacteriological Analytical Manual (BAM) of the U.S. FDA (1) and a study by Bej et al. (2). Panels A-G represent reactions with different combinations of trh, tlh, and tdh primers (3). Panels H-L depict the sensitivity of the PCR method tested with various bacterial DNA concentrations (3). M: Molecular weight marker. The electrophoreses were run for 30, 60, and 90 min to separate three bands through a 1.5% TBE agarose gel.
Figure 1.
Multiplex PCR analysis of Vibrio parahaemolyticus tlh, trh and tdh genes. The protocols were based on the Bacteriological Analytical Manual (BAM) of the U.S. FDA (1) and a study by Bej et al. (2). Panels A-G represent reactions with different combinations of trh, tlh, and tdh primers (3). Panels H-L depict the sensitivity of the PCR method tested with various bacterial DNA concentrations (3). M: Molecular weight marker. The electrophoreses were run for 30, 60, and 90 min to separate three bands through a 1.5% TBE agarose gel.
Figure 2.
Multiplex PCR to enhance the discrimination of three genes of
Vibrio parahaemolyticus. The primers for amplification of
trh (486 bp) and
tdh (270 bp) were adopted from the Bacteriological Analytical Manual (BAM) and a study by Bej et al [
10,
11]. The multiplex PCR included various combination of primers to amplify trh gene (A: 403 bp, B: 359 bp, and C: 369 bp). The amplicons were electrophorized onto 1.5% TBE agarose gel for 30 min (A), 60 min (B), and 90 min (C). M denotes the 100 bp molecular marker.
Figure 2.
Multiplex PCR to enhance the discrimination of three genes of
Vibrio parahaemolyticus. The primers for amplification of
trh (486 bp) and
tdh (270 bp) were adopted from the Bacteriological Analytical Manual (BAM) and a study by Bej et al [
10,
11]. The multiplex PCR included various combination of primers to amplify trh gene (A: 403 bp, B: 359 bp, and C: 369 bp). The amplicons were electrophorized onto 1.5% TBE agarose gel for 30 min (A), 60 min (B), and 90 min (C). M denotes the 100 bp molecular marker.
Figure 3.
Optimization of primer concentrations of enhanced multiplex PCR. Various concentrations of the trh, tlh, and tdh primers were tested. Panel (1) shows the concentration of primers for set A, B, C, D, and E. The amplicons were separated using gel electrophoresis on either a 1.5% TBE agarose gel for 30 min or 60 min (2) or a 2% agarose TBE gel for 15 min, 30 min or 45 min (3). Band intensities across the five tested concentration (lanes A-E) on the 2% TBE agarose gel for 45 min were compared to determine the optimal conditions for simultaneous amplification of all three target genes (4). Lanes M and O denote the 100 bp molecular marker and PCR conducted according to Bej et al (ref), respectively. Data represents the means of three independent replicates (One-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
Figure 3.
Optimization of primer concentrations of enhanced multiplex PCR. Various concentrations of the trh, tlh, and tdh primers were tested. Panel (1) shows the concentration of primers for set A, B, C, D, and E. The amplicons were separated using gel electrophoresis on either a 1.5% TBE agarose gel for 30 min or 60 min (2) or a 2% agarose TBE gel for 15 min, 30 min or 45 min (3). Band intensities across the five tested concentration (lanes A-E) on the 2% TBE agarose gel for 45 min were compared to determine the optimal conditions for simultaneous amplification of all three target genes (4). Lanes M and O denote the 100 bp molecular marker and PCR conducted according to Bej et al (ref), respectively. Data represents the means of three independent replicates (One-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
Figure 4.
Optimization of multiplex PCR cycle number and detection limit. The PCR reactions were conducted with varying cycle numbers: 25, 30, and 35 (1). Panel (2) shows the corresponding band intensities for the amplified tlh, trh, and tdh genes at each cycles number. Panel (3) exhibits the detection limit (sensitivity) of the multiplex PCR with 35 cycles. Data represent the means of three independent replicates (One-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
Figure 4.
Optimization of multiplex PCR cycle number and detection limit. The PCR reactions were conducted with varying cycle numbers: 25, 30, and 35 (1). Panel (2) shows the corresponding band intensities for the amplified tlh, trh, and tdh genes at each cycles number. Panel (3) exhibits the detection limit (sensitivity) of the multiplex PCR with 35 cycles. Data represent the means of three independent replicates (One-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
Figure 5.
Specificity of the enhanced multiplex PCR. Vibrio parahaemolyticus F11-3A exhibited positive results for trh, tlh and tdh genes (1). V. parahaemolyticus ATCC 17802 was only positive for tlh (2). V. parahaemolyticus ATCC 35118 showed positivity for tlh and tdh genes (3). None of the other tested Vibrio strains and foodborne pathogenic bacteria displayed amplification of three genes. (4): Vibrio vulnificus ATCC 33147, (5): Vibrio vulnificus ATCC 27562, (6): Vibrio vulnificus ATCC 33815, (7): Vibrio metschnikovii ATCC 7708, (8): Vibrio fluvialis ATCC 33809, (9): Vibrio mimicus ATCC 33655, (10): Vibrio furnissii ATCC 35627, (11): Vibrio cholerae ATCC 39315, (12): Vibrio alginolyticus ATCC 33840, (13): Escherichia coli ATCC 51739, (14): Escherichia coli K-12, (15): Escherichia coli O157:H7 ATCC 43895, (16): Listeria monocytogenes F5069, (17): Lactobacillus buchneri ATCC 12936, (18): Listeria innocua ATCC 33090, (19): Salmonella enterica Serovar Typhimurium 14028, (20): Salmonella enterica Serovar Gaminara F2712, (21): Salmonella enterica Serovar Montevideo ATCC BAA-1735, (22): Salmonella enterica Serovar Senftenburg ATCC 43845, (23): Salmonella enterica Serovar Enteritidis E190-88, (24): Salmonella enterica Serovar Choleraesuis ATCC 10708, (25): Bacillus subtilis ATCC 9372, (26): Clostridium perfringens ATCC 13124, (27): Enterococcus faecalis ATCC 344, (28): Lactobacillus acidophilus NRRL B1910, (29): Staphylococcus aureus ATCC 25923, (30): Shigella flexineri ATCC 12022. M denotes the 100 bp molecular marker.
Figure 5.
Specificity of the enhanced multiplex PCR. Vibrio parahaemolyticus F11-3A exhibited positive results for trh, tlh and tdh genes (1). V. parahaemolyticus ATCC 17802 was only positive for tlh (2). V. parahaemolyticus ATCC 35118 showed positivity for tlh and tdh genes (3). None of the other tested Vibrio strains and foodborne pathogenic bacteria displayed amplification of three genes. (4): Vibrio vulnificus ATCC 33147, (5): Vibrio vulnificus ATCC 27562, (6): Vibrio vulnificus ATCC 33815, (7): Vibrio metschnikovii ATCC 7708, (8): Vibrio fluvialis ATCC 33809, (9): Vibrio mimicus ATCC 33655, (10): Vibrio furnissii ATCC 35627, (11): Vibrio cholerae ATCC 39315, (12): Vibrio alginolyticus ATCC 33840, (13): Escherichia coli ATCC 51739, (14): Escherichia coli K-12, (15): Escherichia coli O157:H7 ATCC 43895, (16): Listeria monocytogenes F5069, (17): Lactobacillus buchneri ATCC 12936, (18): Listeria innocua ATCC 33090, (19): Salmonella enterica Serovar Typhimurium 14028, (20): Salmonella enterica Serovar Gaminara F2712, (21): Salmonella enterica Serovar Montevideo ATCC BAA-1735, (22): Salmonella enterica Serovar Senftenburg ATCC 43845, (23): Salmonella enterica Serovar Enteritidis E190-88, (24): Salmonella enterica Serovar Choleraesuis ATCC 10708, (25): Bacillus subtilis ATCC 9372, (26): Clostridium perfringens ATCC 13124, (27): Enterococcus faecalis ATCC 344, (28): Lactobacillus acidophilus NRRL B1910, (29): Staphylococcus aureus ATCC 25923, (30): Shigella flexineri ATCC 12022. M denotes the 100 bp molecular marker.
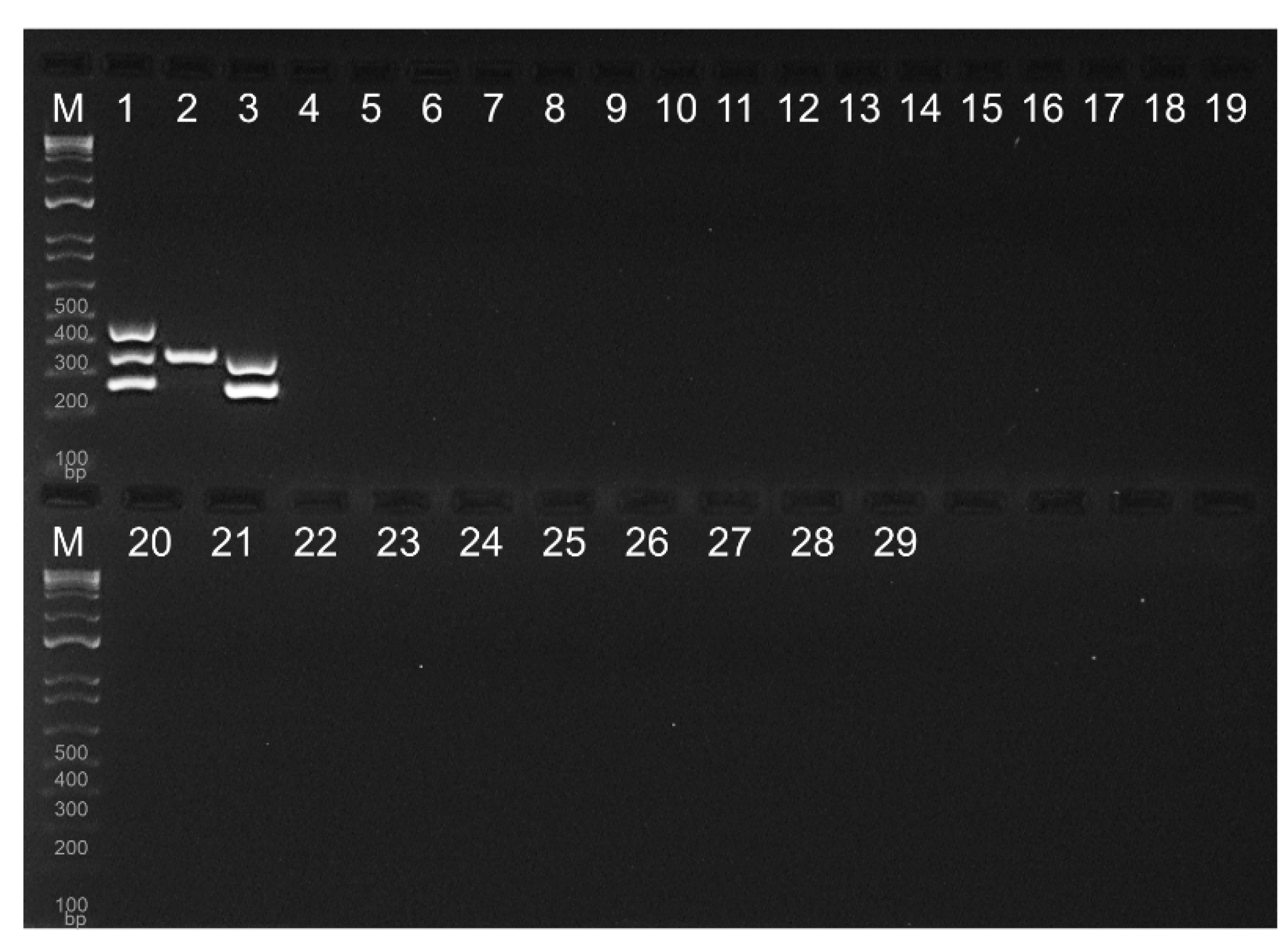
Table 1.
Primers for the amplification of tlh, trh and tdh genes using the multiplex PCR.
Table 1.
Primers for the amplification of tlh, trh and tdh genes using the multiplex PCR.
| Names |
Sequences (5'-3') |
size (bp) |
References |
| VP_TLH_L |
AAAGCGGATTATGCAGAAGCACTG |
450 |
[11] |
| VP_TLH_R |
GCTACTTTCTAGCATTTTCTCTGC |
| VP_TRH_L |
TTGGCTTCGATATTTTCAGTATCT |
486 |
| VP_TRH_R |
CATAACAAACATATGCCCATTTCCG |
| VP_TDH_L |
GTAAAGGTCTCTGACTTTTGGAC |
270 |
| VP_TDH_R |
TGGAATAGAACCTTCATCTTCACC |
| VP_TLH_F2 |
CTCAGTTTAAGTACTCAACACAAGAAGAGAT |
369 |
This study |
| VP_TLH_R2 |
CTAAGTTGTTGCTACTTTCTAGCATTTTCT |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).