Submitted:
12 July 2024
Posted:
12 July 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Materials and Methods
Ethics Statement
Cells and Viruses
Antigen Design and Virus Production
Phylogenetic Analysis
Western Blot
Hemagglutination (HA) Assay
Animal Studies for Vaccine Efficacy Evaluation
Animal Study
Immunization and Challenge
Virus Isolation
Hemagglutination Inhibition (HI) Assay
Generation of Pseudovirus Bearing HA and NA of H5N1 Avian Influenza Virus
HA Pseudovirus Neutralization Assay
Results
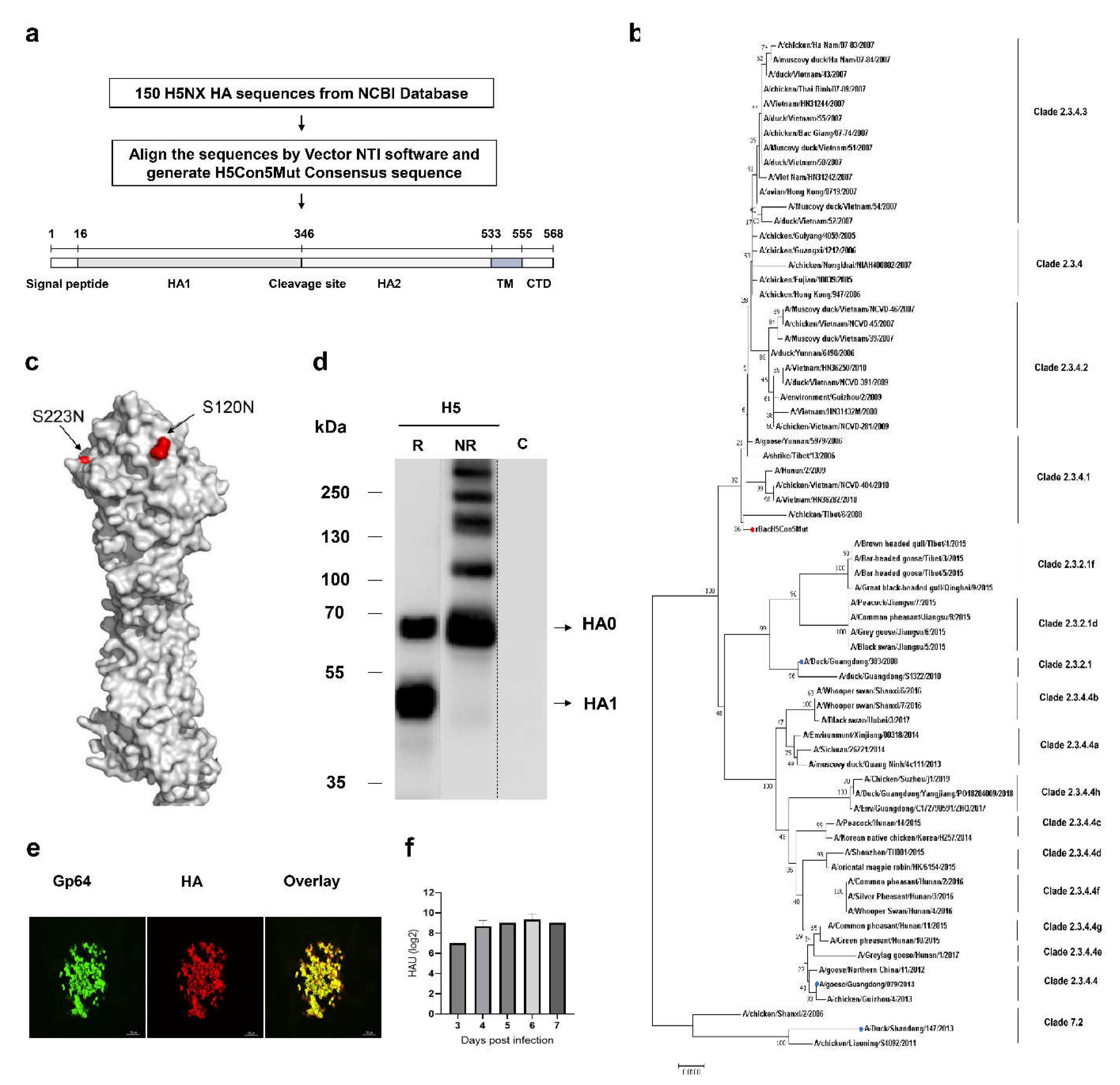
Characterization of H5Nx HA Expressed by Recombinant Baculovirus
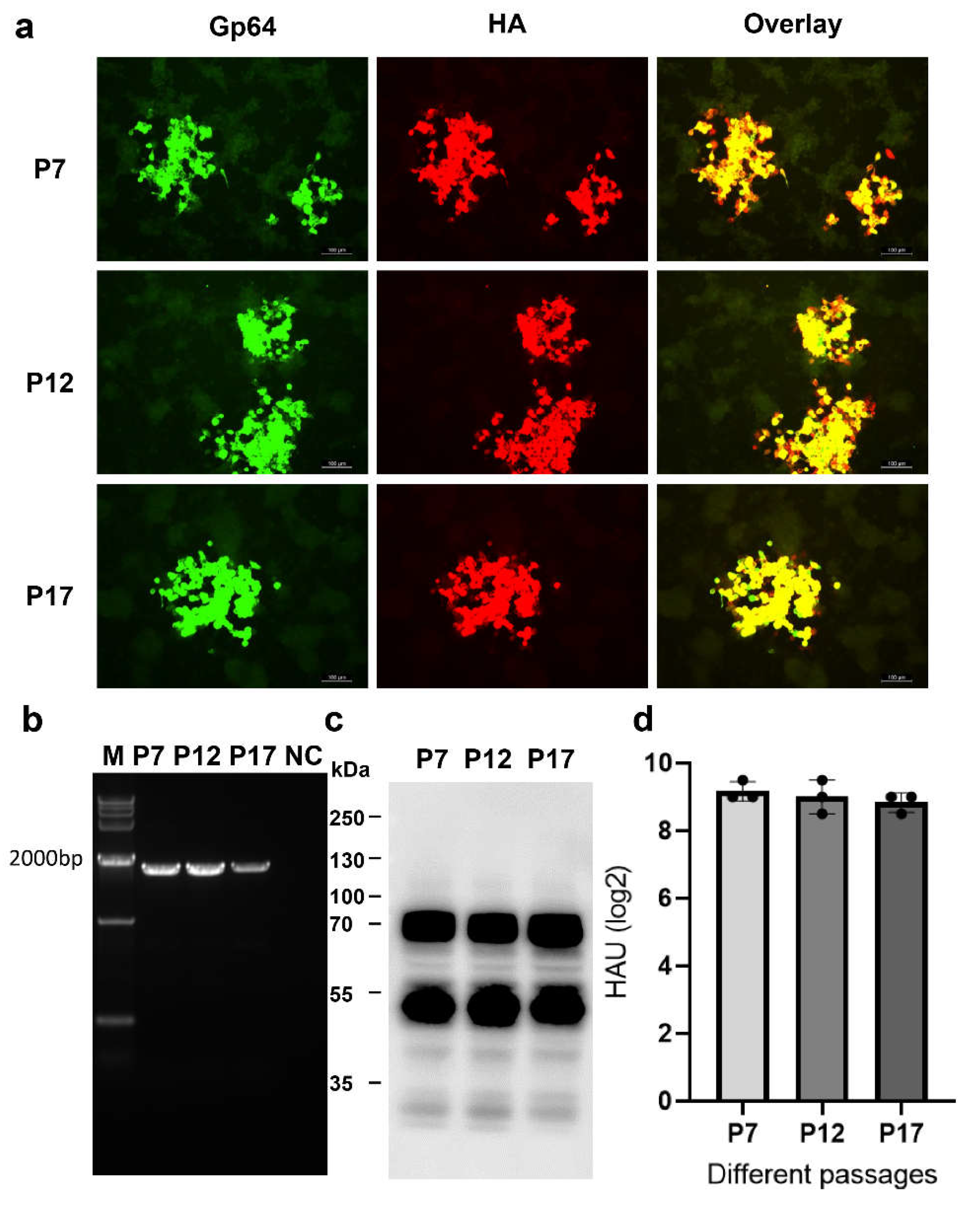
Genetic Stability of rBacH5Con5Mut Virus
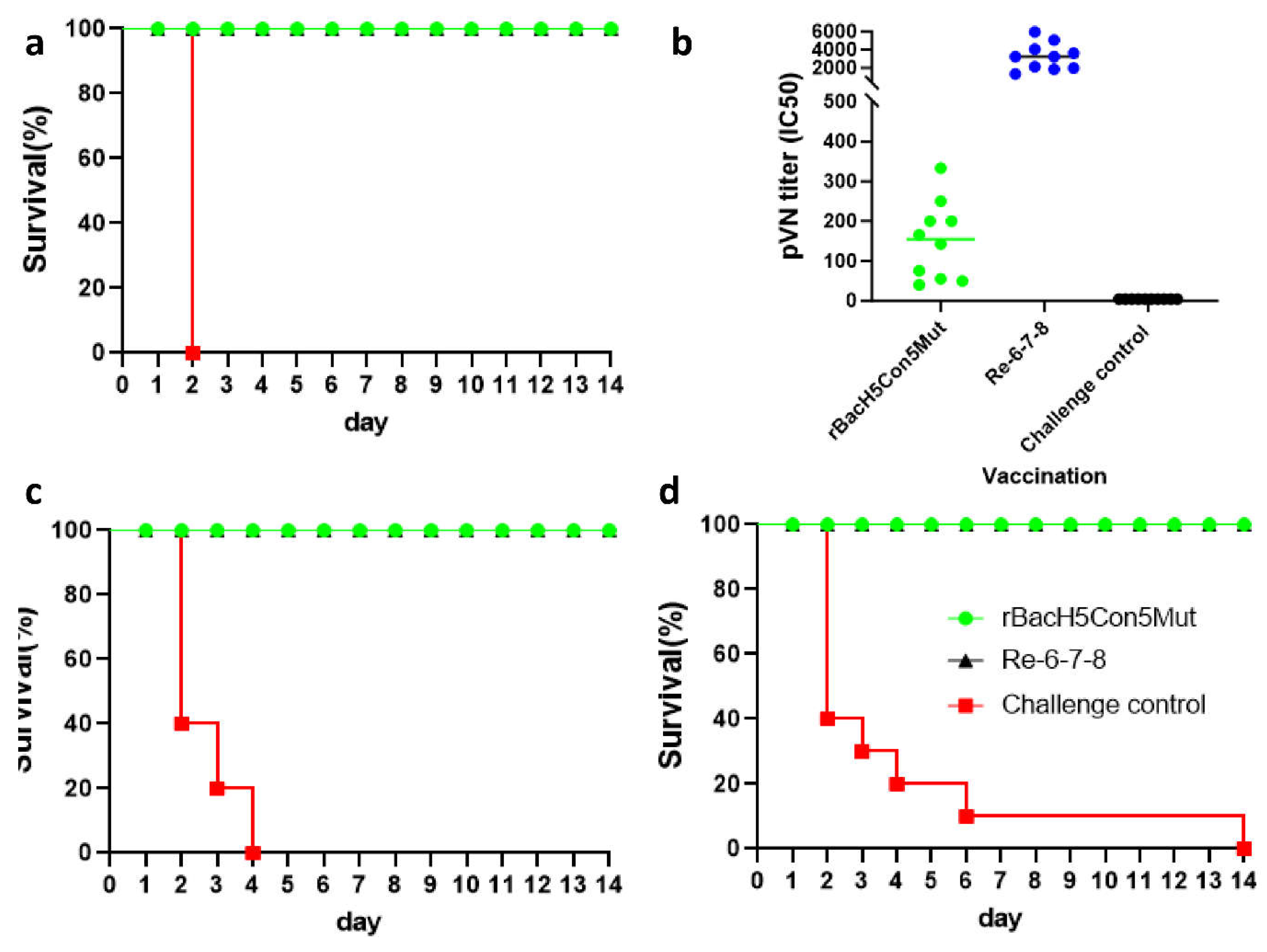
A single Immunization with rBacH5Con5Mut Vaccine Prevented Infection from 2.3.4.4 Clade H5N1 Virus
Pseudovirus-Based Neutralization Assay
The rBacH5Con5Mut Vaccine Was Cross-Protective against Clade 2.3.2.1 H5N1 and Clade 7.2 H5N2 Viruses
Protective Efficacy of rBacH5Con5Mut Vaccine Delivered to One-Day-Old SPF Chickens against 2.3.4.4 Clade H5N1 Virus
Discussion
References
- Smith, G. J. D., Donis, R. O. & Group, W. H. O. O. for A. H. and A. O. (WHO/OIE/FAO) H. E. W. Nomenclature updates resulting from the evolution of avian influenza A(H5) virus clades 2.1.3.2a, 2.2.1, and 2.3.4 during 2013–2014. Influ. Other Respir. Viruses 9, 271–276 (2015). [CrossRef]
- Claes, F., Morzaria, S. P. & Donis, R. O. Emergence and dissemination of clade 2.3.4.4 H5Nx influenza viruses—how is the Asian HPAI H5 lineage maintained. Curr. Opin. Virol. 16, 158–163 (2016).
- Sun, H. et al. Characterization of clade 2.3.4.4 highly pathogenic H5 avian influenza viruses in ducks and chickens. Vet. Microbiol. 182, 116–122 (2016). [CrossRef]
- SWAYNE, D. E., PAVADE, G., HAMILTON, K., VALLAT, B. & MIYAGISHIMA, K. Assessment of national strategies for control of high-pathogenicity avian influenza and lowpathogenicity notifiable avian influenza in poultry, with emphasis on vaccines and vaccination: -EN- -FR- Évaluation des stratégies nationales de lutte contre l’influenza aviaire hautement pathogène et l’influenza aviaire faiblement pathogène à déclaration obligatoire chez les volailles, et plus particulièrement du recours aux vaccins et à la vaccination -ES- Evaluación de estrategias nacionales de lucha contra la influenza aviar altamente patógena y la influenza aviar levemente patógena de declaración obligatoria en aves de corral, haciendo hincapié en las vacunas y la vacunación. Rev. Sci. Tech. l’OIE 30, 839–870 (2011).
- Swayne, D. E. Impact of Vaccines and Vaccination on Global Control of Avian Influenza. Avian Diseases 56, 818–828 (2012). [CrossRef]
- Chen, M.-W. et al. A consensus–hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proc. Natl. Acad. Sci. 105, 13538–13543 (2008). [CrossRef]
- Bi, Y. et al. Highly pathogenic avian influenza H5N1 Clade 2.3.2.1c virus in migratory birds, 2014–2015. Virol. Sin. 31, 300–305 (2016). [CrossRef]
- Tian, H. et al. Spatial, temporal and genetic dynamics of highly pathogenic avian influenza A (H5N1) virus in China. BMC Infect. Dis. 15, 54 (2015). [CrossRef]
- Li, C., Bu, Z. & Chen, H. Avian influenza vaccines against H5N1 ‘bird flu’ Trends in Biotechnology 32, 147–156 (2014).
- Shi, J., Zeng, X., Cui, P., Yan, C. & Chen, H. Alarming situation of emerging H5 and H7 avian influenza and effective control strategies. Emerg. Microbes Infect. 12, 2155072 (2023).
- Ge, J. et al. Newcastle Disease Virus-Based Live Attenuated Vaccine Completely Protects Chickens and Mice from Lethal Challenge of Homologous and Heterologous H5N1 Avian Influenza Viruses. J. Virol. 81, 150–158 (2007). [CrossRef]
- Jiang, Y. et al. Enhanced protective efficacy of H5 subtype avian influenza DNA vaccine with codon optimized HA gene in a pCAGGS plasmid vector. Antivir. Res. 75, 234–241 (2007). [CrossRef]
- Prabakaran, M. et al. Neutralizing Epitopes of Influenza Virus Hemagglutinin: Target for the Development of a Universal Vaccine against H5N1 Lineages. J. Virol. 84, 11822–11830 (2010). [CrossRef]
- Yewdell, J. W., Bennink, J. R., Smith, G. L. & Moss, B. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. 82, 1785–1789 (1985).
- Ekiert, D. C. et al. Antibody Recognition of a Highly Conserved Influenza Virus Epitope. Science 324, 246–251 (2009). [CrossRef]
- Tan, G. S. et al. A Pan-H1 Anti-Hemagglutinin Monoclonal Antibody with Potent Broad-Spectrum Efficacy In Vivo. J. Virol. 86, 6179–6188 (2012). [CrossRef]
- Kamlangdee, A., Kingstad-Bakke, B., Anderson, T. K., Goldberg, T. L. & Osorio, J. E. Broad Protection against Avian Influenza Virus by Using a Modified Vaccinia Ankara Virus Expressing a Mosaic Hemagglutinin Gene. J. Virol. 88, 13300–13309 (2014).
- Wang, T. T. et al. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc. Natl. Acad. Sci. 107, 18979–18984 (2010). [CrossRef]
- Sautto, G. A. & Ross, T. M. Hemagglutinin consensus-based prophylactic approaches to overcome influenza virus diversity. Vet. Ital. 55, 195–201 (2019).
- Ma, N. et al. Development of an mRNA vaccine against a panel of heterologous H1N1 seasonal influenza viruses using a consensus hemagglutinin sequence. Emerg. Microbes Infect. 12, 2202278 (2023). [CrossRef]
- Giles, B. M. & Ross, T. M. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 29, 3043–3054 (2011).
- Wang, G., Yin, R., Zhou, P. & Ding, Z. Combination of the immunization with the sequence close to the consensus sequence and two DNA prime plus one VLP boost generate H5 hemagglutinin specific broad neutralizing antibodies. PLoS ONE 12, e0176854 (2017).
- Zhou, H. et al. Sequential immunization with consensus influenza hemagglutinins raises cross-reactive neutralizing antibodies against various heterologous HA strains. Vaccine 35, 305–312 (2017). [CrossRef]
- Song, J.-M., Rooijen, N. V., Bozja, J., Compans, R. W. & Kang, S.-M. Vaccination inducing broad and improved cross protection against multiple subtypes of influenza A virus. Proc. Natl. Acad. Sci. 108, 757–761 (2011).
- Harada, Y. et al. Inactivated and adjuvanted whole-virion clade 2.3.4 H5N1 pre-pandemic influenza vaccine possesses broad protective efficacy against infection by heterologous clades of highly pathogenic H5N1 avian influenza virus in mice. Vaccine 29, 8330–8337 (2011). [CrossRef]
- Wu, C.-Y. et al. A VLP Vaccine Induces Broad-Spectrum Cross-Protective Antibody Immunity against H5N1 and H1N1 Subtypes of Influenza A Virus. PLoS ONE 7, e42363 (2012). [CrossRef]
- Cavalcanti, M. O. et al. A genetically engineered H5 protein expressed in insect cells confers protection against different clades of H5N1 highly pathogenic avian influenza viruses in chickens. Avian Pathol. 46, 224–233 (2017). [CrossRef]
- Zeng, X. et al. Protective Efficacy of the Inactivated H5N1 Influenza Vaccine Re-6 Against Different Clades of H5N1 Viruses Isolated in China and the Democratic People’s Republic of Korea. Avian Diseases 60, 238–240 (2015). [CrossRef]
- Tian, G., Zeng, X., Li, Y., Shi, J. & Chen, H. Protective Efficacy of the H5 Inactivated Vaccine Against Different Highly Pathogenic H5N1 Avian Influenza Viruses Isolated in China and Vietnam. Avian Diseases 54, 287–289 (2010).
- Zeng, X. et al. Protective Efficacy of an H5N1 Inactivated Vaccine Against Challenge with Lethal H5N1, H5N2, H5N6, and H5N8 Influenza Viruses in Chickens. Avian Diseases 60, 253–255 (2016). [CrossRef]
- Yang, D.-G. et al. Avian Influenza Virus Hemagglutinin Display on Baculovirus Envelope: Cytoplasmic Domain Affects Virus Properties and Vaccine Potential. Mol. Ther. 15, 989–996 (2007). [CrossRef]
- Laddy, D. J. et al. Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine 25, 2984–2989 (2007). [CrossRef]
- Carter, D. M. et al. Design and Characterization of a Computationally Optimized Broadly Reactive Hemagglutinin Vaccine for H1N1 Influenza Viruses. J. Virol. 90, 4720–4734 (2016). [CrossRef]
- Muthumani, K. et al. Immunogenicity of novel consensus-based DNA vaccines against Chikungunya virus. Vaccine 26, 5128–5134 (2008). [CrossRef]
- Obeng-Adjei, N. et al. Synthetic DNA immunogen encoding hepatitis B core antigen drives immune response in liver. Cancer Gene Ther. 19, 779–787 (2012). [CrossRef]
- Latimer, B. et al. A polyantigenic genotype 1a/1b consensus hepatitis C virus DNA vaccine induces broadly reactive HCV-specific cellular immune responses in both mice and non-human primates (VAC7P.985). The Journal of Immunology (2014).
- Yan, J. et al. Immunogenicity of a novel engineered HIV-1 clade C synthetic consensus-based envelope DNA vaccine. Vaccine 29, 7173–7181 (2011). [CrossRef]
- Muthumani, K. et al. A synthetic consensus anti–spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci. Transl. Med. 7, 301ra132 (2015). [CrossRef]
- Hoffmann, E., Lipatov, A. S., Webby, R. J., Govorkova, E. A. & Webster, R. G. Role of specific hemagglutinin amino acids in the immunogenicity and protection of H5N1 influenza virus vaccines. Proc. Natl. Acad. Sci. 102, 12915–12920 (2005).
- Forrest, H. L. et al. Single- and multiple-clade influenza A H5N1 vaccines induce cross protection in ferrets. Vaccine 27, 4187–4195 (2009). [CrossRef]
- Wang, T. T. et al. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc. Natl. Acad. Sci. 107, 18979–18984 (2010). [CrossRef]
- He, F., Prabakaran, M., kumar, S. R., Tan, Y. & Kwang, J. Monovalent H5 vaccine based on epitope-chimeric HA provides broad cross-clade protection against variant H5N1 viruses in mice. Antivir. Res. 105, 143–151 (2014).
- Tang, Y. et al. Chimaeric VP2 proteins from infectious bursal disease virus containing the N-terminal M2e of H9 subtype avian influenza virus induce neutralizing antibody responses to both viruses. Avian Pathol. 42, 260–267 (2013). [CrossRef]
- Bernstein, D. I. et al. Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine candidates: interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect. Dis. 20, 80–91 (2020). [CrossRef]
- Spackman, E. et al. Development of a Real-Time Reverse Transcriptase PCR Assay for Type A Influenza Virus and the Avian H5 and H7 Hemagglutinin Subtypes. J. Clin. Microbiol. 40, 3256–3260 (2002). [CrossRef]
- Kumar, M., Chu, H.-J., Rodenberg, J., Krauss, S. & Webster, R. G. Association of Serologic and Protective Responses of Avian Influenza Vaccines in Chickens. Avian Diseases 51, 481–483 (2007).
- Liu, M. et al. Preparation of a standardized, efficacious agricultural H5N3 vaccine by reverse genetics. Virology 314, 580–590 (2003). [CrossRef]
- Temperton, N. J. et al. A sensitive retroviral pseudotype assay for influenza H5N1-neutralizing antibodies. Influ. Other Respir. Viruses 1, 105–112 (2007). [CrossRef]
- Hyseni, I. et al. Characterisation of SARS-CoV-2 Lentiviral Pseudotypes and Correlation between Pseudotype-Based Neutralisation Assays and Live Virus-Based Micro Neutralisation Assays. Viruses 12, 1011 (2020). [CrossRef]
- Tolah, A. M. K. et al. Evaluation of a Pseudovirus Neutralization Assay for SARS-CoV-2 and Correlation with Live Virus-Based Micro Neutralization Assay. Diagnostics 11, 994 (2021). [CrossRef]
- Rowe, T. et al. Detection of Antibody to Avian Influenza A (H5N1) Virus in Human Serum by Using a Combination of Serologic Assays. J. Clin. Microbiol. 37, 937–943 (1999). [CrossRef]
- Schultsz, C. et al. Prevalence of Antibodies against Avian Influenza A (H5N1) Virus among Cullers and Poultry Workers in Ho Chi Minh City, 2005. PLoS ONE 4, e7948 (2009). [CrossRef]
- Hu, Z. et al. Hemagglutinin-Specific Non-neutralizing Antibody Is Essential for Protection Provided by Inactivated and Viral-Vectored H7N9 Avian Influenza Vaccines in Chickens. Front. Vet. Sci. 6, 482 (2020). [CrossRef]
- Reemers, S., Verstegen, I., Basten, S., Hubers, W. & Zande, S. van de. A broad spectrum HVT-H5 avian influenza vector vaccine which induces a rapid onset of immunity. Vaccine 39, 1072–1079 (2021).
- BUBLOT, M. et al. Development and Use of Fowlpox Vectored Vaccines for Avian Influenza. Ann. N. York Acad. Sci. 1081, 193–201 (2006). [CrossRef]
| Experiment | Vaccination | Challenge virus | Clade | HI titer with different antigens (log2)a | Survival/total | Virus shedding from chickens at five days post challenge | |||
| rBacH5 | CVb | Oropharyngeal | Cloacal | ||||||
| 1 | rBacH5Con5Mut | A/Goose/Guangdong/079/2013 (H5N1, 14079) | 2.3.4.4 | 9.1 | 3.6 | 10/10 | 0/10 | 0/10 | |
| Re6-7-8 | 6.5 | 8 | 10/10 | 0/10 | 0/10 | ||||
| Control | 3.3 | 0 | 0/10 | N/ac | N/a | ||||
| 2 | rBacH5Con5Mut | A/Duck/Guangdong/383/2008 (H5N1, 383) | 2.3.2.1 | 8.0 | 2.1 | 10/10 | 0/10 | 0/10 | |
| Re6-7-8 | 5.7 | 5.8 | 10/10 | 0/10 | 0/10 | ||||
| Control | 3.2 | 0 | 0/10 | N/a | N/a | ||||
| 3 | rBacH5Con5Mut | A/Duck/Shandong/147/2013 (H5N2, 147) | 7.2 | 8.7 | 1.6 | 10/10 | 1/10 | 6/10 | |
| Re6-7-8 | 6.2 | 6.3 | 10/10 | 1/10 | 0/10 | ||||
| Control | 3.3 | 0 | 0/10 | 2/2 | 2/2 | ||||
| Vaccination | Challenge virus | Clade | Days of age | HI titer with different antigens (log2)a | Survival/total | Virus shedding from chickens at five days post challenge | |||
| rBacH5 | CVb | Oropharyngeal | Cloacal | ||||||
| rBacH5Con5Mut | A/Goose/Guangdong/079/2013 (H5N1, 14079) | 2.3.4.4 | 1 | 9.1 | 2.5 | 10/10 | 0/10 | 0/10 | |
| Challenge Control | 1 | 2 | 0 | 0/10 | N/ac | N/a | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




