Submitted:
16 July 2024
Posted:
17 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Description of Salmonella Isolates
2.2. Nanopore Sequencing Bioinformatic Pipeline Performance
2.3. Overall Distribution of Phenotypic Antimicrobial Resistance among Salmonella Isolates
2.4. Antimicrobial Resistance Genes Detected in Salmonella Isolates
2.5. Mapping Antimicrobial Resistance Genes to Plasmids
2.6. Antimicrobial Resistance Genes Significantly Associated with Serotypes
2.7. Antimicrobial Resistance Genes Associated with Salmonella Isolates from Beef and Dairy Operations
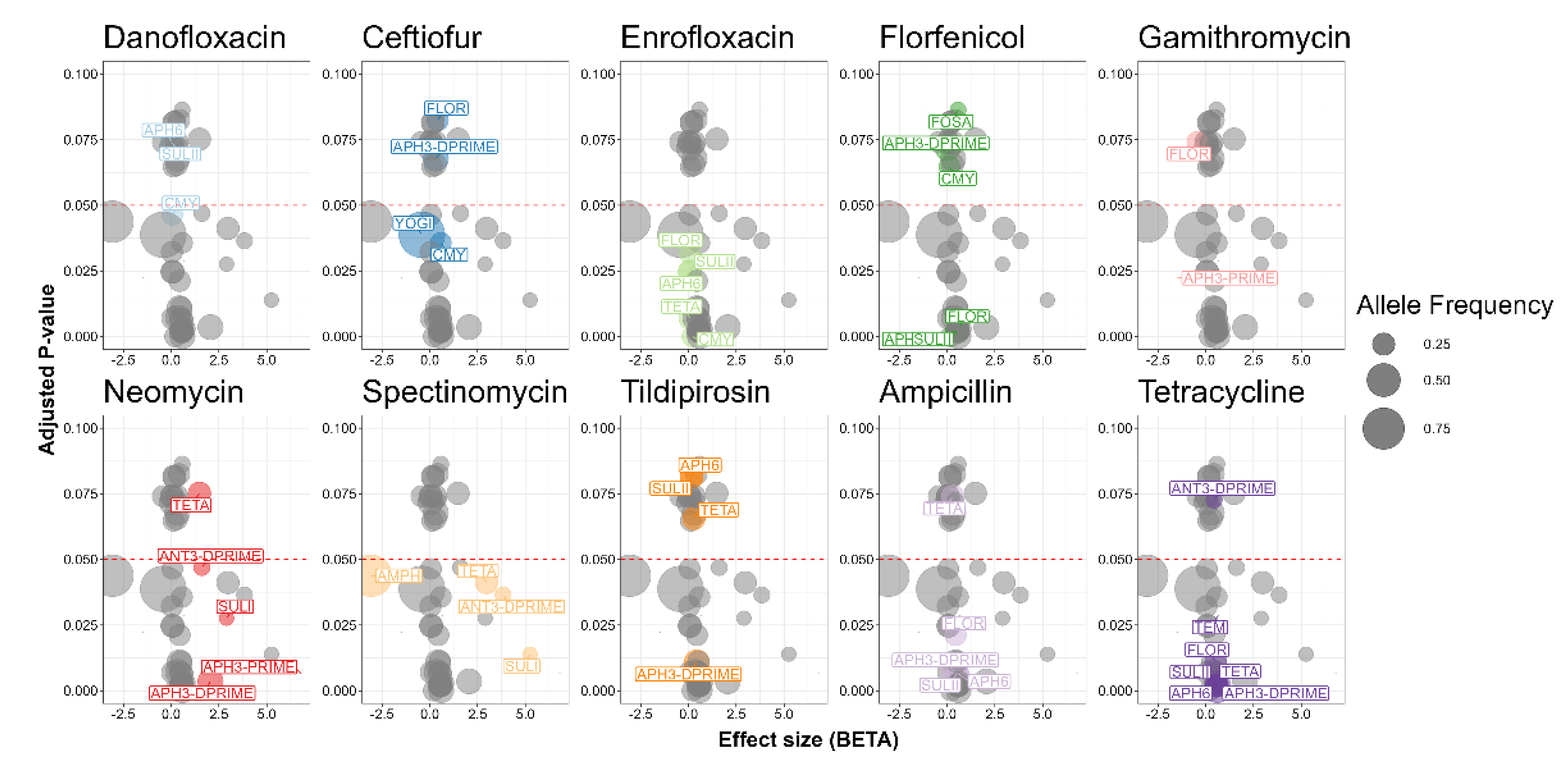
2.8. Association between Antimicrobial Resistance Genes and Antimicrobial Susceptibility Testing
2.9. Miscellaneous Associations
3. Discussion
4. Materials and Methods
4.1. Isolate Selection and Antimicrobial Susceptibility Testing
4.2. Bacteriological Culture
4.3. DNA Extraction
4.4. Whole Genome Sequencing
4.5. Bioinformatics and Statistical Analysis
5. Conclusion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- CDC. Salmonella. Available online: https://www.cdc.gov/salmonella/index.html (accessed on 05/05/2024).
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: a Meta-analysis. Appl Environ Microbiol 2019, 85. [Google Scholar] [CrossRef] [PubMed]
- Razafindrazoto, C.I.; Rakotomalala, J.A.; Randriamifidy, N.H.; Ralaizanaka, B.M.; Maherison, S.; Hasina Laingonirina, D.H.; Rakotomaharo, M.; Rasolonjatovo, A.S.; Rakotozafindrabe, A.L.R.; Rabenjanahary, T.H.; et al. Acute infectious pancreatitis due to Salmonella typhi: Case report and literature review. JGH Open 2021, 5, 1106–1107. [Google Scholar] [CrossRef] [PubMed]
- Elouali, A.; Ouerradi, N.; Ayad, G.; Babakhouya, A.; Rkain, M. Salmonella Meningitis in a Young Infant: A Case Report. Cureus 2023, 15, e44147. [Google Scholar] [CrossRef] [PubMed]
- Hull, D.M.; Harrell, E.; Harden, L.; Thakur, S. Multidrug resistance and virulence genes carried by mobile genomic elements in Salmonella enterica isolated from live food animals, processed, and retail meat in North Carolina, 2018-2019. Int J Food Microbiol 2022, 378, 109821. [Google Scholar] [CrossRef] [PubMed]
- Medalla, F.; Gu, W.; Friedman, C.R.; Judd, M.; Folster, J.; Griffin, P.M.; Hoekstra, R.M. Increased Incidence of Antimicrobial-Resistant Nontyphoidal Salmonella Infections, United States, 2004-2016. Emerg Infect Dis 2021, 27, 1662–1672. [Google Scholar] [CrossRef] [PubMed]
- Scharff, R.L. Food Attribution and Economic Cost Estimates for Meat- and Poultry-Related Illnesses. J Food Prot 2020, 83, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Response to Questions Posed by the Food Safety and Inspection Service: Enhancing Salmonella Control in Poultry Products. J Food Prot 2024, 87, 100168. [CrossRef] [PubMed]
- Tate, H.; Li, C.; Nyirabahizi, E.; Tyson, G.H.; Zhao, S.; Rice-Trujillo, C.; Jones, S.B.; Ayers, S.; M’Ikanatha N, M.; Hanna, S.; et al. A National Antimicrobial Resistance Monitoring System Survey of Antimicrobial-Resistant Foodborne Bacteria Isolated from Retail Veal in the United States. J Food Prot 2021, 84, 1749–1759. [Google Scholar] [CrossRef]
- Carroll, L.M.; Wiedmann, M.; den Bakker, H.; Siler, J.; Warchocki, S.; Kent, D.; Lyalina, S.; Davis, M.; Sischo, W.; Besser, T.; et al. Whole-Genome Sequencing of Drug-Resistant Salmonella enterica Isolates from Dairy Cattle and Humans in New York and Washington States Reveals Source and Geographic Associations. Appl Environ Microbiol 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Pornsukarom, S.; van Vliet, A.H.M.; Thakur, S. Whole genome sequencing analysis of multiple Salmonella serovars provides insights into phylogenetic relatedness, antimicrobial resistance, and virulence markers across humans, food animals and agriculture environmental sources. BMC Genomics 2018, 19, 801. [Google Scholar] [CrossRef] [PubMed]
- Carroll, L.M.; Huisman, J.S.; Wiedmann, M. Twentieth-century emergence of antimicrobial resistant human- and bovine-associated Salmonella enterica serotype Typhimurium lineages in New York State. Sci Rep 2020, 10, 14428. [Google Scholar] [CrossRef] [PubMed]
- Nichols, M.; Gollarza, L.; Sockett, D.; Aulik, N.; Patton, E.; Francois Watkins, L.K.; Gambino-Shirley, K.J.; Folster, J.P.; Chen, J.C.; Tagg, K.A.; et al. Outbreak of Multidrug-Resistant Salmonella Heidelberg Infections Linked to Dairy Calf Exposure, United States, 2015-2018. Foodborne Pathog Dis 2022, 19, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Carroll, L.M.; Buehler, A.J.; Gaballa, A.; Siler, J.D.; Cummings, K.J.; Cheng, R.A.; Wiedmann, M. Monitoring the Microevolution of Salmonella enterica in Healthy Dairy Cattle Populations at the Individual Farm Level Using Whole-Genome Sequencing. Front Microbiol 2021, 12, 763669. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.L.; Fenton, R.J.; Moriyama, E.N.; Loy, J.D.; Moxley, R.A. Association of ISVsa3 with Multidrug Resistance in Salmonella enterica Isolates from Cattle (Bos taurus). Microorganisms 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Nickodem, C.; Arnold, A.; Gehring, K.B.; Gill, J.J.; Richeson, J.T.; Samuelson, K.L.; Scott, H.; Smith, J.; Taylor, T.; Vinasco, J.; Norman, K.N. A longitudinal study on the dynamics of Salmonella enterica prevalence and serovar composition in beef cattle feces and lymph nodes and potential contributing sources from the feedlot environment. Applied and Environmental Microbiology 2023, 89. [Google Scholar] [CrossRef] [PubMed]
- Velasquez-Munoz, A.; Castro-Vargas, R.; Cullens-Nobis, F.M.; Mani, R.; Abuelo, A. Review: Salmonella Dublin in dairy cattle. Front Vet Sci 2023, 10, 1331767. [Google Scholar] [CrossRef] [PubMed]
- Cornell University CVM. Salmonellosis: Background, Management and Control. Available online: https://www.vet.cornell.edu/animal-health-diagnostic-center/programs/nyschap/modules-documents/salmonellosis-background-management-and-control#:~:text=Overall%2C%2028%25%20of%20dairy%20farms,a%20short%20period%20of%20time. (accessed on 05/05/2024).
- Eyler, A.B.; M’Ikanatha N, M.; Xiaoli, L.; Dudley, E.G. Whole-genome sequencing reveals resistome of highly drug-resistant retail meat and human Salmonella Dublin. Zoonoses Public Health 2020, 67, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Srednik, M.E.; Morningstar-Shaw, B.R.; Hicks, J.A.; Mackie, T.A.; Schlater, L.K. Antimicrobial resistance and genomic characterization of Salmonella enterica serovar Senftenberg isolates in production animals from the United States. Front Microbiol 2022, 13, 979790. [Google Scholar] [CrossRef] [PubMed]
- Marshall, K.E.H.; Tewell, M.; Tecle, S.; Leeper, M.; Sinatra, J.; Kissler, B.; Fung, A.; Brown, K.; Wagner, D.; Trees, E.; et al. Protracted Outbreak of Salmonella Newport Infections Linked to Ground Beef: Possible Role of Dairy Cows - 21 States, 2016-2017. MMWR Morb Mortal Wkly Rep 2018, 67, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Levent, G.; Schlochtermeier, A.; Ives, S.E.; Norman, K.N.; Lawhon, S.D.; Loneragan, G.H.; Anderson, R.C.; Vinasco, J.; Scott, H.M. Population Dynamics of Salmonella enterica within Beef Cattle Cohorts Followed from Single-Dose Metaphylactic Antibiotic Treatment until Slaughter. Appl Environ Microbiol 2019, 85. [Google Scholar] [CrossRef] [PubMed]
- Manishimwe, R.; Moncada, P.M.; Bugarel, M.; Scott, H.M.; Loneragan, G.H. Antibiotic resistance among Escherichia coli and Salmonella isolated from dairy cattle feces in Texas. PLoS One 2021, 16, e0242390. [Google Scholar] [CrossRef] [PubMed]
- Levent, G.; Schlochtermeier, A.; Ives, S.E.; Norman, K.N.; Lawhon, S.D.; Loneragan, G.H.; Anderson, R.C.; Vinasco, J.; den Bakker, H.C.; Scott, H.M. High-Resolution Genomic Comparisons within Salmonella enterica Serotypes Derived from Beef Feedlot Cattle: Parsing the Roles of Cattle Source, Pen, Animal, Sample Type, and Production Period. Appl Environ Microbiol 2021, 87, e0048521. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.A.; Ossa-Trujillo, C.; Vinasco, J.; Jordan, E.R.; García Buitrago, J.A.; Hagevoort, R.; Norman, K.N.; Lawhon, S.D.; Piñeiro, J.M.; Levent, G.; Scott, H.M. Use of critically important antimicrobial classes early in life may adversely impact bacterial resistance profiles during adult years: potential co-selection for plasmid-borne fluoroquinolone and macrolide resistance via extended-spectrum beta-lactam use in dairy cattle. Lett Appl Microbiol 2021, 72, 220–224. [Google Scholar] [CrossRef] [PubMed]
- USDA. Annual State Agricultural Exports Interactive Chart. Available online: https://www.ers.usda.gov/data-products/state-agricultural-trade-data/annual-state-agricultural-exports/ (accessed on 05/05/2024).
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 6th ed.; Clinical and Laboratory Standards Institute: 2023; Volume CLSI Supplement VET01S.
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res 2020, 30, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, C.E.; Kruczkiewicz, P.; Laing, C.R.; Lingohr, E.J.; Gannon, V.P.; Nash, J.H.; Taboada, E.N. The Salmonella In Silico Typing Resource (SISTR): An Open Web-Accessible Tool for Rapidly Typing and Subtyping Draft Salmonella Genome Assemblies. PLoS One 2016, 11, e0147101. [Google Scholar] [CrossRef] [PubMed]
- Veeraraghavan, B.; Jacob, J.J.; Prakash, J.A.J.; Pragasam, A.K.; Neeravi, A.; Narasimman, V.; Anandan, S. Extensive drug resistant Salmonella enterica serovar Senftenberg carrying bla(NDM) encoding plasmid p5558 (IncA/C) from India. Pathog Glob Health 2019, 113, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Power, R.A.; Parkhill, J.; de Oliveira, T. Microbial genome-wide association studies: lessons from human GWAS. Nat Rev Genet 2017, 18, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Sul, J.H.; Martin, L.S.; Eskin, E. Population structure in genetic studies: Confounding factors and mixed models. PLoS Genet 2018, 14, e1007309. [Google Scholar] [CrossRef] [PubMed]
- Paudyal, N.; Pan, H.; Elbediwi, M.; Zhou, X.; Peng, X.; Li, X.; Fang, W.; Yue, M. Characterization of Salmonella Dublin isolated from bovine and human hosts. BMC Microbiol 2019, 19, 226. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.; Wickramasingha, D.; Abdelfattah, E.M.; Pereira, R.V.; Okello, E.; Maier, G. Prevalence of antimicrobial resistance in fecal Escherichia coli and Enterococcus spp. isolates from beef cow-calf operations in northern California and associations with farm practices. Front Microbiol 2023, 14, 1086203. [Google Scholar] [CrossRef] [PubMed]
- Berge, A.C.; Hancock, D.D.; Sischo, W.M.; Besser, T.E. Geographic, farm, and animal factors associated with multiple antimicrobial resistance in fecal Escherichia coli isolates from cattle in the western United States. J Am Vet Med Assoc 2010, 236, 1338–1344. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Iwabuchi, E.; Hasegawa, M.; Esaki, H.; Muramatsu, M.; Hirayama, N.; Hirai, K. Prevalence and molecular epidemiological characterization of antimicrobial-resistant Escherichia coli isolates from Japanese black beef cattle. J Food Prot 2013, 76, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Markland, S.; Weppelmann, T.A.; Ma, Z.; Lee, S.; Mir, R.A.; Teng, L.; Ginn, A.; Lee, C.; Ukhanova, M.; Galindo, S.; et al. High Prevalence of Cefotaxime Resistant Bacteria in Grazing Beef Cattle: A Cross Sectional Study. Front Microbiol 2019, 10, 176. [Google Scholar] [CrossRef] [PubMed]
- University of Georgia Extension. Mineral supplements for beef cattle. Available online: https://extension.uga.edu/publications/detail.html?number=B895&title=mineral-supplements-for-beef-cattle (accessed on 05/05/2024).
- Folster, J.P.; Pecic, G.; Bolcen, S.; Theobald, L.; Hise, K.; Carattoli, A.; Zhao, S.; McDermott, P.F.; Whichard, J.M. Characterization of extended-spectrum cephalosporin-resistant Salmonella enterica serovar Heidelberg isolated from humans in the United States. Foodborne Pathog Dis 2010, 7, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Mangat, C.S.; Bekal, S.; Avery, B.P.; Côté, G.; Daignault, D.; Doualla-Bell, F.; Finley, R.; Lefebvre, B.; Bharat, A.; Parmley, E.J.; et al. Genomic Investigation of the Emergence of Invasive Multidrug-Resistant Salmonella enterica Serovar Dublin in Humans and Animals in Canada. Antimicrob Agents Chemother 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Srednik, M.E.; Lantz, K.; Hicks, J.A.; Morningstar-Shaw, B.R.; Mackie, T.A.; Schlater, L.K. Antimicrobial resistance and genomic characterization of Salmonella Dublin isolates in cattle from the United States. PLoS One 2021, 16, e0249617. [Google Scholar] [CrossRef] [PubMed]
- Fricke, W.F.; Welch, T.J.; McDermott, P.F.; Mammel, M.K.; LeClerc, J.E.; White, D.G.; Cebula, T.A.; Ravel, J. Comparative genomics of the IncA/C multidrug resistance plasmid family. J Bacteriol 2009, 191, 4750–4757. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, R.L.; Fedorka-Cray, P.J.; Frye, J.G.; Meinersmann, R.J. Inc A/C plasmids are prevalent in multidrug-resistant Salmonella enterica isolates. Appl Environ Microbiol 2009, 75, 1908–1915. [Google Scholar] [CrossRef] [PubMed]
- Welch, T.J.; Fricke, W.F.; McDermott, P.F.; White, D.G.; Rosso, M.L.; Rasko, D.A.; Mammel, M.K.; Eppinger, M.; Rosovitz, M.J.; Wagner, D.; et al. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2007, 2, e309. [Google Scholar] [CrossRef] [PubMed]
- Akinyemi, K.O.; Fakorede, C.O.; Linde, J.; Methner, U.; Wareth, G.; Tomaso, H.; Neubauer, H. Whole genome sequencing of Salmonella enterica serovars isolated from humans, animals, and the environment in Lagos, Nigeria. BMC Microbiol 2023, 23, 164. [Google Scholar] [CrossRef] [PubMed]
- Raufu, I.A.; Ahmed, O.A.; Aremu, A.; Ameh, J.A.; Timme, R.E.; Hendriksen, R.S.; Ambali, A.G. Occurrence, antimicrobial resistance and whole genome sequence analysis of Salmonella serovars from pig farms in Ilorin, North-central Nigeria. Int J Food Microbiol 2021, 350, 109245. [Google Scholar] [CrossRef] [PubMed]
- Aworh, M.K.; Nilsson, P.; Egyir, B.; Owusu, F.A.; Hendriksen, R.S. Rare serovars of non-typhoidal Salmonella enterica isolated from humans, beef cattle and abattoir environments in Nigeria. PLoS One 2024, 19, e0296971. [Google Scholar] [CrossRef]
- Cummings, K.J.; Rodriguez-Rivera, L.D.; Norman, K.N.; Ohta, N.; Scott, H.M. Identification of a Plasmid-Mediated Quinolone Resistance Gene in Salmonella Isolates from Texas Dairy Farm Environmental Samples. Zoonoses Public Health 2017, 64, 305–307. [Google Scholar] [CrossRef] [PubMed]
- Slowey, R.; Kim, S.W.; Prendergast, D.; Madigan, G.; Van Kessel, J.A.S.; Haley, B.J. Genomic diversity and resistome profiles of Salmonella enterica subsp. enterica serovar Kentucky isolated from food and animal sources in Ireland. Zoonoses Public Health 2022, 69, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cloeckaert, A.; Chaslus-Dancla, E. Mechanisms of quinolone resistance in Salmonella. Vet Res 2001, 32, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci 2015, 1354, 12–31. [Google Scholar] [CrossRef] [PubMed]
- McDermott, P.F.; Tyson, G.H.; Kabera, C.; Chen, Y.; Li, C.; Folster, J.P.; Ayers, S.L.; Lam, C.; Tate, H.P.; Zhao, S. Whole-Genome Sequencing for Detecting Antimicrobial Resistance in Nontyphoidal Salmonella. Antimicrob Agents Chemother 2016, 60, 5515–5520. [Google Scholar] [CrossRef]
- Day, M.R.; Doumith, M.; Do Nascimento, V.; Nair, S.; Ashton, P.M.; Jenkins, C.; Dallman, T.J.; Stevens, F.J.; Freedman, J.; Hopkins, K.L.; et al. Comparison of phenotypic and WGS-derived antimicrobial resistance profiles of Salmonella enterica serovars Typhi and Paratyphi. J Antimicrob Chemother 2018, 73, 365–372. [Google Scholar] [CrossRef]
- Katz, T.S.; Harhay, D.M.; Schmidt, J.W.; Wheeler, T.L. Identifying a list of Salmonella serotypes of concern to target for reducing risk of salmonellosis. Front Microbiol 2024, 15, 1307563. [Google Scholar] [CrossRef] [PubMed]
- Fritz, H.M.; Pereira, R.V.; Toohey-Kurth, K.; Marshall, E.; Tucker, J.; Clothier, K.A. Salmonella enterica Serovar Dublin from Cattle in California from 1993-2019: Antimicrobial Resistance Trends of Clinical Relevance. Antibiotics (Basel) 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Campioni, F.; Vilela, F.P.; Cao, G.; Kastanis, G.; Dos Prazeres Rodrigues, D.; Costa, R.G.; Tiba-Casas, M.R.; Yin, L.; Allard, M.; Falcão, J.P. Whole genome sequencing analyses revealed that Salmonella enterica serovar Dublin strains from Brazil belonged to two predominant clades. Sci Rep 2022, 12, 10555. [Google Scholar] [CrossRef] [PubMed]
- Afema, J.A.; Mather, A.E.; Sischo, W.M. Antimicrobial Resistance Profiles and Diversity in Salmonella from Humans and Cattle, 2004-2011. Zoonoses Public Health 2015, 62, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rivera, L.D.; Moreno Switt, A.I.; Degoricija, L.; Fang, R.; Cummings, C.A.; Furtado, M.R.; Wiedmann, M.; den Bakker, H.C. Genomic characterization of Salmonella Cerro ST367, an emerging Salmonella subtype in cattle in the United States. BMC Genomics 2014, 15, 427. [Google Scholar] [CrossRef] [PubMed]
- Cummings, K.J.; Warnick, L.D.; Elton, M.; Rodriguez-Rivera, L.D.; Siler, J.D.; Wright, E.M.; Gröhn, Y.T.; Wiedmann, M. Salmonella enterica serotype Cerro among dairy cattle in New York: an emerging pathogen? Foodborne Pathog Dis 2010, 7, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Fluit, A.C. Towards more virulent and antibiotic-resistant Salmonella? FEMS Immunol Med Microbiol 2005, 43, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mølbak, K. Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin Infect Dis 2005, 41, 1613–1620. [Google Scholar] [CrossRef]
- Burciaga, S.; Trachsel, J.M.; Sockett, D.; Aulik, N.; Monson, M.S.; Anderson, C.L.; Bearson, S.M.D. Genomic and phenotypic comparison of two variants of multidrug-resistant Salmonella enterica serovar Heidelberg isolated during the 2015-2017 multi-state outbreak in cattle. Front Microbiol 2023, 14, 1282832. [Google Scholar] [CrossRef] [PubMed]
- McMillan, E.A.; Weinroth, M.D.; Frye, J.G. Increased Prevalence of Salmonella Infantis Isolated from Raw Chicken and Turkey Products in the United States Is Due to a Single Clonal Lineage Carrying the pESI Plasmid. Microorganisms 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, M.; Heider, L.C.; Stryhn, H.; McClure, J.T.; Léger, D.; Rizzo, D.; Dufour, S.; Roy, J.P.; Kelton, D.F.; Renaud, D.L.; et al. Frequency of isolation and phenotypic antimicrobial resistance of fecal Salmonella enterica recovered from dairy cattle in Canada. J Dairy Sci 2024, 107, 2357–2373. [Google Scholar] [CrossRef] [PubMed]
- Aleri, J.W.; Sahibzada, S.; Harb, A.; Fisher, A.D.; Waichigo, F.K.; Lee, T.; Robertson, I.D.; Abraham, S. Molecular epidemiology and antimicrobial resistance profiles of Salmonella isolates from dairy heifer calves and adult lactating cows in a Mediterranean pasture-based system of Australia. J Dairy Sci 2022, 105, 1493–1503. [Google Scholar] [CrossRef] [PubMed]
- Noyes, N.R.; Yang, X.; Linke, L.M.; Magnuson, R.J.; Cook, S.R.; Zaheer, R.; Yang, H.; Woerner, D.R.; Geornaras, I.; McArt, J.A.; et al. Characterization of the resistome in manure, soil and wastewater from dairy and beef production systems. Sci Rep 2016, 6, 24645. [Google Scholar] [CrossRef] [PubMed]
- Carey, A.M.; Capik, S.F.; Giebel, S.; Nickodem, C.; Piñeiro, J.M.; Scott, H.M.; Vinasco, J.; Norman, K.N. Prevalence and Profiles of Antibiotic Resistance Genes mph(A) and qnrB in Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli Isolated from Dairy Calf Feces. Microorganisms 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Pradhan, A.K.; Karns, J.S.; Hovingh, E.; Wolfgang, D.R.; Vinyard, B.T.; Kim, S.W.; Salaheen, S.; Haley, B.J.; Van Kessel, J.A.S. Age-Associated Distribution of Antimicrobial-Resistant Salmonella enterica and Escherichia coli Isolated from Dairy Herds in Pennsylvania, 2013-2015. Foodborne Pathog Dis 2019, 16, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.V.; Siler, J.D.; Ng, J.C.; Davis, M.A.; Grohn, Y.T.; Warnick, L.D. Effect of on-farm use of antimicrobial drugs on resistance in fecal Escherichia coli of preweaned dairy calves. J Dairy Sci 2014, 97, 7644–7654. [Google Scholar] [CrossRef] [PubMed]
- Hille, K.; Ruddat, I.; Schmid, A.; Hering, J.; Hartmann, M.; von Münchhausen, C.; Schneider, B.; Messelhäusser, U.; Friese, A.; Mansfeld, R.; et al. Cefotaxime-resistant E. coli in dairy and beef cattle farms-Joint analyses of two cross-sectional investigations in Germany. Prev Vet Med 2017, 142, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.A.; Hancock, D.D.; Besser, T.E.; Daniels, J.B.; Baker, K.N.; Call, D.R. Antimicrobial resistance in Salmonella enterica serovar Dublin isolates from beef and dairy sources. Vet Microbiol 2007, 119, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Lipkens, Z.; Piepers, S.; De Vliegher, S. Impact of Selective Dry Cow Therapy on Antimicrobial Consumption, Udder Health, Milk Yield, and Culling Hazard in Commercial Dairy Herds. Antibiotics (Basel) 2023, 12. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.; Piepers, S.; Supré, K.; Dewulf, J.; De Vliegher, S. Quantification of antimicrobial consumption in adult cattle on dairy herds in Flanders, Belgium, and associations with udder health, milk quality, and production performance. J Dairy Sci 2016, 99, 2118–2130. [Google Scholar] [CrossRef] [PubMed]
- Tello, M.; Ocejo, M.; Oporto, B.; Hurtado, A. Prevalence of Cefotaxime-Resistant Escherichia coli Isolates from Healthy Cattle and Sheep in Northern Spain: Phenotypic and Genome-Based Characterization of Antimicrobial Susceptibility. Appl Environ Microbiol 2020, 86. [Google Scholar] [CrossRef]
- USDA. Can antibiotics be used in cattle raising? Available online: https://ask.usda.gov/s/article/Can-antibiotics-be-used-in-cattle-raising#:~:text=Antibiotics%20may%20be%20given%20to,can%20exit%20the%20animal’s%20system. (accessed on 05/05/2024).
- Rovira, P.; McAllister, T.; Lakin, S.M.; Cook, S.R.; Doster, E.; Noyes, N.R.; Weinroth, M.D.; Yang, X.; Parker, J.K.; Boucher, C.; et al. Characterization of the Microbial Resistome in Conventional and “Raised Without Antibiotics” Beef and Dairy Production Systems. Front Microbiol 2019, 10, 1980. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wei, X.; Wu, L.; Shang, X.; Cheng, F.; Li, B.; Zhou, X.; Zhang, J. The occurrence of antibiotic resistance genes in the microbiota of yak, beef and dairy cattle characterized by a metagenomic approach. J Antibiot (Tokyo) 2021, 74, 508–518. [Google Scholar] [CrossRef] [PubMed]
- Warburton, D.W.; Bowen, B.; Konkle, A.; Crawford, C.; Durzi, S.; Foster, R.; Fox, C.; Gour, L.; Krohn, G.; LaCasse, P.; et al. A comparison of six different plating media used in the isolation of Salmonella. Int J Food Microbiol 1994, 22, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Institution, B.S. Methods for microbiological examination of food and animal feeding stuffs: Part 4. Detection of Salmonella:. British Standards Institution: 1990.
- Arroyo, G.; Arroyo, J.A. Efficiency of different enrichment and isolation procedures for the detection of Salmonella serotypes in edible offal. J Appl Bacteriol 1995, 79, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A.; McAllister, T.A. Antimicrobial usage and resistance in beef production. J Anim Sci Biotechnol 2016, 7, 68. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Silley, P.; Simjee, S.; Woodford, N.; van Duijkeren, E.; Johnson, A.P.; Gaastra, W. Editorial: assessing the antimicrobial susceptibility of bacteria obtained from animals. J Antimicrob Chemother 2010, 65, 601–604. [Google Scholar] [CrossRef] [PubMed]
- D’Amato, R.F.; Hochstein, L.; Vernaleo, J.R.; Cleri, D.J.; Wallman, A.A.; Gradus, M.S.; Thornsberry, C. Evaluation of the BIOGRAM antimicrobial susceptibility test system. J Clin Microbiol 1985, 22, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Fader, R.C.; Weaver, E.; Fossett, R.; Toyras, M.; Vanderlaan, J.; Gibbs, D.; Wang, A.; Thierjung, N. Multilaboratory study of the Biomic automated well-reading instrument versus MicroScan WalkAway for reading MicroScan antimicrobial susceptibility and identification panels. J Clin Microbiol 2013, 51, 1548–1554. [Google Scholar] [CrossRef] [PubMed]
- Fukasawa, Y.; Ermini, L.; Wang, H.; Carty, K.; Cheung, M.S. LongQC: A Quality Control Tool for Third Generation Sequencing Long Read Data. G3 (Bethesda) 2020, 10, 1193–1196. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Nanoporetech. Medaka. Available online: https://github.com/nanoporetech/medaka (accessed on 07/05/2023).
- Huang, Y.T.; Liu, P.Y.; Shih, P.W. Homopolish: a method for the removal of systematic errors in nanopore sequencing by homologous polishing. Genome Biol 2021, 22, 95. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol Biol 2019, 1962, 227–245. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Snippy.
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Abricate. Available online: https://github.com/tseeman/abricate (accessed on 07/05/2024).
- Doster, E.; Lakin, S.M.; Dean, C.J.; Wolfe, C.; Young, J.G.; Boucher, C.; Belk, K.E.; Noyes, N.R.; Morley, P.S. MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res 2020, 48, D561–D569. [Google Scholar] [CrossRef] [PubMed]
- Brynildsrud, O.; Bohlin, J.; Scheffer, L.; Eldholm, V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol 2016, 17, 238. [Google Scholar] [CrossRef] [PubMed]
- Lees, J.A.; Galardini, M.; Bentley, S.D.; Weiser, J.N.; Corander, J. pyseer: a comprehensive tool for microbial pangenome-wide association studies. Bioinformatics 2018, 34, 4310–4312. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [PubMed]
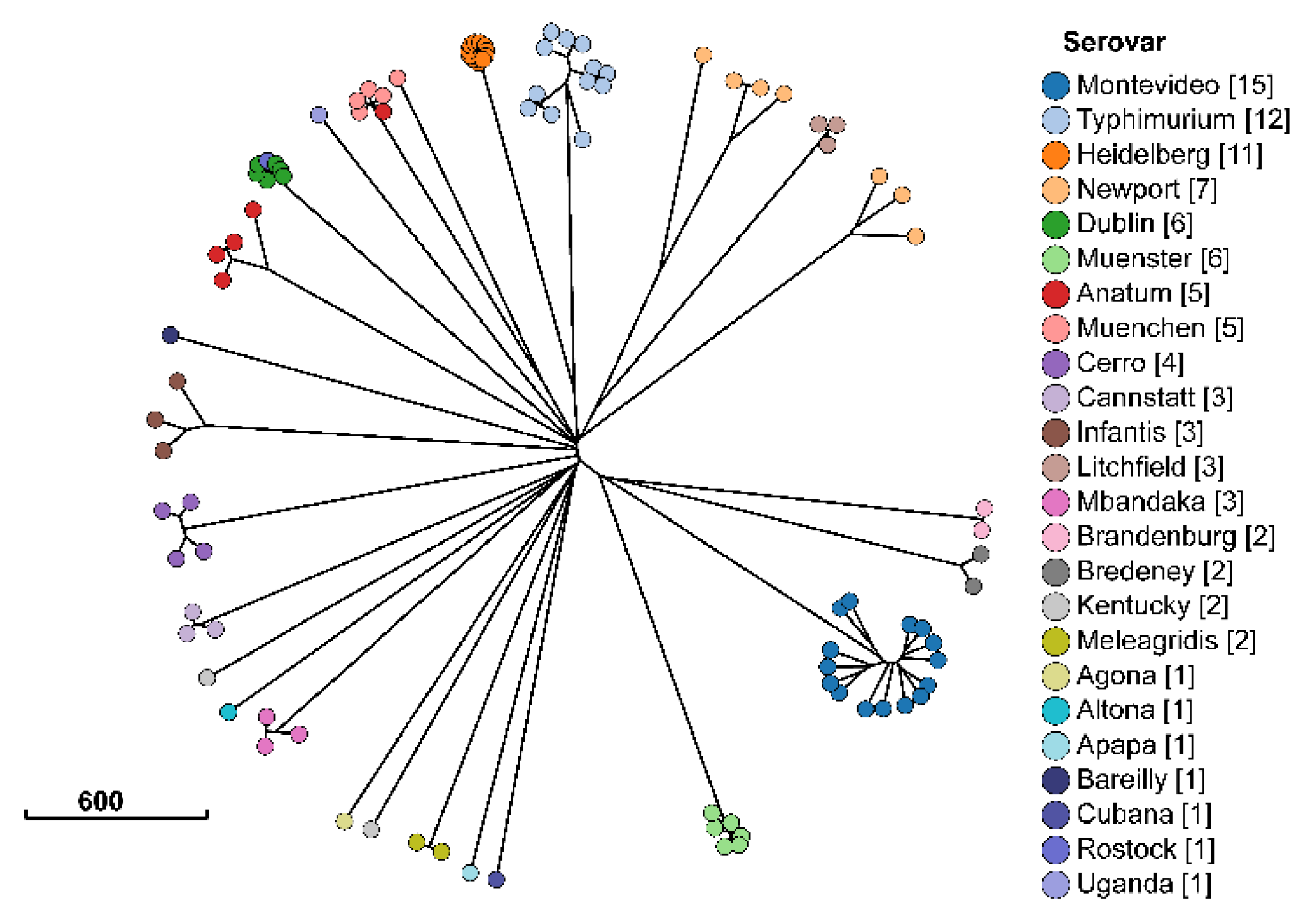
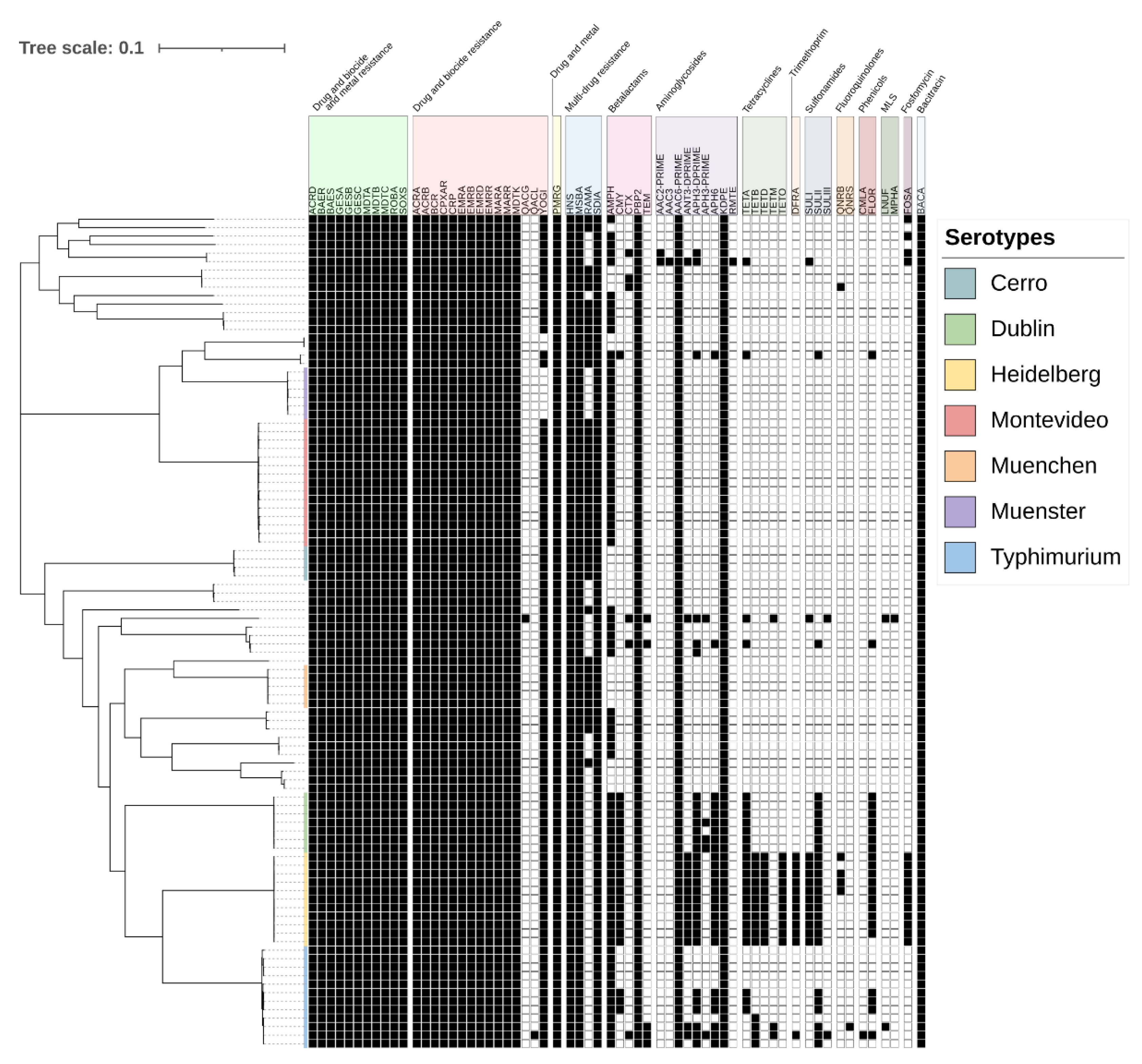
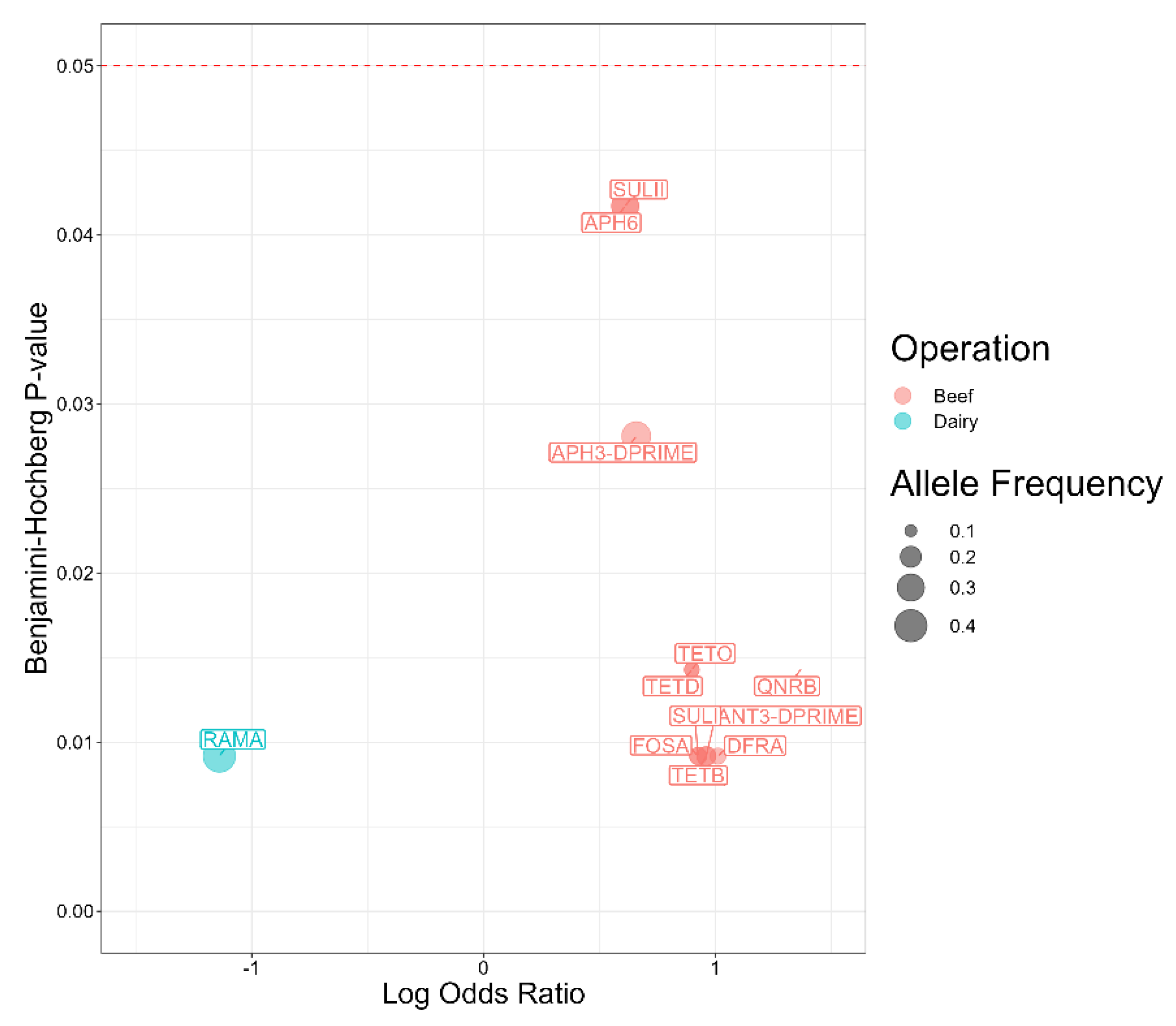
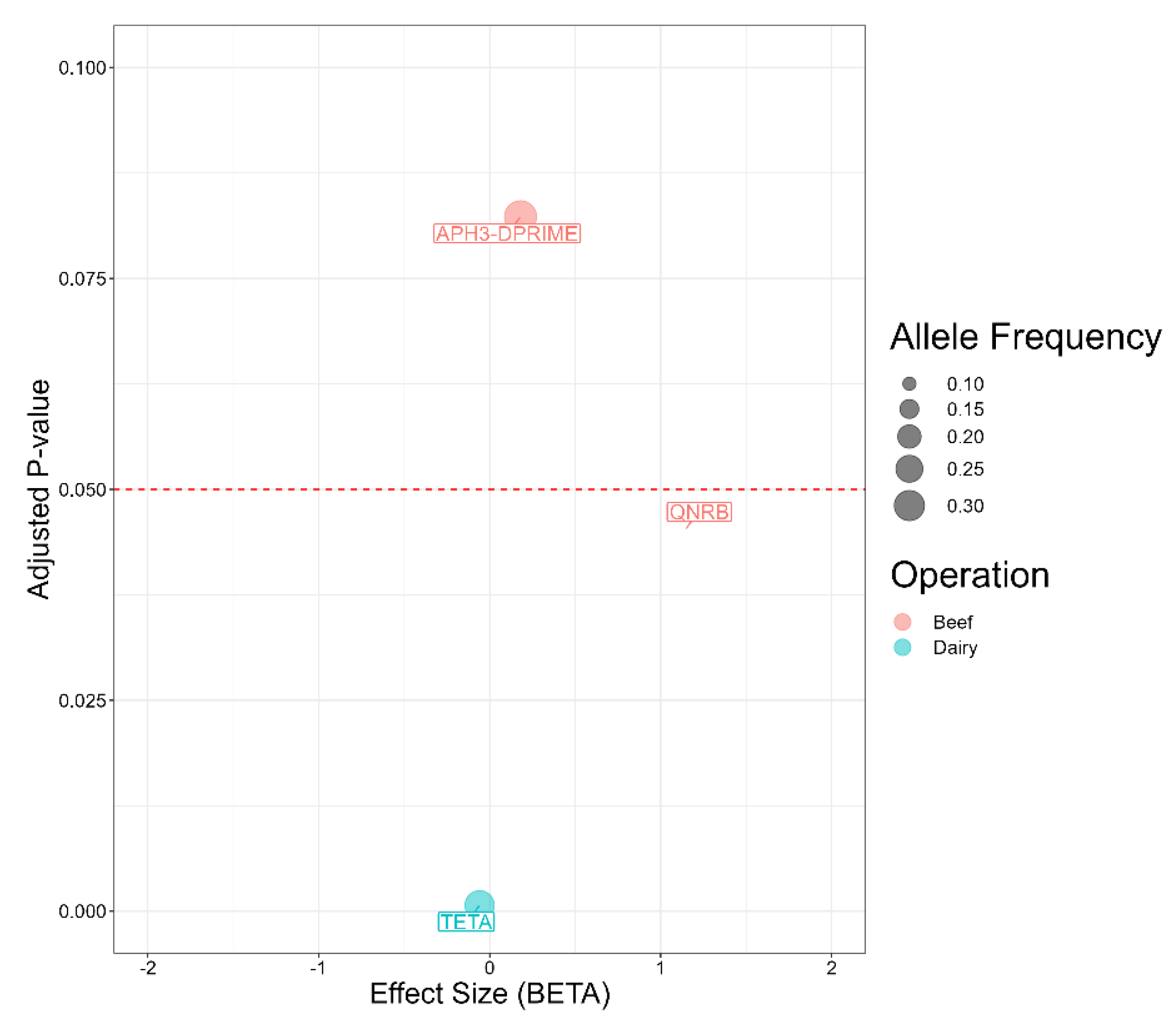
| Antimicrobial | Susceptible | Intermediate | Resistant |
|---|---|---|---|
| Ampicillin | 39 (40) | 1 (1) | 58 (59) |
| Gentamicin | 94 (96) | 1 (1) | 3 (3) |
| Tetracycline | 38 (39) | 3 (3) | 57 (58) |
| Trimethoprim-sulfamethoxazole | 82 (84) | 0 (0) | 16 (16) |
| Serotype | N | Tetracycline | Ampicillin | Gentamicin | Trimethoprim-sulfamethoxazole |
|---|---|---|---|---|---|
| Montevideo | 15 | 7 (47) | 7 (47) | 1 (7) | 2 (13) |
| Typhimurium | 12 | 8 (67) | 9 (75) | 0 | 10 (83) |
| Heidelberg | 11 | 11 (100) | 11 (100) | 0 | 9 (82) |
| Dublin | 7 | 7 (100) | 7 (100) | 0 | 1 (14) |
| Newport | 7 | 2 (29) | 2 (29) | 0 | 0 |
| Muenchen | 6 | 4 (67) | 4 (67) | 0 | 1 (17) |
| Muenster | 6 | 3 (50) | 2 (33) | 0 | 1 (17) |
| Cerro | 4 | 4 (100) | 4 (100) | 0 | 0 |
| Anatum | 4 | 1 (25) | 1 (25) | 0 | 0 |
| Litchfield | 3 | 1 (33) | 1 (33) | 0 | 0 |
| Cannstatt | 3 | 1 (33) | 1(33) | 1 (33) | 0 |
| Infantis | 3 | 0 | 0 | 0 | 0 |
| Mbandaka | 3 | 1 (33) | 1 (33) | 0 | 1 (33) |
| Bredeney | 2 | 2 (100) | 2 (100) | 0 | 0 |
| Brandenburg | 2 | 0 | 0 | 0 | 0 |
| Kentucky | 2 | 1 (50) | 1 (50) | 0 | 0 |
| Meleagridis | 2 | 2 (100) | 1 (50) | 1 (50) | 0 |
| Uganda | 1 | 1 (100) | 1 (100) | 0 | 0 |
| Cubana | 1 | 1 (100) | 1 (100) | 0 | 0 |
| Apapa | 1 | 0 | 1 (100) | 0 | 0 |
| Altona | 1 | 0 | 1 (100) | 0 | 0 |
| Algona | 1 | 0 | 0 | 0 | 0 |
| Bareilly | 1 | 0 | 0 | 0 | 0 |
| Resistance pattern | Serotype | Plasmid replicon | Antimicrobial resistance genes |
|---|---|---|---|
| 1 | Heidelberg | IncA/C2 | ant(3”), aph(3”), aph(3’), aph(6), blaBIL, blaCFE, blaCMY, blaLAT, dfrA, qacE, qacEdelta1, sul1, sul2, tet(A), tet(B), tet(D), tet(O) |
| 2 | Heidelberg | IncA/C2 | ant(3”), aph(3”), aph(3’), aph(6), blaBIL, blaCFE, blaCMY, blaLAT, dfrA, floR, qacE, qacEdelta1, sul1, sul2, tet(A), tet(B), tet(D), tet(O) |
| 3 | Heidelberg | IncA/C2 | aac(6’), ant(2”), ant(3”), aph(3”), aph(3’), aph(6), blaBIL, blaCARB, blaCFE, blaCMH, blaCMY, blaCTX-M, blaIMP, blaLAT, blaMOX, blaOXA, blaVEB, blaVIM, dfrA, floR, ges, qacE, qacEdelta1, sul1, sul2, tet32, tet(A), tet(B), tet(D), tet(O), tet(W) |
| 4 | Heidelberg | IncA/C2 | ant(3”), aph(3”), aph(6), blaBIL, blaCFE, blaCMY, blaLAT, dfrA, floR, qacE, qacEdelta1, sul1, sul2, tet(A), tet(B), tet(D), tet(O) |
| 5 | Heidelberg | IncA/C2 | ant(3”), aph(3”), aph(6), blaCFE, blaCMY, blaLAT, dfrA, floR, qacE, qacEdelta1, sul1, sul2, tet(A), tet(B), tet(D), tet(O) |
| 6 | Typhimurium | IncFIB | ant(3”), aph(3”), aph(3’), blaTEM, cmlA, cmlB, dfrA, floR, qacF, qacL, sul1, sul2, sul2, tet(M) |
| 7 | Typhimurium Dublin |
IncA/C2 | aph(3”), aph(6), blaBIL, blaCFE, blaCMY, blaTEM, floR, sul1, sul2, tet(A) |
| 8 | Typhimurium | IncFIB | ant(3”), lnuF, qnrS, tet(M), tet(O), tet(S) |
| 9 | Dublin | IncA/C2 | aph(3”), aph(6), blaBIL, blaCFE, blaCMY, blaLAT, blaTEM, floR, sul1, sul2, tet(A) |
| 10 | Dublin | IncA/C2 | aph(3”), aph(3’), aph(6), blaBIL, blaCFE, blaCMY, blaLAT, blaTEM, floR, sul1, sul2, tet(A) |
| 11 | Meleagridis | IncHI2 | aac(3), ant(3”), aph(3”), aph(3’), qacE, qacEdelta1, rmtE, sul1, sul2, tet(A) |
| 12 | Bredeney Anatum |
IncA/C2 IncR |
aph(3”), aph(3’), aph(6), blaBIL, blaCFE, blaCMY, blaLAT, floR, sul1, sul2, tet(A) |
| 13 | Uganda | IncHI2 | ant(3”), aph(3”), aph(3’), blaCTX-M, blaTEM, lnuF, mphA, qacE, qacEdelta1, qacG, sul1, sul2, sul2I, tet(A), tet(M), tet(S) |
| Salmonella serotype | ARGs | Odds ratio* |
|---|---|---|
| Dublin |
aph(3”) aph(6) aph(3’) blaCMY floR sul2 tet(A) |
Inf Inf 33.4 Inf Inf Inf Inf |
| Heidelberg |
ant(3”) aph(3”) aph(6) blaCMY dfrA floR fosA qnrB sul1 sul2 tet(A) tet(B) tet(D) tet(O) |
Inf Inf Inf Inf Inf 56.9 Inf 49.1 Inf Inf Inf Inf Inf Inf |
| Meleagridis | aac(2’) | Inf |
| Montevideo | ramA | Inf |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





