1. Introduction
Lung cancer remains among the most widespread and malignant tumours globally, representing the leading cause of cancer-related mortality [
1]. Recent years have witnessed ground-breaking advancements in cancer treatment with immune checkpoint inhibitors (ICIs), sparking growing interest [
2,
3]. Nonetheless, ICIs do not uniformly produce positive outcomes for all patients. Response rates to ICIs can differ significantly among patients due to their unique molecular, histological, or genetic profiles. It is important to note that durable benefits from ICIs are observed in only approximately 20-30% of patients diagnosed with non-small cell lung cancer (NSCLC) [
4,
5].
Despite the presence of biomarkers like PD-L1 expression, tumour mutation burden (TMB), and microsatellite instability, which can predict the effectiveness of ICIs, nearly half of the patients do not benefit from ICIs even when these biomarkers indicate a positive result [
5,
6,
7]. However, the utility of biomarkers is limited due to factors such as invasiveness, limited tissue availability, and intratumoral heterogeneity [
8]. Radiographic images offer a comprehensive assessment of lung lesions and the surrounding environment. Moreover, they possess the advantages of being low risk and non-invasive. Hence, there is a need to explore additional prognostic factors for patients undergoing ICIs treatment.
Recently, artificial intelligence techniques have gained considerable attention in the medical and healthcare sectors, demonstrating significant potential to improve medical services across various domains such as diagnosis, risk analysis, lifestyle monitoring, and beyond [
9,
10].
While several studies have showcased the efficacy of radiomics and deep learning models in lung cancer, few have focused on utilizing time-series CT images to develop models. Additionally, there is a scarcity of articles applying artificial intelligence specifically for cancer ICIs in lung cancer [
11,
12].
Conventional deep learning methods for CT scan processing are mainly based on recurrent neural networks (RNNs) [
13] and convolutional neural networks (CNNs) [
14], where RNNs involve representing features along the time dimension and CNNs involve modelling the spatial features. However, RNNs are constrained by their forgetting mechanism, while CNNs benefit from spatial locality and translation invariance. However, neither RNNs nor CNNs effectively capture inter-feature connections. When using CNNs to extract features from a series of CT scans, weight sharing and the spatial locality inherent in CNNs can lead to incorrect inferences on out-of-distribution data, as focus points may vary. Moreover, regardless of the model or expertise, multiple CT scans must typically be aggregated for accurate diagnosis.
Based on the described analysis, the hope of the present authors is for the model to dynamically learn the significance of both temporal and spatial features without overlooking any critical aspect. This led to the use of a self-attention-based approach for feature extraction and modelling of CT scan sequences [
15]. Originating from transformers, self-attention, as described in Equation 1, aims to capture correlations between features, ensuring comprehensive representation of each feature in the sequence.
where
,
and
represents the feature matrix multiplied with respective weight matrices.
is the dimension of the feature matrix while
being the transpose operation. Recently, there has been significant research interest focused on computer vision [
16,
17,
18]. In the field of video processing, Video Vision Transformer (ViViT) [
19] is the first deep learning model based on multi-head self-attention mechanism, fusing both temporal and spatial features in the global context.
The aim of the present study was to uncover how deep learning can enable a connection between serial CT images and clinical outcomes among NSCLC patients undergoing ICIs.
2. Materials and Methods
2.1. Patient Cohort
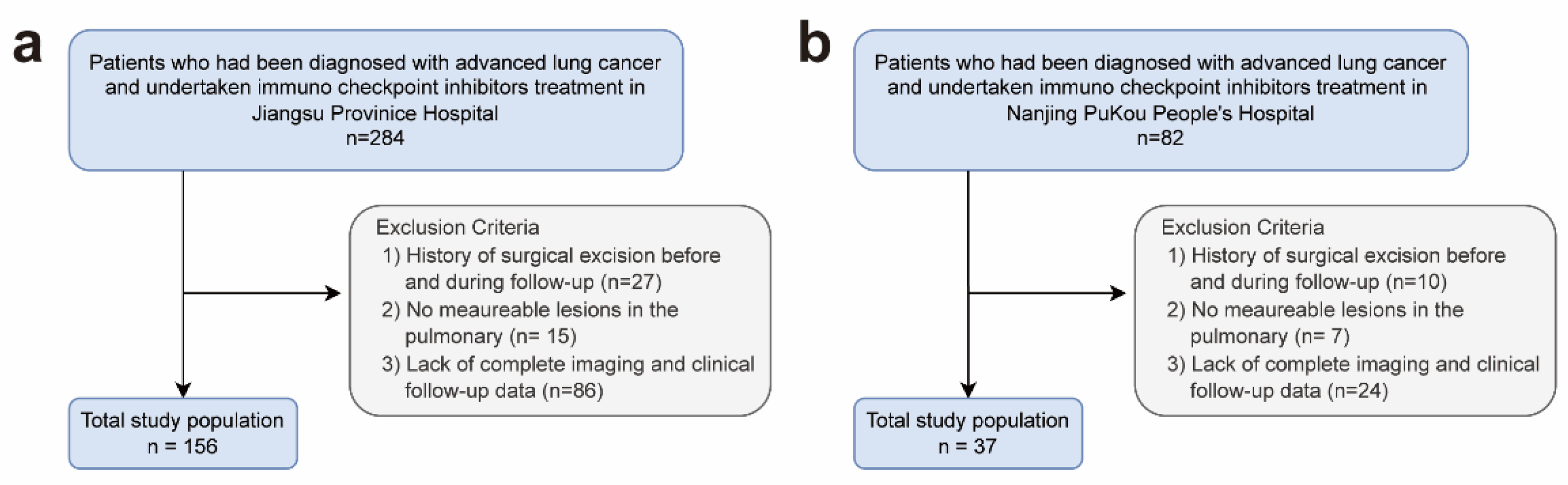
Retrospectively, a series of CT images and clinical follow-up information were collected for 156 patients (comprising a total of 684 CT images) at Jiangsu Province Hospital (the first affiliated hospital with Nanjing Medical University) during the period from December 2018 to July 2022. All eligible patients underwent initial chest CT scans up to 8 weeks before commencing ICIs, with subsequent follow-up occurring at intervals of 6 to 12 weeks in accordance with National Comprehensive Cancer Network guidelines. These images were then randomly assigned to three groups: a training cohort (n=496), a validation cohort (n=64), and a test cohort (n=64). Due to practical limitations, not all patients could adhere to the standard review schedule, necessitating an extended follow-up period by 2 weeks before and after the standard review time. Additionally, external validation data were retrospectively collected from 37 patients (comprising 37 CT images and corresponding clinical follow-up data) at Nanjing Pukou People's Hospital between July 2019 and July 2023.
Clinical data, encompassing age, gender, stage, histology type, smoking status, and progression-free survival (PFS), were extracted from medical records. All patients underwent restaging in accordance with the eighth edition criteria of the American Joint Committee on Cancer [
20]. Post-treatment tumour response to ICIs was evaluated following with RECIST version 1.1 [
21]. Disease progression was defined as either a 20% increase in the volume of targeted lesions or the appearance of new lesions. The primary endpoint of the study is PFS, measuring the duration from the start of ICIs to either disease progression or death. Instances involving death, loss to follow-up, or absence of outcome occurrence were appropriately censored.
In the present study, patients with PFS greater than 7 months were categorized as the ICIs responded group, while those with PFS less than or equal to 7 months were classified as the ICIs non-responded group. Progression was determined through imaging reports indicating tumour growth or the appearance of new disease sites, coupled with assessments conducted by the treating physician. The compilation of clinical data was finalized for outcome analysis on September 10, 2020.
The flow chart in
Figure 1 illustrates the patient selection process:
The admission criteria for patients included:
- (1)
Histologically confirmed primary lung cancer.
- (2)
Patients diagnosed with stage III or IV lung cancer.
- (3)
Patients received first-line treatment with either ICIs monotherapy (200 mg every 3 weeks at the approved dose) or a combination of ICIs and chemotherapy.
- (4)
Endpoint events and status were clearly recorded.
- (5)
A full set of pre-treatment and three consecutive follow-up CT scans.
- (6)
Comprehensive clinical data, encompassing gender, age, smoking status, TNM stage, pathological type, and tumour treatment regimen.
Exclusion criteria encompassed patients who fulfilled any of the subsequent criteria:
- (1)
History of surgical excision before ICIs and during follow-up.
- (2)
No measurable lesions in the pulmonary window according to RECIST 1.1 criteria.
Figure 1.
Flow chart of patient recruitment.
Figure 1.
Flow chart of patient recruitment.
2.2. Kaplan-Meier Analysis
KM curves were employed to depict the PFS rates at Jiangsu Province Hospital and Nanjing Pukou People's Hospital. Differences in survival curves were compared using the log-rank test.
2.3. CT Scans Acquisition and Selection
- (1)
The patients underwent scans using CT machines including SIEMENS SOMATOM Definition AS+, SIEMENS SOMATOM Definition Force, SIEMENS SOMATOM go.Up, SIEMENS Emotion 16, GE MEDICAL SYSTEMS Revolution CT, GE MEDICAL SYSTEMS Optima CT520 Series, and Philips iCT. The pulmonary window CT images had a section thickness of 1.5 mm.
- (2)
The images were pre-processed to manually exclude the layers of the cervical spine and abdomen.
- (3)
Image pre-processing and data enhancement.
Each CT image was resized to 320×320 pixels, and the CT images for each patient were standardized to ensure consistent feedforward into the neural network. Due to the limited sample size, random data augmentation was applied to each CT image in the series to enhance data diversity, alleviate the overfitting problem, and improve the model's generalization. The augmentation techniques included order shuffling, flipping, edge clipping, and adding Gaussian noise, as depicted in
Figure 2.
2.4. Model Construction
Leveraging the concept of a global self-attention mechanism, the ViViT model was adeptly adapted to classify the treatment response of ICIs through analysis of an individual's 3D CT volume. The proposed deep learning approach is depicted in
Figure 3. The methodology involves utilizing a series of 3D CT scans as input, subsequently generating binary prognosis outcomes: favourable and unfavourable. For the present analysis, 3D CT scans acquired via a CT scanner were employed.
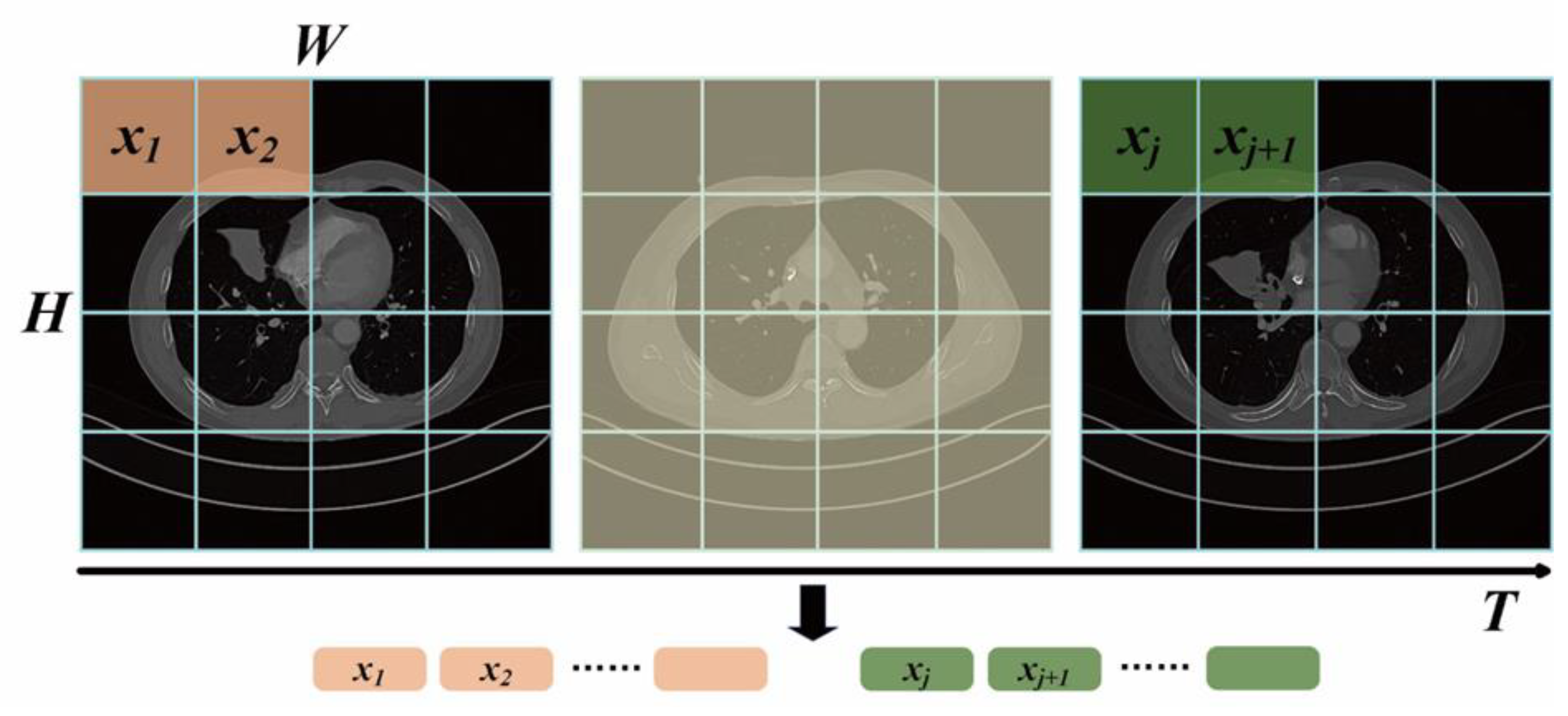
After random data augmentation, as described in
Section 4(c), the 3D CT scan series was input into the ViViT model. Initially, features of the CT scans are embedded as depicted in
Figure 4. The scan series from
need to be mapped to the token series
. The input series
is sampled uniformly for the
scan image, where the same tokenizing method with Vision Transformer is applied to each scan image, that is, convolution is used to downsample grids of each scan image. These tokens are concatenated to form the token series
. Additionally, due to the loss of spatial information after feeding into ViViT, learnable position embeddings are added to the token series
, and a classification token is appended
at the end to capture global token features. Then, the token series
is reshaped with position information from
to
, where the first three dimensions are flattened. This completes the procedure of tokenizing the scan series.
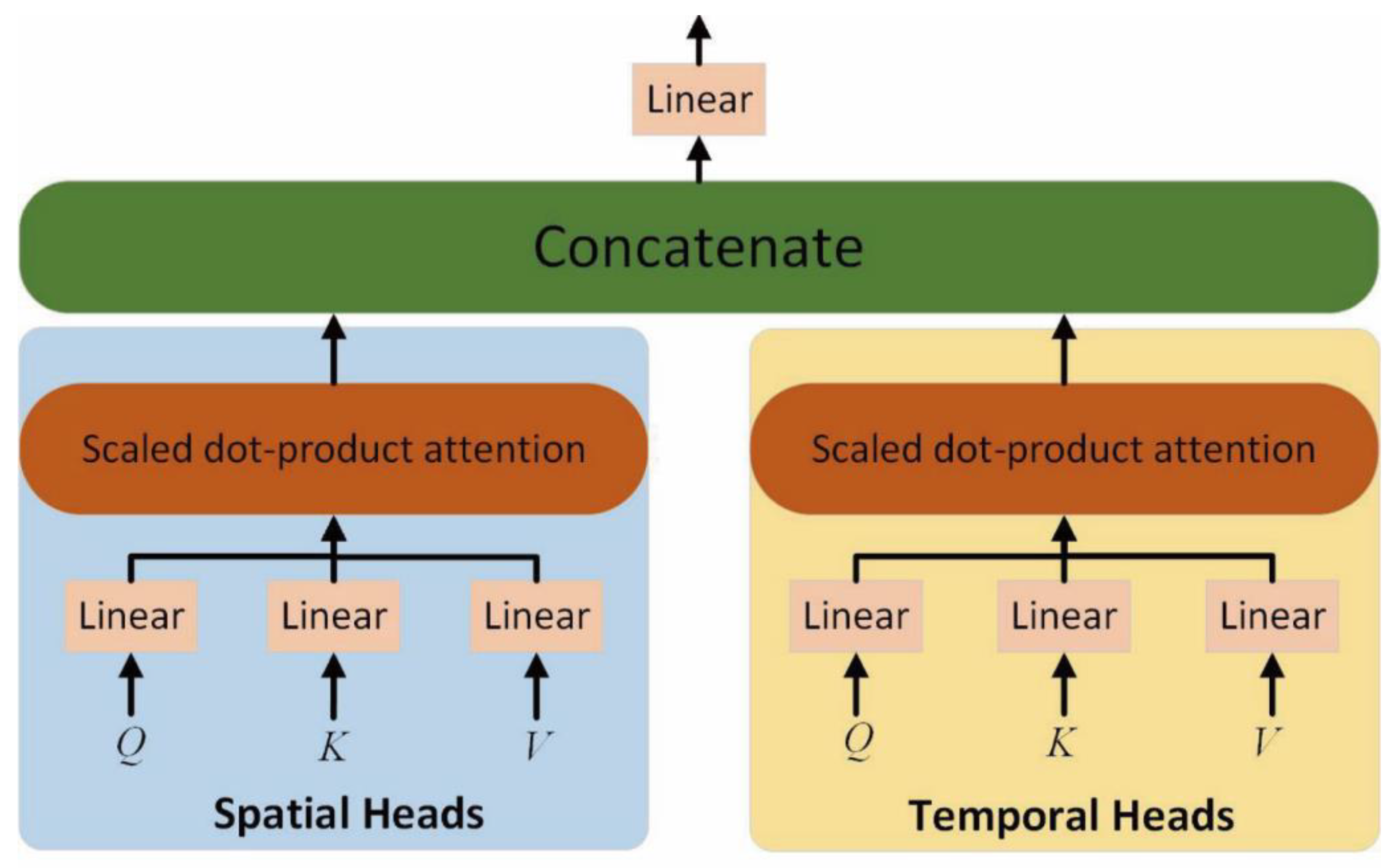
After obtaining the token sequence of CT scan series with position embeddings, the factorized self-attention encoder is adopted to model the spatial and temporal features. As shown in
Figure 5, the factorized self-attention encoder decouples the spatial and temporal self-attention heads. The self-attention weights of spatial and temporal features are calculated separately by multi-head operations. Each head module contains multiple matrices of queries
, keys
and values
, as denoted in Equations (2)–(4), which are the linear projections of
.
where the shape of
,
and
is
.and
.
As presented in
Figure 4, in addition to the learnable query matrix
, for spatial heads, the key and value of the spatial dimension are constructed, where
,
. For the temporal head, the key and value of the temporal dimension are
,
. Therefore, the spatial and temporal features with self-attention mechanisms can be obtained as denoted in Equations (5) and (6):
where spatial and temporal features occupy half of the self-attention heads, respectively.
Finally, the features of spatial self-attention and temporal self-attention can be concatenated as denoted in Equation (7):
After the operations of multi-head
self-attention, layer normalization and fully connected layers are adopted as
the feedforward module. Through the stack of L ViViT modules, a classification head is connected to output the
result of prognosis. During the training procedure, label smoothing is adopted
to alleviate the over-confidence of the model.
Figure 5.
Factorized dot-product self-attention encoder.
Figure 5.
Factorized dot-product self-attention encoder.
2.5. Performance of ViViT Model
The output of the classification model was determined as the predicted probability of response risk to ICIs. To evaluate the model's ability to differentiate between responders and non-responders, we utilized Receiver Operating Characteristic (ROC) curves. The Area Under the Curve (AUC) was computed to compare performance across different cohorts. Additionally, the confusion matrix of our model was analysed to further assess performance in the training, validation, test, and external validation cohorts.
2.6. Statistics Analysis
Statistical analyses were executed utilizing Python 3.11.4 and R 4.2.3 software. All statistical tests were evaluated using a two-sided significance level, with a significance threshold established at 0.05. Patient stratification into distinct risk groups was assessed through Kaplan–Meier analysis and log-rank testing. The confusion matrix module from the sklearn.metrics package was employed to compute the confusion matrix.
3. Results
3.1. Characteristics of Participants
The overall patient cohort consisted of 195 individuals diagnosed with advanced non-small cell lung cancer (NSCLC). Of these, 156 patients were recruited from Jiangsu Provincial People's Hospital, and an additional 37 patients were from Nanjing Pukou People's Hospital. The training, internal validation, test, and external validation cohorts included 496, 64, 64, and 37 CT scans, respectively, with each person having approximately 200 CT images per scan. Among the 156 patients from Jiangsu Provincial People’s Hospital, 127 (81.4%) were male and 29 (18.6%) were female. Histologically, there were 71 cases of squamous cell carcinoma (45.5%), 83 cases of adenocarcinoma (53.2%), and 2 cases of other lung cancer types (1.2%). Of these patients, 84 (53.8%) were classified as responders to ICIs, and 72 (46%) were classified as non-responders. Regarding histological subgroups, the dataset included adenocarcinoma (ADC), squamous cell carcinoma (SCC), and a category labelled as “Other.” In the cohort from Jiangsu Provincial People’s Hospital, stage IV was the predominant tumour pathological stage (69.2%), and adenocarcinoma (ADC) was the most common histological type (50.0%). No significant differences were observed in terms of gender, age, histological subtype, pathological stage, or smoking status between the groups that responded to ICIs and those that did not.
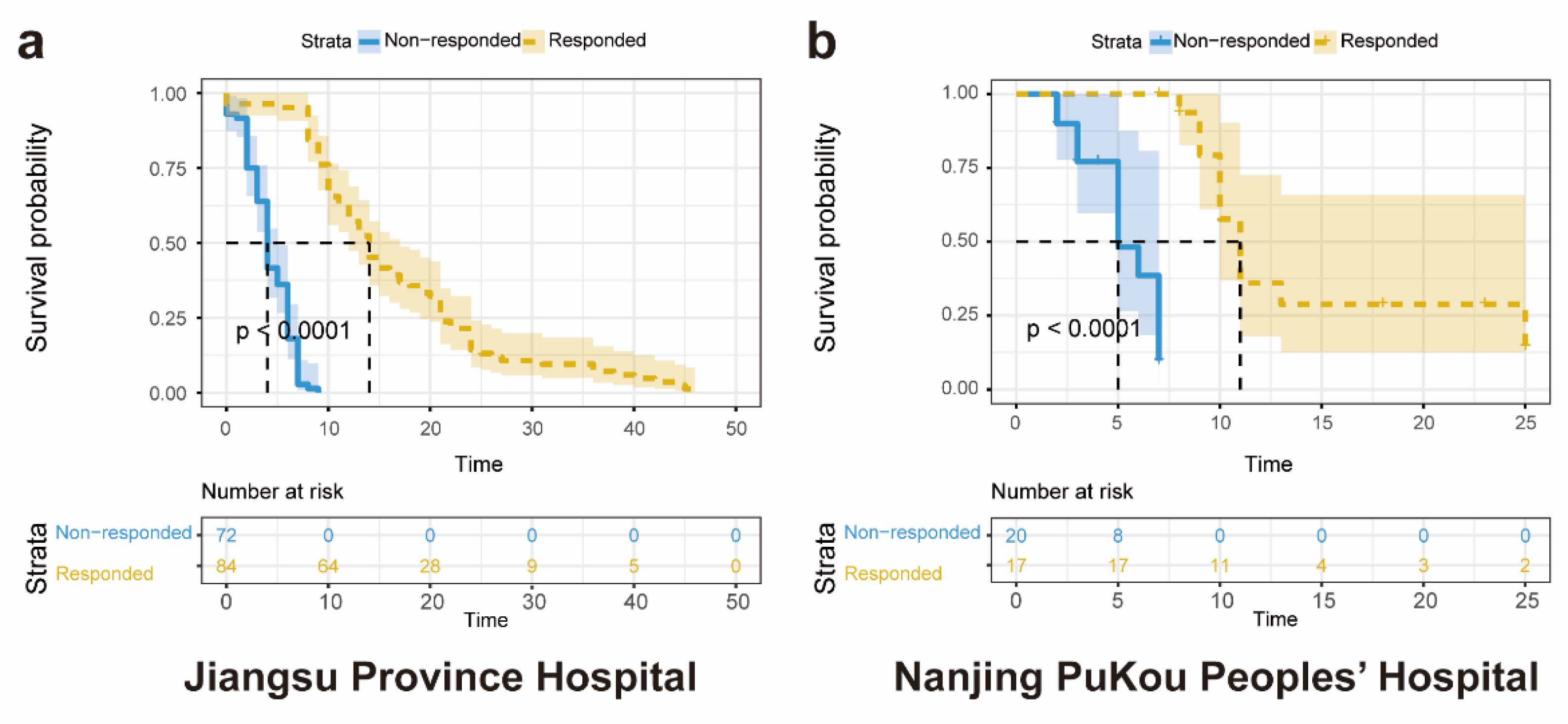
3.2. Kaplan-Meier Analysis of Participants
Analysis was conducted involving a cohort of 156 patients diagnosed with advanced NSCLC from Jiangsu Province Hospital, along with 624 corresponding CT scans. The Kaplan-Meier analysis, stratified by predicted response to ICIs, demonstrated statistically significant differences in overall survival between the responded and non-responded groups (log-rank test, P < 0.01) (
Figure 6A). Median survivals were reported as 14 months for the responded group and 4 months for the non-responded group. In the external validation set, Kaplan-Meier analysis comparing the ICIs responded and non-responded groups showed statistically significant differences in overall survival (log-rank test, P < 0.01) (Fig. 6B). Median survivals were observed as 11 months for the responded group and 7 months for the non-responded group.
3.3. Experiment Settings
The dataset was split into a training set, a validation set, and a test set based on the ratio of 80%, 10% and 10%. The images were resized into 320 × 320 pixels and the model was trained on the training set with the batch size of 2. An initial learning rate of 1e-3 was set with the cosine scheduler. AdamW [
22] was adopted as the optimizer with a weight decay of 5e-4. Cross entropy was used as the loss function (Equation 8).
where
is the label and
is the prediction value.
3.4. Performance of the ViViT Model
3.4.1. ROC curve
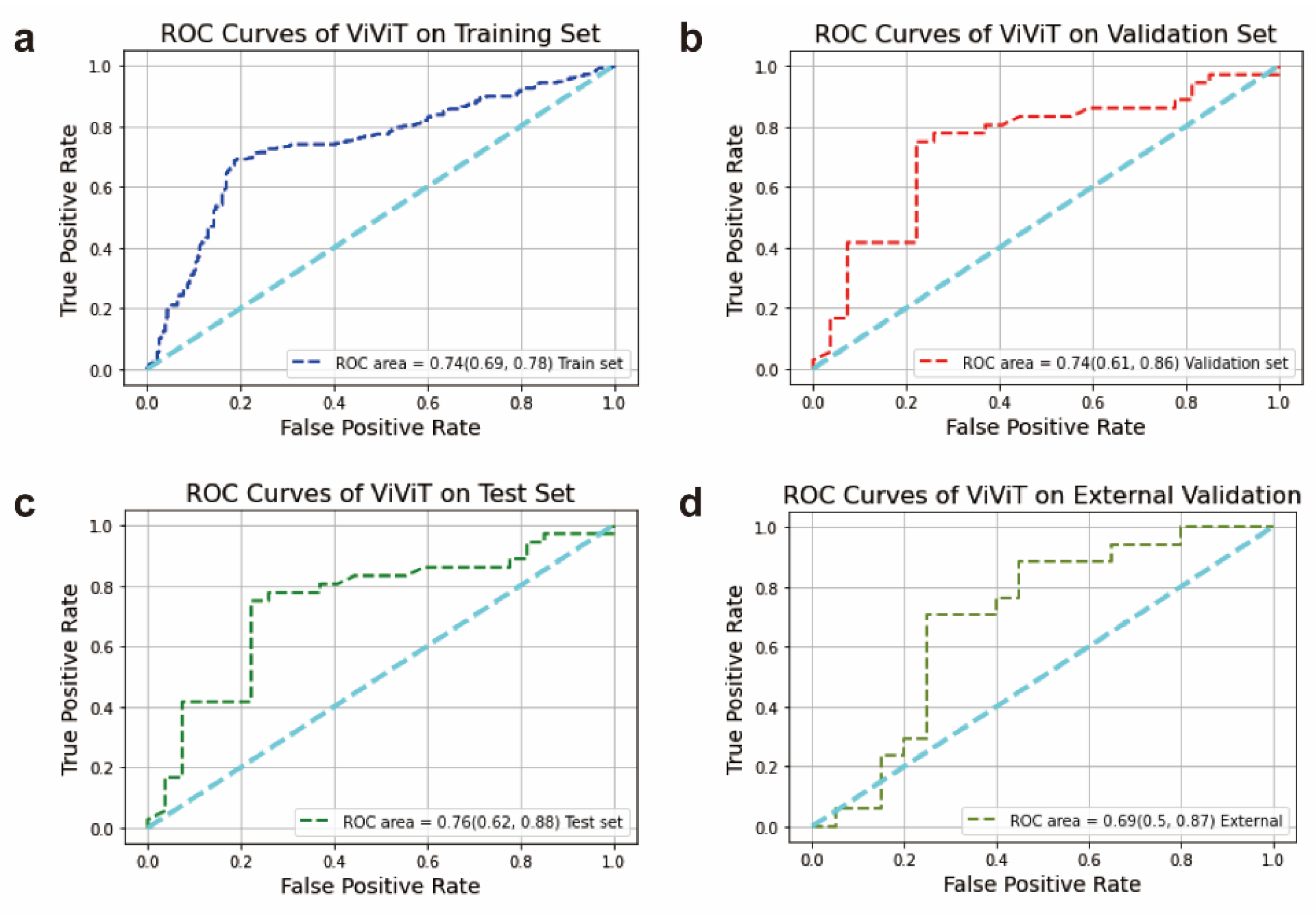
A deep learning model was formulated to anticipate patients' likelihood of progression after immune checkpoint inhibition, employing their successive chest CT images. Following image pre-processing, the CT image data were fed into the ViViT model, thereby forecasting the efficacy of ICIs. For evaluating the predictive capabilities of the models, diverse descriptive indices such as the AUC, accuracy, precision, recall, F1 score, specificity, and confusion matrix were employed. Typically, features or models exhibiting an AUC exceeding 0.60 are widely regarded as predictive in analogous studies [
23]. The ViViT model demonstrated good performance, with the AUC values for the training set, validation set, test set and external validation set being 0.74 (95% CI: 0.69 to 0.78), 0.74 (95% CI: 0.61 to 0.86), 0.76 (95% CI: 0.62 to 0.88), and 0.69 (95% CI: 0.50 to 0.87), respectively (
Figure 7a–d).
3.4.2. Confusion Matrix
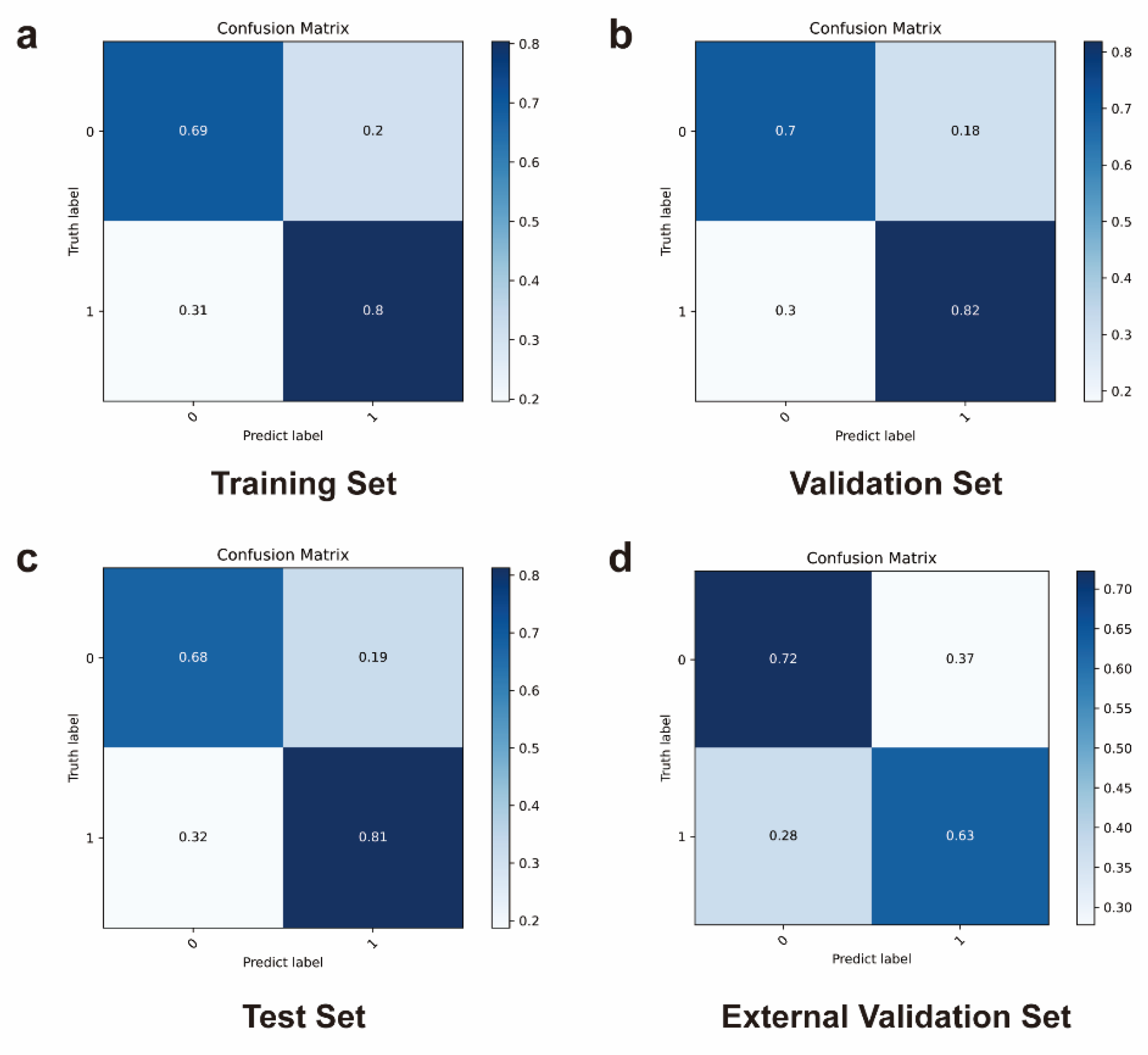
Further, for a more comprehensive assessment of the utility of deep learning in predicting outcomes based on patient images before and after ICIs therapy, the confusion matrix was also obtained.
Figure 8a–d depict the confusion matrix of the ViViT model, portraying the outcomes of the classification between the ICIs-responded and non-responded groups. For ViViT model, the overall accuracy levels in the training set, validation set, test set, and external validation set were 0.74, 0.76, 0.75, and 0.68, precision levels were 0.80, 0.82, 0.81, and 0.63, recall levels were 0.69, 0.75, 0.72, and 0.71, F1 levels were 0.74, 0.78, 0.77, and 0.67, and specificity levels were 0.80, 0.78, 0.78, and 0.65, respectively (
Table 1).
On the external validation set, the ViViT model attained an accuracy of 69%, with the AUC serving as the evaluation metric. Leveraging the evaluation metrics including AUC, accuracy, precision, recall, F1 score, and specificity, the proposed ViViT model demonstrated strong performance in the external validation set.
Table 1.
Prediction performance of ViViT model.
Table 1.
Prediction performance of ViViT model.
| |
Training set
N=496 |
Validation set
N=64 |
Test set
N=64 |
External validation set
N =37 |
| AUC |
0.74 |
0.74 |
0.76 |
0.69 |
| Accuracy |
0.74 |
0.76 |
0.75 |
0.68 |
| Precision |
0.80 |
0.82 |
0.81 |
0.63 |
| Recall |
0.69 |
0.75 |
0.72 |
0.71 |
| F1 |
0.74 |
0.78 |
0.77 |
0.67 |
| Specificity |
0.80 |
0.78 |
0.78 |
0.65 |
3.5. Performance Comparison with Slow-Fast Model
As a classical sequence classification model, Slow-Fast is based on convolutional neural networks (CNN). Compared with the proposed ViViT, Slow-Fast has the characteristics of locality, but is not capable of modelling the global contextual features of the sequence. The performance of Slow-Fast was compared with the proposed ViViT-based model on the validation and test sets. The F1 score was employed as the chosen evaluation metric, representing the harmonic mean of precision and recall.
According to the data presented in
Table 2, the proposed ViViT-based model outperformed the CNN-based Slow Fast model. This outcome underscores the crucial role of the self-attention mechanism in handling sequence data.
4. Discussion
Recently, deep learning in cancer prognostics has shown significant promise [
24]. Nonetheless, the role that conventional diagnostic modalities play in ICIs risk stratification remains inadequately comprehended.
Conventional tumour time monitoring typically relies on size metrics (for example, RECIST1.1) [[
21]. However, tumour size can be influenced by factors such as inflammation, interstitial changes, fibrosis, or pleural effusion. In response to the limitations of RECIST, deep learning models have been employed to analyse multidimensional quantitative characteristics, encompassing tumour size, shape, density, and texture patterns.
In the present study, a deep learning model that anticipates the survival of lung cancer patients receiving ICIs treatment was established, utilizing real-world data gathered from two medical institutions. ViViT demonstrated precise prediction of outcomes, showcasing commendable values in AUC, Accuracy, precision, recall, F1, and specificity across both internal and external validation cohorts.
Several studies have reported that imaging features from lesions hold prognostic value for patients undergoing ICIs [
25,
26]. Advances in AI technology now enable the extraction of quantitative information from CT images, allowing for the capture of not only visual anomalies but also underlying features such as tumour budding and vascular invasion [
27].
Multiple investigations have explored both the tumour region and the peritumoral boundary, recognizing their potential significance in understanding tumour vascularization and the tumour microenvironment [
28]. An association between lung lesions and treatment outcomes was noted, wherein about 85% of these lesions were marked as hotspots in prognostic maps generated by AI, regardless of their size [
29].
Applying a deep learning algorithm to capture image features has shown promise in predicting genetic mutations (such as PD-L1 expression, microsatellite instability or stability, EGFR mutation, and other molecular subtypes) and survival outcomes across various types of malignancies [
12,
30,
31,
32,
33]. In line with such investigations, the proposed ViViT prediction model exhibited promising predictive performance for ICIs efficacy, offering support to oncologists in primary treatment selection, surveillance planning, maintenance therapy decisions, and patient counselling regarding investigative clinical trials.
The advantages of the proposed ViViT model include the following. First, the present dataset includes CT scans obtained from two institutions, enhancing confidence in the results' generalizability.
Second, previous radiomic analyses aimed to predict the effectiveness of ICIs by correlating imaging surrogates with specific molecular biomarkers such as CD8+ T-cell infiltration, TMB expression, or PD-L1 expression [
34,
35]. However, such endeavours face the fundamental constraint that these biological attributes offer limited predictive power for ICIs response [
36]. These attributes only capture a portion of the complex and diverse molecular characteristics underlying responsiveness. Therefore, imaging surrogates for these molecular markers are unlikely to exceed the inherent predictive performance of the markers themselves.
Third, the study introduced an AI model that monitors the entire image, encompassing healthy tissue and tumours. While conventional research often begins by mapping the tumour or peritumour area, it then narrows its focus to the ROI region for subsequent analysis [
37]. Certain articles have concentrated on analysing tumour lesions to elucidate how the tumour’s growth environment impacts patient prognosis, employing factors such as vascularity, oxygenation, and metabolic activity [
38]. The proposed approach introduces a completely automated procedure that eliminates the need for time-consuming segmentations, while also delivering a comprehensive depiction of the patient's status using chest imaging. Nevertheless, this does not undermine the importance of the single-lesion approach; rather, it paves the way for multi-scale solutions. Future advancements will tackle the challenge of manual ROI segmentation by employing automatic segmentation and artificial intelligence techniques. This will enable a comprehensive assessment of the patient's condition based on chest imaging. Unlike traditional radiomics, this model operates more rapidly and conveniently, obviating the need for segmentation. These results are comparable to methods that currently rely on labour-intensive segmentation processes.
Fourth, the ViViT model was trained using a combination of multiple time points, including pre-treatment CT images and the subsequent three follow-up CT images. This approach augments the pool of training CT images and expands the subsequent validation and training sets with additional time points.
Fifth, considering that patients undergoing ICIs may experience initial pseudoprogression within the first 12 weeks, they were categorized based on PFS data instead of RECIST 1.1 criteria [
39].
Currently, the present study exhibits several limitations. Firstly, it was conducted at two centres with a limited patient cohort, and the predictive validity of the model has not yet been corroborated across a broader spectrum of centres. Further, the study was designed retrospectively, encompassing only clinical data, radiological images, post-treatment assessment of ICIs, and PFS data. This design may have introduced potential biases. Additionally, there is uncertainty regarding the predictive efficacy of the model for long-term outcomes of ICIs. Despite utilizing an extended time series for training, the optimal timing for discontinuing ICIs remains elusive. Further endeavours are dedicated to forecasting the appropriate point at which patients should discontinue their use of ICIs.
In future studies, the plan is to encompass a broader range of institutions and a larger cohort of ICIs patients. A combination of retrospective and prospective methodologies will be employed to refine and enhance the model training.
In the future, significant quantities of well-annotated multi-institutional image data will play a crucial role in advancing deep learning predictive models within the medical domain. Serving as potent biomarkers, these decision aids have the potential to be seamlessly integrated into routine clinical care. Yet, further efforts are required to enhance the interpretability of deep learning models. The establishment of an AI solution serving as a clinical decision support system is anticipated. This system will provide supplementary information to treating physicians, augmenting the available data and enhancing clinical decision-making.
5. Conclusions
In the present study, a dataset of patients with NSCLC was assembled, which was utilized to develop deep learning models for risk stratification among patients receiving ICIs treatment. The present findings suggest that analysing sequential CT images captured before and during treatment can contribute to predicting the response to ICIs. This approach presents a practical, non-invasive biomarker that holds potential for risk stratification in advanced lung cancer patients, suitable for integration into routine clinical practice.
Collectively, the present study indicates that utilizing deep learning for predicting ICIs efficacy can advance the goal of achieving precise immunotherapy for lung cancer. The extension of deep learning techniques to encompass a broader spectrum of tumour-related and other disease-associated classification challenges is anticipated.
Author Contributions
Conceptualization, Yuemin Wu, Runwei Guan and Xiao Liang; Data curation, Yuqin Jiang, Wenxin Zhou, Qi Liang, Pengpeng Zhang and Yi Chen; Formal analysis, Yuemin Wu and Runwei Guan; Funding acquisition, Renhua Guo; Investigation, Yuemin Wu; Methodology, Yuemin Wu and Runwei Guan; Project administration, Renhua Guo; Resources, Wei Zhang, Yi Chen, Jiali Xu, Jun Li, Tongfu Yu and Renhua Guo; Software, Runwei Guan; Supervision, Tongfu Yu and Renhua Guo; Validation, Yuemin Wu and Runwei Guan; Visualization, Yuemin Wu and Runwei Guan; Writing – original draft, Yuemin Wu; Writing – review & editing, Runwei Guan, Yanan Cui, Jiali Dai and Chen Zhang.
Funding
This research was funded by the National Natural Science Foundation of China (81972188, 82272669, and 82203010), the Medical Important Talents of Jiangsu Province (ZDRCA2016024), and the Cancer Foundation of China (CFCSSSQ017).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Jiangsu Province Hospital of Nanjing Medical University (No. 2023-SR-111).
Informed Consent Statement
Patient consent was waived due to retrospective nature of the study.
Data Availability Statement
Acknowledgments
The authors extend their gratitude to the data providers for their valuable contributions.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446-454. [CrossRef]
- Shankar, B.; Zhang, J.; Naqash, A.R.; Forde, P.M.; Feliciano, J.L.; Marrone, K.A.; Ettinger, D.S.; Hann, C.L.; Brahmer, J.R.; Ricciuti, B.; et al. Multisystem Immune-Related Adverse Events Associated With Immune Checkpoint Inhibitors for Treatment of Non-Small Cell Lung Cancer. JAMA Oncol 2020, 6, 1952-1956. [CrossRef]
- Osmani, L.; Askin, F.; Gabrielson, E.; Li, Q.K. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): Moving from targeted therapy to immunotherapy. Semin Cancer Biol 2018, 52, 103-109. [CrossRef]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015, 373, 1627-1639. [CrossRef]
- Shitara, K.; Özgüroğlu, M.; Bang, Y.-J.; Di Bartolomeo, M.; Mandalà, M.; Ryu, M.-H.; Fornaro, L.; Olesiński, T.; Caglevic, C.; Chung, H.C.; et al. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): a randomised, open-label, controlled, phase 3 trial. Lancet 2018, 392, 123-133. [CrossRef]
- Yu, H.; Boyle, T.A.; Zhou, C.; Rimm, D.L.; Hirsch, F.R. PD-L1 Expression in Lung Cancer. J Thorac Oncol 2016, 11, 964-975. [CrossRef]
- Ganesh, K.; Stadler, Z.K.; Cercek, A.; Mendelsohn, R.B.; Shia, J.; Segal, N.H.; Diaz, L.A. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol 2019, 16, 361-375. [CrossRef]
- Jia, Q.; Wu, W.; Wang, Y.; Alexander, P.B.; Sun, C.; Gong, Z.; Cheng, J.-N.; Sun, H.; Guan, Y.; Xia, X.; et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun 2018, 9, 5361. [CrossRef]
- van der Velden, B.H.M.; Kuijf, H.J.; Gilhuijs, K.G.A.; Viergever, M.A. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med Image Anal 2022, 79, 102470. [CrossRef]
- Chen, X.; Wang, X.; Zhang, K.; Fung, K.-M.; Thai, T.C.; Moore, K.; Mannel, R.S.; Liu, H.; Zheng, B.; Qiu, Y. Recent advances and clinical applications of deep learning in medical image analysis. Med Image Anal 2022, 79, 102444. [CrossRef]
- Tian, P.; He, B.; Mu, W.; Liu, K.; Liu, L.; Zeng, H.; Liu, Y.; Jiang, L.; Zhou, P.; Huang, Z.; et al. Assessing PD-L1 expression in non-small cell lung cancer and predicting responses to immune checkpoint inhibitors using deep learning on computed tomography images. Theranostics 2021, 11, 2098-2107. [CrossRef]
- Mu, W.; Jiang, L.; Shi, Y.; Tunali, I.; Gray, J.E.; Katsoulakis, E.; Tian, J.; Gillies, R.J.; Schabath, M.B. Non-invasive measurement of PD-L1 status and prediction of immunotherapy response using deep learning of PET/CT images. J Immunother Cancer 2021, 9. [CrossRef]
- Yan, R.; Liao, J.; Yang, J.; Sun, W.; Nong, M.; Li, F. Multi-hour and multi-site air quality index forecasting in Beijing using CNN, LSTM, CNN-LSTM, and spatiotemporal clustering. Expert Systems with Applications 2021, 169, 114513. [CrossRef]
- Nguyen, H.-T.; Nguyen, T.-O. Attention-based network for effective action recognition from multi-view video. Procedia Computer Science 2021, 192, 971-980. [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Advances in neural information processing systems 2017, 30.
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 2020.
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the Proceedings of the IEEE/CVF international conference on computer vision, 2021; pp. 10012-10022.
- Wang, W.; Xie, E.; Li, X.; Fan, D.-P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. Pvt v2: Improved baselines with pyramid vision transformer. Computational Visual Media 2022, 8, 415-424.
- Arnab, A.; Dehghani, M.; Heigold, G.; Sun, C.; Lučić, M.; Schmid, C. Vivit: A video vision transformer. In Proceedings of the Proceedings of the IEEE/CVF international conference on computer vision, 2021; pp. 6836-6846.
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin 2017, 67, 93-99. [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009, 45, 228-247. [CrossRef]
- Loshchilov, I.; Hutter, F. Decoupled weight decay regularization. arXiv preprint arXiv:1711.05101 2017.
- Coroller, T.P.; Agrawal, V.; Narayan, V.; Hou, Y.; Grossmann, P.; Lee, S.W.; Mak, R.H.; Aerts, H.J.W.L. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol 2016, 119, 480-486. [CrossRef]
- Kleppe, A.; Skrede, O.-J.; De Raedt, S.; Liestøl, K.; Kerr, D.J.; Danielsen, H.E. Designing deep learning studies in cancer diagnostics. Nat Rev Cancer 2021, 21, 199-211. [CrossRef]
- Ito, K.; Schöder, H.; Teng, R.; Humm, J.L.; Ni, A.; Wolchok, J.D.; Weber, W.A. Prognostic value of baseline metabolic tumor volume measured on 18 F-fluorodeoxyglucose positron emission tomography/computed tomography in melanoma patients treated with ipilimumab therapy. European journal of nuclear medicine and molecular imaging 2019, 46, 930-939.
- Peeken, J.C.; Bernhofer, M.; Spraker, M.B.; Pfeiffer, D.; Devecka, M.; Thamer, A.; Shouman, M.A.; Ott, A.; Nüsslin, F.; Mayr, N.A. CT-based radiomic features predict tumor grading and have prognostic value in patients with soft tissue sarcomas treated with neoadjuvant radiation therapy. Radiotherapy and Oncology 2019, 135, 187-196.
- Tanaka, M.; Yojiro Hashiguchi, M., Hideki Ueno, MD, Kazuo Hase, MD, Hidetaka Mochizuki, MD. Tumor budding at the invasive margin can predict patients at high risk of recurrence after curative surgery for stage II, T3 colon cancer. Diseases of the colon & rectum 2003, 46, 1054-1059.
- Mehta, T.S.; Raza, S. Power Doppler sonography of breast cancer: does vascularity correlate with node status or lymphatic vascular invasion? AJR. American journal of roentgenology 1999, 173, 303-307.
- Trebeschi, S.; Bodalal, Z.; Boellaard, T.N.; Tareco Bucho, T.M.; Drago, S.G.; Kurilova, I.; Calin-Vainak, A.M.; Delli Pizzi, A.; Muller, M.; Hummelink, K.; et al. Prognostic Value of Deep Learning-Mediated Treatment Monitoring in Lung Cancer Patients Receiving Immunotherapy. Front Oncol 2021, 11, 609054. [CrossRef]
- Bustos, A.; Payá, A.; Torrubia, A.; Jover, R.; Llor, X.; Bessa, X.; Castells, A.; Carracedo, Á.; Alenda, C. xDEEP-MSI: Explainable Bias-Rejecting Microsatellite Instability Deep Learning System in Colorectal Cancer. Biomolecules 2021, 11. [CrossRef]
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jäger, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 2019, 25, 1054-1056. [CrossRef]
- Wang, S.; Shi, J.; Ye, Z.; Dong, D.; Yu, D.; Zhou, M.; Liu, Y.; Gevaert, O.; Wang, K.; Zhu, Y.; et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J 2019, 53. [CrossRef]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018, 24, 1559-1567. [CrossRef]
- He, B.; Dong, D.; She, Y.; Zhou, C.; Fang, M.; Zhu, Y.; Zhang, H.; Huang, Z.; Jiang, T.; Tian, J.; et al. Predicting response to immunotherapy in advanced non-small-cell lung cancer using tumor mutational burden radiomic biomarker. J Immunother Cancer 2020, 8. [CrossRef]
- Chen, Y.; Sun, Z.; Chen, W.; Liu, C.; Chai, R.; Ding, J.; Liu, W.; Feng, X.; Zhou, J.; Shen, X.; et al. The Immune Subtypes and Landscape of Gastric Cancer and to Predict Based on the Whole-Slide Images Using Deep Learning. Front Immunol 2021, 12, 685992. [CrossRef]
- Palmeri, M.; Mehnert, J.; Silk, A.W.; Jabbour, S.K.; Ganesan, S.; Popli, P.; Riedlinger, G.; Stephenson, R.; de Meritens, A.B.; Leiser, A.; et al. Real-world application of tumor mutational burden-high (TMB-high) and microsatellite instability (MSI) confirms their utility as immunotherapy biomarkers. ESMO Open 2022, 7, 100336. [CrossRef]
- Wang, T.; She, Y.; Yang, Y.; Liu, X.; Chen, S.; Zhong, Y.; Deng, J.; Zhao, M.; Sun, X.; Xie, D.; et al. Radiomics for Survival Risk Stratification of Clinical and Pathologic Stage IA Pure-Solid Non-Small Cell Lung Cancer. Radiology 2022, 302, 425-434. [CrossRef]
- Li, J.; Qiu, Z.; Zhang, C.; Chen, S.; Wang, M.; Meng, Q.; Lu, H.; Wei, L.; Lv, H.; Zhong, W.; et al. ITHscore: comprehensive quantification of intra-tumor heterogeneity in NSCLC by multi-scale radiomic features. Eur Radiol 2023, 33, 893-903. [CrossRef]
- Hochmair, M.J.; Schwab, S.; Burghuber, O.C.; Krenbek, D.; Prosch, H. Symptomatic pseudo-progression followed by significant treatment response in two lung cancer patients treated with immunotherapy. Lung Cancer 2017, 113, 4-6. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).