1. Introduction
Campylobacteriosis was the most reported zoonosis and foodborne illness of 2022 in EU. A total of 140.241 cases were reported, compared to 129.960 the previous year, with a notification rate of 46.9. The countries with the highest rates are Czechia and Luxembourg while the lowest rates were reported in Bulgaria, Greece, Poland and Romania. The case fatality rate are low, accounting for 0.04% of the reported cases, compared to Listeriosis and West Nile virus infection with a percentage of 18.1% and 8.3% respectively [
1,
2].
One of the main causing pathogens of gastroenteritis worldwide is
Campylobacter jejuni which is transmitted to humans by consumption of raw or undercooked meat, milk and dairy products or cross contamination events [
3,
4,
5] . Another important risk factor is international travel, especially in the Southeast parts of Asia[
3]. Moreover, pets and contaminated water were also reported as being a source of infection [
6]. Although the illness lasts for only a few days and it’s an acute and self-limiting infection, it may cause severe and prolonged symptoms in young and elderly patients. This can lead to complications such as the post infection immune disorder Guillain-Barre syndrome, Miller-Fisher syndrome, meningitis, brain abscess, sepsis, endocarditis, myocarditis, reactive arthritis, inflammatory bowel disease, celiac disease, cholecystitis, and colon cancer [
7] thus requiring antibiotic treatment [
8]. Most commonly used antibiotics are macrolides and fluoroquinolones. Tetracycline and gentamycin are used as alternative treatment when the conventional treatment is not efficient [
7].
Multidrug resistant
C. jejuni exhibits resistance against erythromycin, fluoroquinolones and tetracycline. The use of antibiotics as prophylaxis in livestock seems to be the main cause of antibiotic resistance acquisition, either through horizontal gene transfer from other bacteria or mutations [
6].
A single-point mutation within the quinolone resistance region – QRDR – gyrA is sufficient to confer resistance to fluroquionolones. One mutation frequently associated with AR is C-257 to T (Thr86Ala). Other single-point mutations, such as Tr86Ala, Ala70Thr, and Asp90Asn were described [
9]. (Furthermore, mutations in gyrA gene are not only responsible for the resistance to fluroquinolones, but they also increase the virulence of strains [
9]. Another mechanism involved in the decreased susceptibility to these antibiotics is the activity of the CmeABC efflux pump [
7].
Most commonly, resistance to macrolides is acquired due to a chromosomal mutation in the V domain of the 23S rRNA gene. Other mechanisms of resistance to macrolide include the presence of the gene ermB (rRNA methyltransferase gene) and point mutations in L4/L22 encoding ribosomal proteins [
7].
To date, more than 60 tetracycline resistance genes have been described, most of them located on plasmids [
10]. Mosaic tet genes are common, a vast majority of them being derived from tet(O), tet(W), tet(S), tet(M) and tet32. The resistance levels of these genes is often similar to the non-mosaic genes, however, tet(O/W/32/o/w/o) confers a higher level of resistance than the original genes [
10]. There are similarities between tet(O/32/O) and tet(O/M/O), namely the length and insertion position of tet32 and tet(M) within tet(O), thus distinguishing between them is rather difficult [
10].
2. Materials and Methods
Bacterial isolates
A total of 66 C. jejuni strains isolated between 2017 and 2020 from stool samples were included in the study. The isolates were provided by The Department of Public Health located in Prahova and Maramures regions of Romania. C. jejuni strains were recovered on Columbia enriched blood agar media (7%) and incubated for 48h at 42°C in a micro aerobic atmosphere (85% N2, 10% CO2, and 5% O2) using Campygen Compact sachets (Thermo Fisher Scientific Oxoid Ltd, Basingstoke UK). The species were confirmed using the MALDI TOF Mass Spectrometry System - Bruker Daltonik GmbH Biotyper Identification.
Antimicrobial susceptibility
The antimicrobial susceptibility testing to tetracycline (TE - 30μg), nalidixic acid (NA - 30μg), ciprofloxacin (CIP - 5μg), erythromycin (E - 15μg) was performed by Disc Diffusion Kirby–Bauer method according to the protocol of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) for fastidious organisms, using the ATCC 33560 C. jejuni as quality control.
Genetic support of resistance and virulence
In order to investigate the genetic support of the antibiotic resistance and virulence, bacterial DNA was extracted with the Invitrogen Pure Link kit, following the producer’s instruction and quantified by Quibit 3.0 (Invitrogen). The primers (DNA Technology Biozyme) used for amplifying virulence and resistance genes and their annealing temperatures are presented in
Table 1. (Probably sup). The reactions were carried out in a final volume of 50µl, using 2µl of DNA and 1.5U Taq polymerase (Promega).
The amplicons were purified with the NucleoSpin Gen kit and PCR Clean-up- Macherey Nagel, as recommended by the producers and quantified with Qubit 3.0 (Invitrigen). DNA sequences were determined using primer pairs gyrA7/gyrA8 and BigDye Terminator v3.1 (Applied Biosystems), according to the manufacturer’s instructions, and purified using DyeEx 2.0 Spin kit colonies (Qiagen). Separation and detection of sequences was performed using a SeqStudio genetic analyzer (Applied Biosystems), the obtained sequences were edited and analyzed with the BioEdit Sequence Alignment Editor program, version 7.0.5.3.
C. jejuni strains both sensitive and resistant to fluoroquinolones were compared and analyzed, the detection of mutations in the amino acid sequence at position 86 in the gyrA gene were compared with the sequence in the database with the accession number L04566 DNA sequence in GenBank [
19].
3. Results
The analysis of Campylobacter jejuni strains isolated from Romania between 2017 and 2020 yielded significant insights into the genetic diversity, virulence, and antibiotic resistance profiles of these pathogens. A combination of phenotypical testing, PCR, and antibiotic susceptibility testing provided a comprehensive overview of the resistance mechanisms and genetic relatedness of the isolates.
Virulence genes
The results obtained for the seven genes encoding virulence factors and toxins specific to
Campylobacter jejuni strains isolated from co-cultures originating from human faeces are presented in
Table 2.
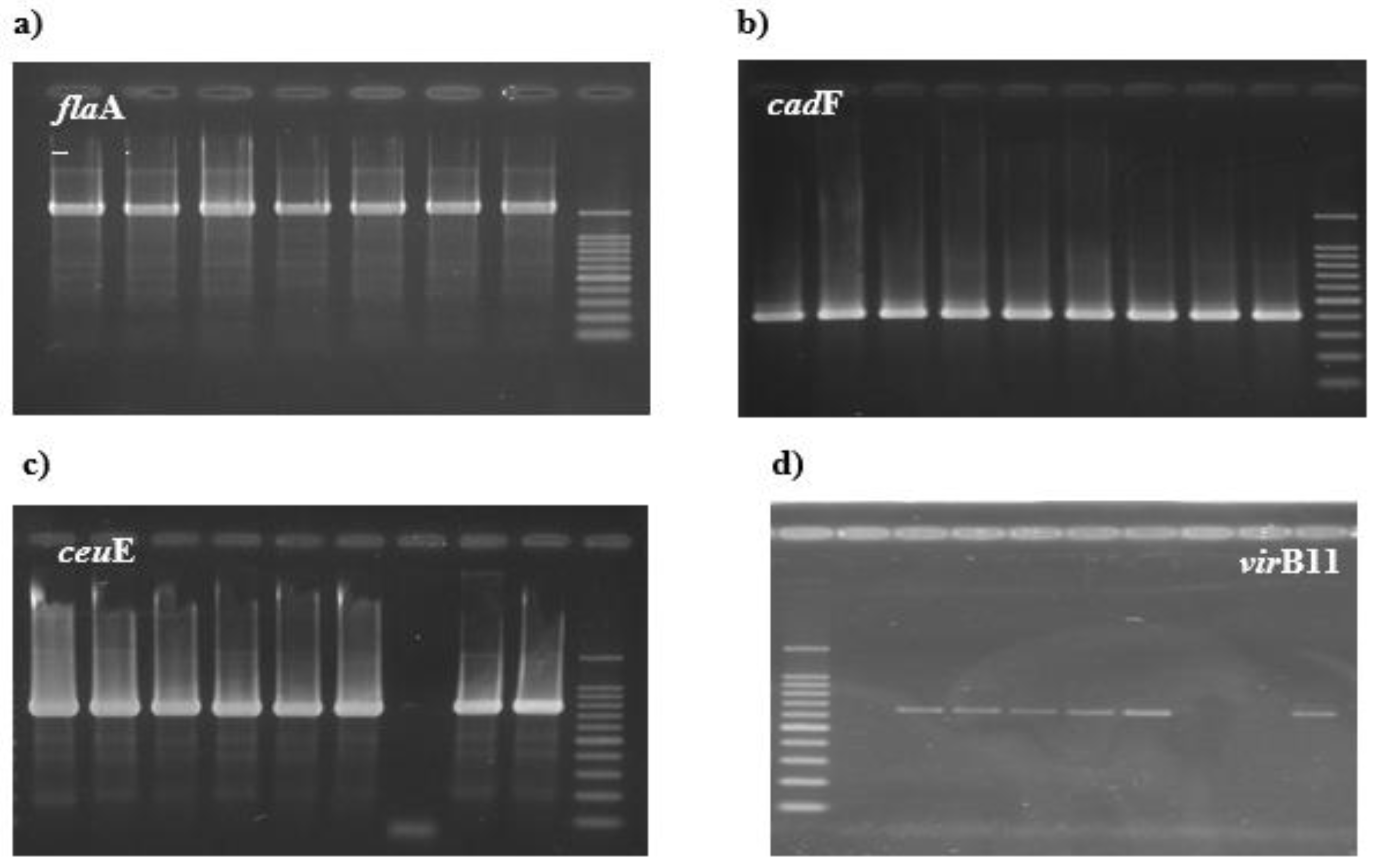
A single specific amplicon for the
flaA gene corresponding to the expected size of 1728bp was generated for all 66
C. jejuni strains (100% )
Figure 1a), similar results were also obtained for the cadF (100%) and ceuE genes respectively (100%)
Figure 1b,c. For the
virB11 gene, 14 isolates (21%) from the 66 strains
of C. jejuni were positive (
Figure 1d).
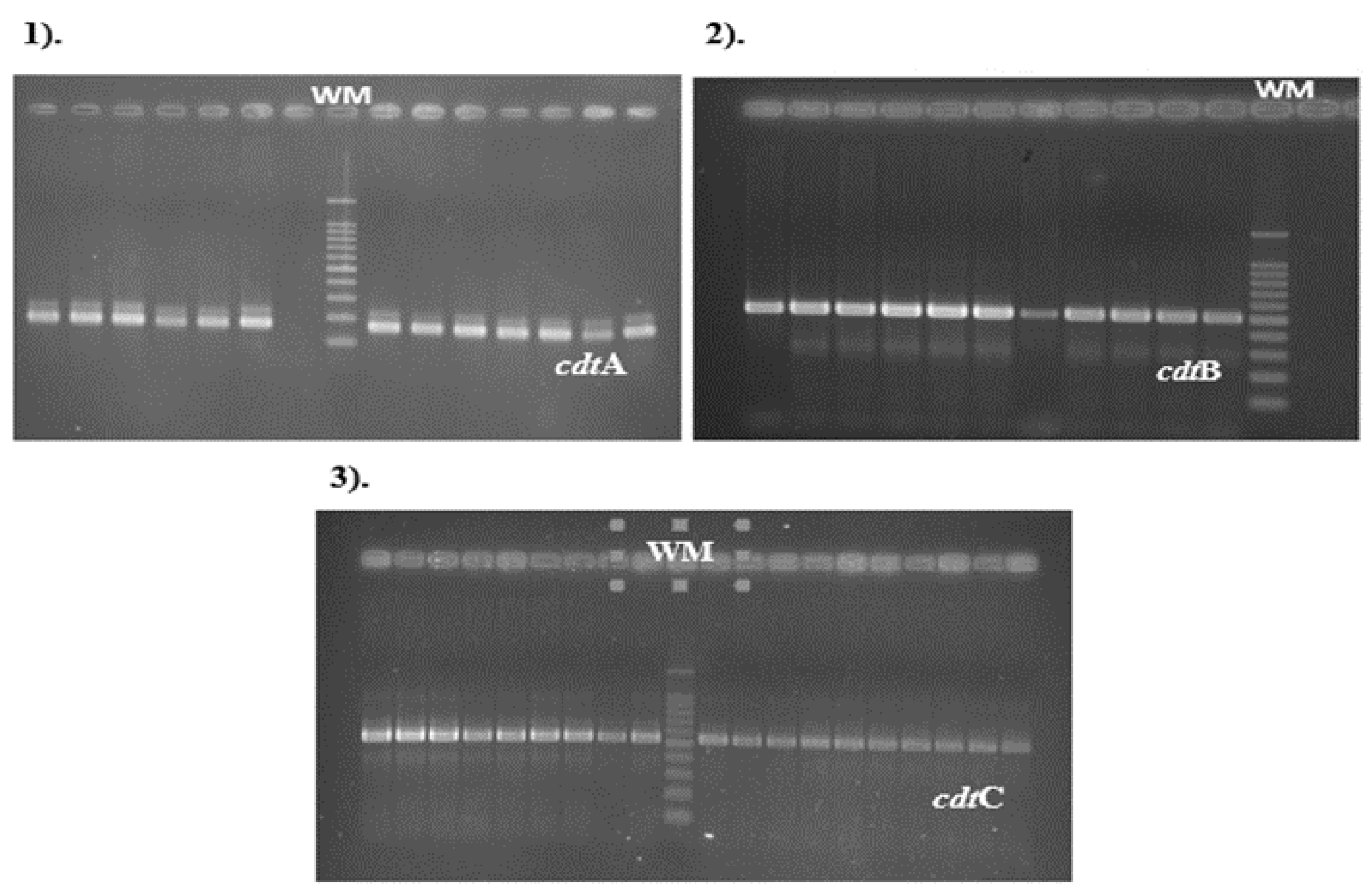
Primers used for the detection of cdt (cytolethal distending toxin) genes cdtA, cdtB and cdtC, were obtained in all
C. jejuni isolates analyzed (
Figure 2).The cdtA gene PCR product with an expected size of 165bp was observed in 62/66
C. jejuni isolates (93%), cdtB gene sequences were detected in 98% and cdtC gene amplicons with an expected size of 555 bp were also observed in a percentage of 98% (65/66).
Resistance. In this study, the data analysis of the obtained sequences determined the substitution in position 86 in the amino acid sequence or in position 257 in the nucleotide sequence of the gyrA gene for the 47
C. jejuni isolates selected with increased resistance to fluoroquinolones with inhibition diameter <26mm, due to a DNA codon mutation from ACA (Thr-threonine) to ATA (Ile-isoleucine) (see
Supplementary Figures S1 and S2)).
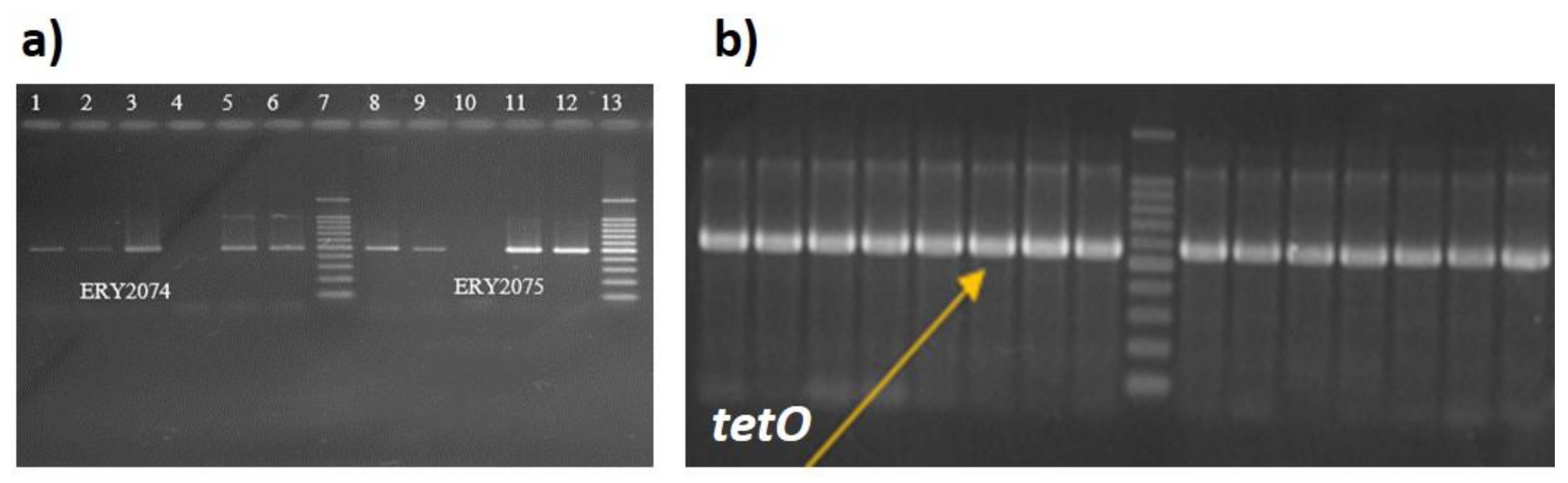
Out of 12 strains (18%) that were phenotypically resistant to Erythromycin (see
Figure 3), 5 isolates harbour a double mutation (2074 A-C, 2075 A-G), while in other 5 only the 2075 single point mutation was detected. (
Supplementary Figure S3).
Results of tetracycline susceptibility testing determined that 55% of clinical isolates (36 out of 66) showed high-level resistance to tetracycline, whereas 45% of isolates showed sensitivity to this class of antibiotics. Primers DMT1 and DMT2 (Invitrogen) produced a 559 bp amplicon for
36 C. jejuni strains specific for the tetO gene responsible for tetracycline resistance, phenotypically sensitive strains did not generate amplification for this gene (
Figure 3b).
In this study, virulence and resistance profiles to antibiotics such as fluoroquinolones, macrolides, and tetracyclines, classes of antibiotics that are often used first-line in the treatment of infections with C. jejuni strains, were observed.
Sequence typing. Sequence typing (7-gene MLST) was performed in order to determine the genetic relatedness between the isolates, by analysing the following genes: aspA, gln, gltA, glyA, pgm, tkt and uncA [
20].
The 66 Campylobacter jejuni strains from sporadic cases of gastroenteritis from children aged between 3 months and 9 years provided by the Public Health in our country, were analyzed and compared. PCR products were amplified with oligonucleotide primer pairs, designated after the published sequences for C.jejuni [
21].
Sequences were determined using internal primer pairs of approximately 400–500 bp and BigDye Terminator v3.1 (Applied Biosystems) according to the manufacturer’s instructions and purified using the DyEx 2.0 Spin kit colonies (Qiagen).
Sequence separation and detection was performed using a 3130 ABI Prism Genetic Analyzer (Applied Biosystems).
Assignment of alleles and ST type (sequence type)
The length and frequency of the alleles defined by the MLST scheme are shown in
Table 4, namely: aspA 477bp, glnA 477bp, gltA 402bp, glyA 507bp, pgm 498bp, tkt 459bp and uncA 489bp. From the 66 C.jejuni isolates, 10 strains were included in CC (clonal complex) 257 (9 strains ST824, 1 strain ST2254), 10 strains in CC353 (4 strains ST400, 4 strains ST353 and 2 strains in ST356), 5 strains in CC21 included in ST50, 4 strains in CC 443 (ST51) and the rest of the strains were included in other types of clonal complexes. The allelic profiles obtained generated 38 types of ST sequences (sequence type) of which 3 new STs of Campylobacter jejuni (10589, 10595, 10686) absent in the database used to establish the ST type (Table 5) and entered in the database international (pubmlst.org) having accession number 106266, 106267 and 106791.
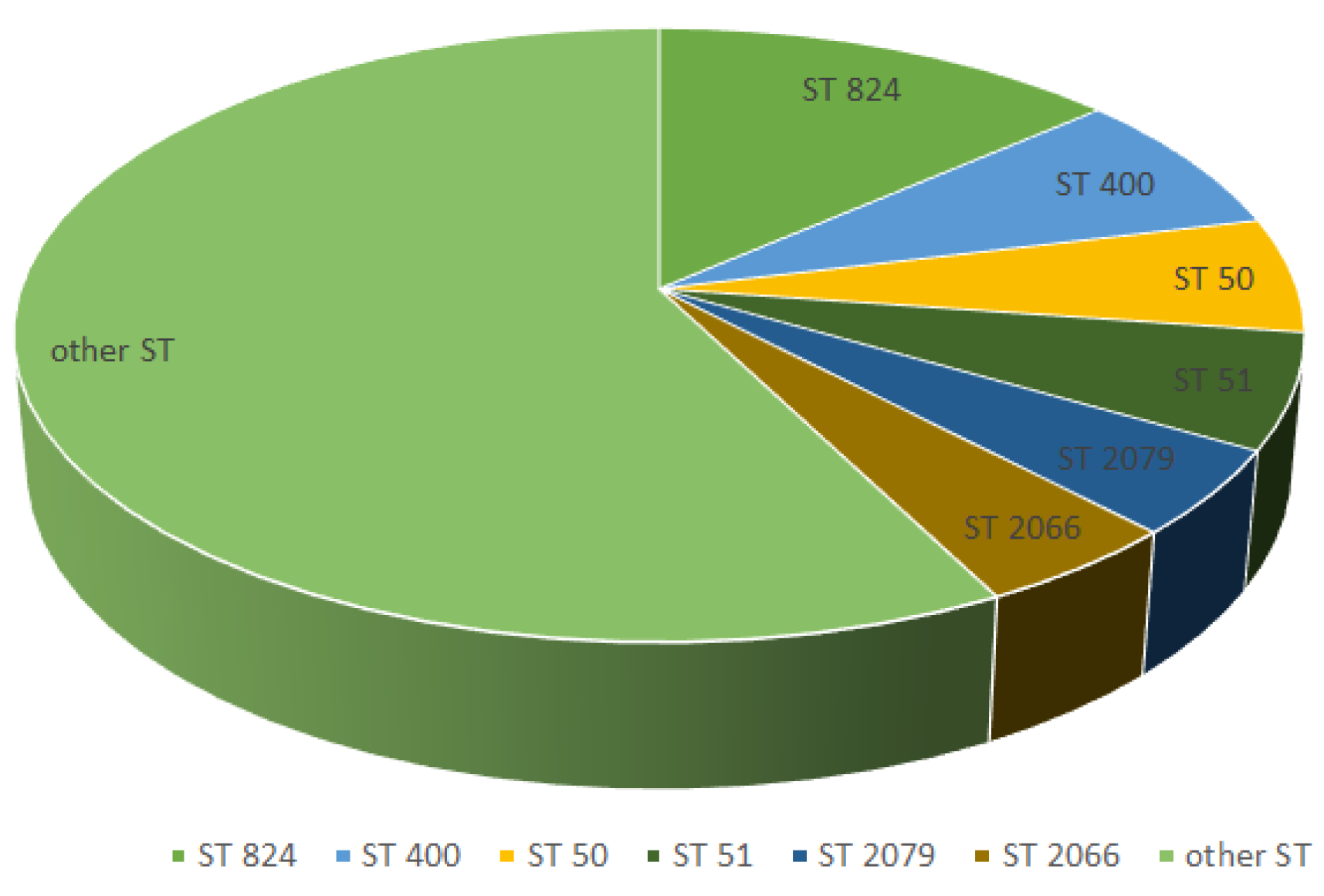
The most common type sequences found among
C. jejuni isolates in this study were ST824 (9 strains), ST400 (4 strains), ST50 (5 strains), ST51 (4 strains), ST2079 and ST2066 (3 strains) (
Figure 4 ).
The strains of C. jejuni in this study were typed by this method, the allelic profiles obtained generated various types of ST sequences (sequence type) already known, these data indicated a genetically diverse population, having a weak clonal structure.
The advantages of MLST include high discrimination among isolates, reproducibility, simplicity of interpretation, and generate data that are comparable to other laboratories by accessing them from international databases.
4. Discussion
In this study, we analysed the sequence data of antibiotic resistance and virulence determinants collected in Romania. Furthermore, the relatedness of the strains was assessed.
To our knowledge, data available on the genetic analysis of antibiotic resistant C. jejuni strains recovered from human campylobacteriosis cases in Romania is very scarce. Annually, up to 10% of the population is affected by foodborne diseases according to WHO.
The prevalence for the seven virulence genes and toxins of the analyzed C. jejuni strains was determined by PCR. The sequences for the three virulence genes (cadF, flaA, ceuE) as well as the cdtC toxin were detected in 98% - 100% of the isolates analyzed in this study.
A high prevalence of 100% of cadF, ceuE and flaA gene was consistent with similar results of previous studies using identical PCR detection assays [
11] (, this high prevalence may be explained by that the cadF gene represents an important virulence factor as well as the flaA gene that plays an important role in
Campylobacter pathogenesis, because Campylobacter motility requires flagellum production.
The low prevalence of 21% of the virB11 gene among the isolates analyzed compares with studies reported by Bacon et al., 2000, the low virulence being due to isogenic mutations. VirB-D – plasmid-mediated – associated with plasmid pVir [
22]. The detection of mutations in the gyrA gene in the QRDR (quinolone resistance determining region) region associated with fluoroquinolone resistance (for resistance to nalidixic acid and ciprofloxacin - widely used antibiotics) was determined in 47 strains of the 66 C.jejuni isolates that have shown resistance to nalidixic acid and ciprofloxacin, widely used antibiotics. The data analysis of the obtained sequences determined the substitution in position 86 in the amino acid sequence or in position 257 in the nucleotide sequence of the gyrA gene for the 47 C. jejuni isolates selected with increased resistance to fluoroquinolones with inhibition diameter < 26mm, due to a DNA codon mutation from ACA (Thr-threonine) to ATA (Ile-isoleucine). Also tested were 3 sensitive strains having inhibition diameter ≥50mm around antibiotic discs and did not show mutation at position 86 in the amino acid sequences for
C. jejuni strains. The quinolone resistance mutation at amino acid position 86 is due to environmental events, these unique DNA sequences in the QRDR region may be useful during the investigation of an epidemiological outbreak.
An alarmingly increasing rate of resistance to quinolone resistance was reported in a neighbouring country, Turkey, registering up to 74.3% (2013) resistant isolates [
7,
24].
The detection of mutations in the rRNA23S gene at position 2074 and 2075 associated with macrolide resistance (for erythromycin resistance) was determined in 12 strains (18%) of the 66 C. jejuni isolates. The data obtained by sequencing were compared and analyzed with the sequence from the C. jejuni NCTC 11168 23Sribosomal database, which confirmed the presence of a point mutation at position 2075 (A-G) in all 12 isolates of C.jejuni with high erythromycin resistance, 5 isolates showed double mutation in both position 2074 (A-C) and position 2075, 5 strains showed mutation in position 2074 and 7 strains did not show this mutation.
The results are consistent with similar results due to the presence of the mutation in position 2075 for C. jejuni strains with increased resistance, therefore the validation of these tests for the detection of point mutations in the rRNA23S gene associated with erythromycin resistance was confirmed.
Results of tetracycline susceptibility testing determined that 55% of
C. jejuni clinical isolates (36 of 66 isolates) showed high level resistance, whereas 45% of isolates showed sensitivity to this class of antibiotics. Due to the long widespread use of tetracyclines, a number of determinants of resistance to this class of antibiotics have been observed in a variety of bacteria, generally this resistance is mediated by one of four mechanisms: efflux pumps, chemical modification of tetracyclines, ribosomal protection proteins and by mutations in rRNA [
23]. Efflux pump and ribosomal protection genes are the most important mechanisms of tetracycline resistance, the acquisition of new tetracycline resistance genes is mostly associated with mobile components such as plasmids or transposons which are often conjugative elements [
25].
Increasing antibiotic resistance of Campylobacter spp. strains isolated from both humans and animals is expected to become a significant public health problem in both developed and developing countries [
26]. Most Campylobacter infections have been found to be self-limiting, antibiotics may be prescribed in severe cases or in immunocompromised patients [
27], virulence and antibiotic resistance profiles such as fluoroquinolones were observed in this study, macrolides and tetracyclines, classes of antibiotics that are often used first-line in the treatment of infections with
C. jejuni strains.
An ECDC surveillance reported that the highest level of resistant C. jejuni isolated from broilers were detected in Romania and Cyprus.
Also, an increased level to resistance was detected in pig isolates, but the resistance to colistin in broilers decreased [
27].
Moreover, ECDC reported in disease surveillance that the European countries with the highest percentage of reported
C. jejuni infections in 2022 are Germany followed by Spain and Czechia, Romanian being located on the 21st place. The mean rate of notification reported by the 30 EU/EEA countries was 46.83 cases per 100.000 population with the highest rate in children under 5 years old. Romania was the only country where all the reported cases were hospitalised [
28]. It is therefore necessary to undertake ongoing monitoring of both the prevalence and molecular characteristics of bacterial virulence and resistance regarding effective treatment information for
Campylobacter infections.
The diversity of house keeping genes for the 66 isolates of C. jejuni was analyzed by MLST, the allelic profiles obtained generated 38 types of ST sequences (sequence type) of which 3 new Campylobacter jejuni STs (10589, 10595, 10686) were absent in the database used for ST typing and entered in the international database under accession numbers 106266, 106267 and 106791. Schemes for the characterization and discrimination of various bacterial isolates are essential for epidemiological, genetic and evolutionary studies.
The study provides the first information on the circulation of a polyclonal population of Campylobacter jejuni at the local level. The molecular typing technique by MLST discriminates between C. jejuni isolates effectively and generates data that can be applied to investigate population structure and its evolutionary mechanisms.
Schemes for the characterization and discrimination of diverse bacterial isolates are essential for epidemiological, genetic and evolutionary studies. Ideally these schemes generate data that are relevant to all these domains, but before the advent of high-throughput nucleotide sequencing technology, this goal proved elusive [
29]
The need for appropriate molecular typing schemes for
C. jejuni isolates is essential because this human pathogen possesses extensive reservoirs from animals and the environment, and the relationships between diseases associated with the animal population or the environment remain completely unclear [
30].
This study demonstrates that the molecular typing technique by MLST discriminates between C. jejuni isolates effectively and generates data that can be applied to investigate population structure and its evolutionary mechanisms.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org.
Author Contributions
Conceptualization, M.B. and I.S.; methodology, M.B.;D.C.; validation, M.B., G.G.P and C.U.; formal analysis, M.O.; investigation, M.B. and M.S.; writing—original draft preparation, M.B.; writing—review and editing, G.G.P.;
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest
References
-
https://www.ecdc.europa.eu/sites/default/files/documents/CAMP_AER_2022_final.pdf.
-
https://www.ecdc.europa.eu/sites/default/files/documents/EFS2_8442.pdf.
- Kaakoush, NO., Castaño-Rodríguez N., Mitchell HM., Man SM., 2015. Global epidemiology of Campylobacter infection. Clin Microbiol Rev,Vol 28, no 3. [CrossRef]
- Cardoso, MJ., Ferreira V., Truninger M., Maia R., Teixeira P., 2021. Cross-contamination events of Campylobacter spp. in domestic kitchens associated with consumer handling practices of raw poultry. International Journal of Food Microbiology, Volume 338. [CrossRef]
- Santos-Ferreira, N., Alves A., Cardoso MJ., Langsrud S., Malheiro AR., Fernandes R., et al., 2021. Cross-contamination of lettuce with Campylobacter spp. via cooking salt during handling raw poultry. PLoS ONE 16 (5). [CrossRef]
- Noreen, Z., Siddiqui F., Javed S., Wren BW., Bokhari H., 2020. Transmission of multidrug-resistant Campylobacter jejuni to children from different sources in Pakistan. Journal of Global Antimicrobial Resistance,Volume 20 Pages 219-224. [CrossRef]
- Kayman, T., Abay S., Aydin F., Şahin O., 2019. Antibiotic resistance of Campylobacter jejuni isolates recovered from humans with diarrhoea in Turkey. J Med Microbiol. 68 (2):136-142. [CrossRef]
- Myintzaw, P., Jaiswal AK., Jaiswal S., 2021. A Review on Campylobacteriosis Associated with Poultry Meat Consumption. Food Reviews International, vol. 39, no. 4, 2107–2121. [CrossRef]
- Whelan MVX., Ardill L., Koide K. et al.,2019. Acquisition of fluoroquinolone resistance leads to increased biofilm formation and pathogenicity in Campylobacter jejuni. Sci Rep 9. [CrossRef]
- Hormeño, L., Campos MJ., Vadillo S., Quesada A.,2020. Occurrence of tet(O/M/O) Mosaic Gene in Tetracycline-Resistant Campylobacter. Microorganisms. 31; 8(11):1710. [CrossRef]
- Konkel, M.E., Gray S.A., Kim B.J., Garvis S.T. and Yoon J., 1999. Identification of the enteropathogens Campylobacter jejuni and Campylobacter coli based on the cadF virulence gene and its products. Journal of Clinical Microbiology 37, 510–517. [CrossRef]
- Gonzalez, I., Grant K.A., Richardson P.T., Park S.F. and Collins M.D., 1997. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. Journal of Clinical Microbiology 35, 759–763. [CrossRef]
- Bacon, DJ., Alm RA., Burr DH., Hu L., Kopecko DJ., Ewing CP., Trust TJ. and Guerry P., 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81–176. Infection and Immunity 68, 4384–4390. [CrossRef]
- Nachamkin, I., Bohachick K. and Patton CM., 1993a. Flagellin gene typing of Campylobacter jejuni by restriction fragment length polymorphism analysis. Journal of Clinical Microbiology 31, 1531– 1536. [CrossRef]
- Pickett, C.L., Pesci E.C., Cottle D.L., Russell G., Erdem A.N. and Zeytin H., 1996. Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter spp. cdtB genes. Infection and Immunity 64, 2070–2078. [CrossRef]
- Zirnstein, G., LI Y., Swaminathan B. and Angulo F., 1999. Ciprofloxacin Resistance in Campylobacter jejuni Isolates: Detection of gyrA Resistance Mutations by Mismatch Amplification Mutation Assay PCR and DNA Sequence Analysis. Journal of Clinical Microbiology,Vol.37,Nr.10p.3276–3280. [CrossRef]
- Alonso, R., Mateo E., Churruca E., Martinez I, Girbau, Fernandez-Astorga A., 2005. MAMA-PCR assay for the detection of point mutations associated with high-level erythromycin resistance in Campylobacter jejuni and Campylobacter coli strains. Journal of Microbiological Methods 63 99– 103. ttps://doi.org/10.1016/j.mimet.2005.03.013.
- Gibreel, A., Tracz DM., Nonaka L., Ngo TM., Connell SR., and Taylor DE., 2004. Incidence of antibiotic resistance in Campylobacter jejuni isolated in Alberta, Canada, from 1999 to 2002, with special reference to tet (O)-mediated tetracycline resistance. Antimicrobial Agents and Chemotherapy, vol. 48, no. 9, pp. 3442–3450. [CrossRef]
- Wang, Y., Wai Mun Huang W.M.H. and Taylor DE., 1993. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrobial Agents and Chemotherapy, vol. 37, no. 3, pp. 457– 463. [CrossRef]
- Dingle, KE., Colles FM., Wareing DRA., Ure R., Fox A.J., Bolton FE., Bootsma HJ., Willems RJL., Urwin R., and Maiden MCJ., 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14–23. [CrossRef]
- Parkhill, J., Wren BW., Mungall K., Ketley JM., Churcher C., Basham D., Chillingworth T., Davies RM., Feltwell T., Holroyd S., Jagels K., Karlyshev AV., Moule S., Pallen MJ., Penn CW., Quall MA., Rajandream MA., Rutherford KM.,van Vliet AHM., Whitehead S. and Barrell BG., 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. [CrossRef]
- Panzenhagen, P., Portes A.B., dos Santos AMP, Silva Duque S. and Conte Junior CA., 2021.The Distribution of Campylobacter jejuni Virulence Genes in Genomes Worldwide Derived from the NCBI PathogenDetection Database. Genes 12, 1538. [CrossRef]
- Kayman, T., Abay S., Aydin F., Şahin O.,2019. Antibiotic resistance of Campylobacter jejuni isolates recovered from humans with diarrhoea in Turkey. J Med Microbiol.,68(2):136-142. [CrossRef]
- Connell, SR., Trieber CA., Dinos GP., Einfeldt E., Taylor DE. and Nierhaus KH., 2003. Mechanism of Tet(O)-mediated tetracycline resistance. EMBO Journal, vol. 22, no. 4, pp. 945– 953. [CrossRef]
- Roberts, MC., 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol lett 245: 195-203. [CrossRef]
- Luangtongkum, T., Jeon B., Han J., Plummer P., Logue C., Zhang Q., 2009. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol 4: 189-200. [CrossRef]
- Ge, B., Wang F, Sjolund-Karlsson M., McDermott P., 2013. Antimicrobial resistance in Campylobacter: susceptibility testing methods and resistance trends. J Microbiol Methods 95: 57-67.
-
http://atlas.ecdc.europa.eu/public/index.aspx.
- Maiden MCJ., 2000. High-throughput sequencing in the population analysis of bacterial pathogens. Int. J.Med. Microbiol. 290:183–190. [CrossRef]
- Altekruse, SF., Stern NJ., Fields PI., Swerdlow DL., 1999. Campylobacter jejuni an emerging foodborne pathogen. Emerg. Infect. Dis. 5:28–35. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).