1. Introduction
Autism Spectrum Disorder as described by American Psychiatric Association (APA) in the Diagnostic Statistical Manual (DSM-V) [
1] is a neurological disorder characterized with lack of communication , restrained social skills and visible signs of repetitive behaviours like stacking of objects in those children who have ASD. Prevalence of ASD worldwide is around 1% [
2] while estimates are even higher in the high-income countries , for example in the US [
3] about 2.3% of the children aged 8 years and about 2.2% of the adults are suffering with the ASD which makes ASD a global economic burden .Early diagnosis of ASD is considered crucial because early diagnosis and timely start of therapies and interventions in ASD subjects have shown improvement in the quality of life of people suffering with ASD [
4] .The challenging issue is that the early diagnosis of ASD is difficult because of lack of trained ASD specialists world-wide , demographic variations across countries, lack of universally ASD screening tests and most importantly the interventions and therapies that exist around the world to target ASD are mostly designed for adult population [
5,
6]. In the light of these evidences it is of significant interest to find psychological interventions , biological drugs and artificial intelligence based methods to tackle and treat the ASD disease in general and finding those neuro-markers and genetic factors related to the ASD that can reduce the economic burden of ASD and improve the quality of lives of people suffering with ASD in particular.
1.1. Motivations
The functional Magnetic Resonance Imaging (fMRI) modality is getting more attention from researchers in the field of neuro-science due to it is non-invasiveness , high spatial resolution and lesser cost over the time , fMRI works by measuring the magnitude of the Blood Oxygenated Level Dependent (BOLD) signal which in turn acts as the proxy of the underlying neural activity in the brain region.[
7,
8]. A range of brain disorders including Alzheimer [
9] , Attention Deficit Hyperactivity Disorder [
10] , Autism Spectrum Disorder [
11], Epilepsy [
12] , Major Depressive Disorders [
13] and Parkinson [
14] have now been explored using fMRI which makes fMRI a promising futuristic modality to study various brain disorders. Therefore , A plethora of fMRI studies on various brain disorders , non-invasive nature of fMRI and high spatial resolution of fMRI make this imaging modality a promising research direction to pursue and explore brain disorders.
Of late , Deep Learning (DL) [
15] based approaches have revolutionized the field of Artificial Intelligence and are being used in variety of tasks such as classification , detection , segmentation and natural language processing. The underlying idea of the DL which is also called a representational learning is to map the underlying data distribution to a non-linear latent dimension by using first a multi-layered neural network , then train the network using the back-propagation algorithm and after training , testing the trained neural networks on the unseen or testing data . DL based approaches have shown huge potential in the fields of Image classification [
16] , Brain Tumor Detection [
17] , Drug discovery [
18] and Natural Language Processing (NLP) [
19] which makes DL an active area of research for many open problems motivating a community of researchers to use the ideas of DL in their respective fields. In the following we briefly review the related articles specific to our proposed approach and with which we are to compare our results with.
1.2. Related State-of-the-Art
RFE-GNN [
20] is proposed to classify the ASD using the spatial features extracted from recusrive features elimination and then concatenating them with the phenotypic information on the subjects. The author’s achieved promising results as their fusion based approach achieved an overall accuracy of 80%. A potential drawback of their approach is the use of features selection approach and then a naive contaentated approach to fuse two set of features . Our proposed approach neither relies on those hand-crafted features and nor use the phenotypic information and still outperforms this approach which shows the robustness of our approach.
MHSA-FC [
21] is a promising approach that focused on the architecture of transformer based multi-head attention and a data augmentation module. Authors achieved promising results using their End-To-End approach that did not require hand-crafted features with 81.47% accuracy , 83.8% sensitivity and 80.16% specificity . But the main weakness of their approach is the augment module where the authors used a sliding window approach that require extensive selection of parameters and experimentation. To this end our proposed approach’s GAN powered data augment module solves this problem by not relying on sliding window to augment dataset.
MVES [
22] is proposed to tackle the classification of ASD by first extracting the mean time series of the fMRI data and then low/high level functional connectivity features using PCA and autoencoder and finally an ensemble based model was used for the classification.Authors reported the classification accuracy of 72% and a highest classification accuracy of 92.9% was reported on the CMU site. In MVES authors used a fixed length window approach to augment the data which is the main limitation of their approach because by using the fixed length window the inherent meaningful structure of the full length fMRI gets ignored. To this end we again firmly believe that our proposed approach is superior to their fixed length approach because our GAN based data augmentation approach is tackling this augmentation of data by generating fixed length correlation features that do not depend on the varying time series of a subject.
NVS-VA [
23] is a features selection based approach that by using a pre-trained variational autoencoder achieved a 10-fold accuracy of 78.12% on the overall dataset.The authors also proposed an innovative activation function and devised a normalization pipeline in their approach. But their approach used a hand-crafted features selection approach that has become obsolete and our End-To-End approach tackles this limitation well , Secondly , authors did not report site-wise metrics for the 17 sites which again limits the usefulness of the their approach as it might not work well on site wise data because some sites have significantly smaller data points which make classification a challenging task on sites.
MSC-FS [
24] is proposed to tackle the classification task using nested features selection where the authors first divided the dataset into two categories of ASD and HC and then the features got selected from the relevant sites using SVD resulting in an accuracy of 68.42%. The major limitations of their work is the hand-crafted features and reporting of accuracy on limited sites instead of all the 17 sites.
DeepGCN [
25] is a promising End-To-End approach using Graph Convolution Networks (GCN) that achieved promising results on the ABIDE dataset as the authors reported a cross validated accuracy of 73.7%. The main limitation of their approach is that the authors , although used a End-To-End approach for the classification of ABIDE , yet authors did not report site wise comparison of the accuracy which limits usefulness of their approach as already discussed site wise accuracy reporting is considered more challenging due to lesser data points.
1.3. Contributions
The main contributions of the proposed approach are as follows.
A GAN based synthetic dataset generator is proposed to augment the connectivity features of a subject . The reason for the generation of fixed length connectivity features per subject using Pearson correlation ,is to avoid the variable time length based subject time series data.
A Multi-Head based attention based deep learning model is proposed and the motivation for this is because we do not want to use hand-crafted features for the classification as multi-head based attention mechanism with skip connections tackle the task of selecting useful features in an End-To-End manner.
A pre-trained Autoencoder trained on the training subjects data is used to train the GAN because we want to generate the subject’s features which are as close to the original subjects distribution as possible , therefore when training the GAN , In addition to the losses of discriminator and generator we also focus on the pre-trained Autoencoder loss.
1.4. Objectives
We have used in this study correlation based connectivity features to tackle the task of varying time series of brain AAL template per subject. The connectivity features have dimension of 6670 and they are still a challenge to handle because of the noise in the time series data. The main objectives of the proposed Deep Learning based approach is to first come up with a synthetic subjects data on ASD and HC and and then using an End-To-End framework for the classification of subjects into ASD and HC without using hand-crafted and obsolete features selection approaches. Since the ASD dataset on some sites contain too few subjects which make accurate classification very challenging , our GAN based approach of data augmentation tackles this problem in an ingenious way . Moreover , The Multi-Head Attention mechanism that we proposed in the model tackles the problem of over-fitting by focusing on only those areas of the brain regions which actually play their roles in the ASD underlying neural activity patterns.
2. Materials and Methods
In the following two sections we now explain the used Dataset, challenges related to the Dataset and the proposed methodology in detail.
2.1. Dataset
Autim Brain Imaging Data Exchange (ABIDE) [
26] is a public consortium that has provided the resting state fMRI datasets on the individual with ASD which was preprocessed using the procedures described in [
27] where pre-processing includes steps like slice-timing correction , motion realignment , normalization and registration . The original dataset included 1112 individuals with 539 suffering from the ASD and 573 are typical controls but as per latest update the provided dataset contains now 884 total subjects as shown in
Table 1. Moreover, we have used the provided Automated Anatomical Labeling (AAL) [
28] based regions of interests .
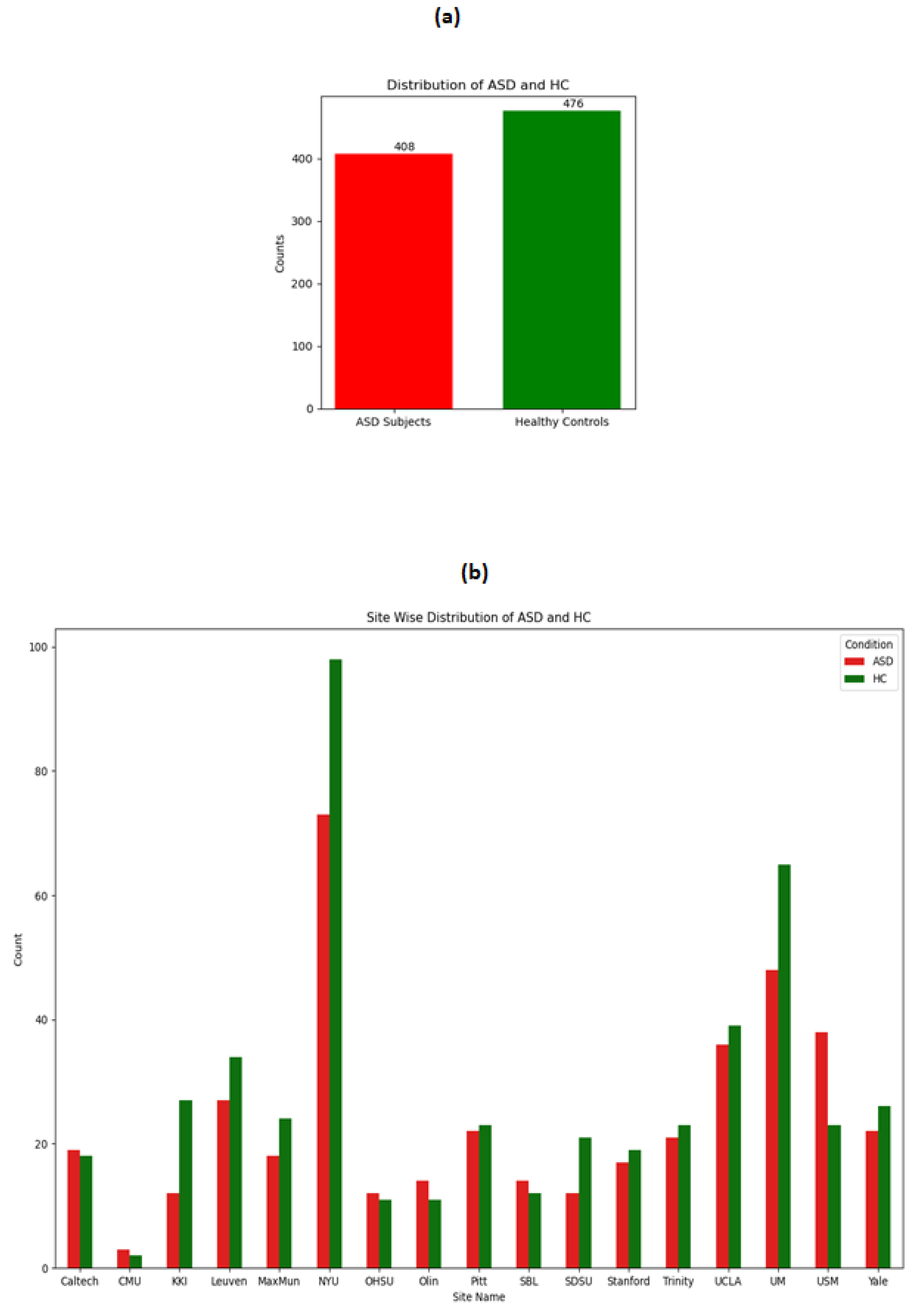
The distribution of ASD and HC on the whole dataset and across the sites is shown in following
Figure 1.It can be seen from the figure that although the overall distribution of ASD and HC is not much imbalanced but when observing the distribution site wise , there are drastic variations. For example site CMU has not even a total of 10 data points whereas the NYU site has more than 100 data points. These observations make stratified split of training and testing more logical because if not then it might be possible that no training data gets included from the CMU site leading to biased in the model.
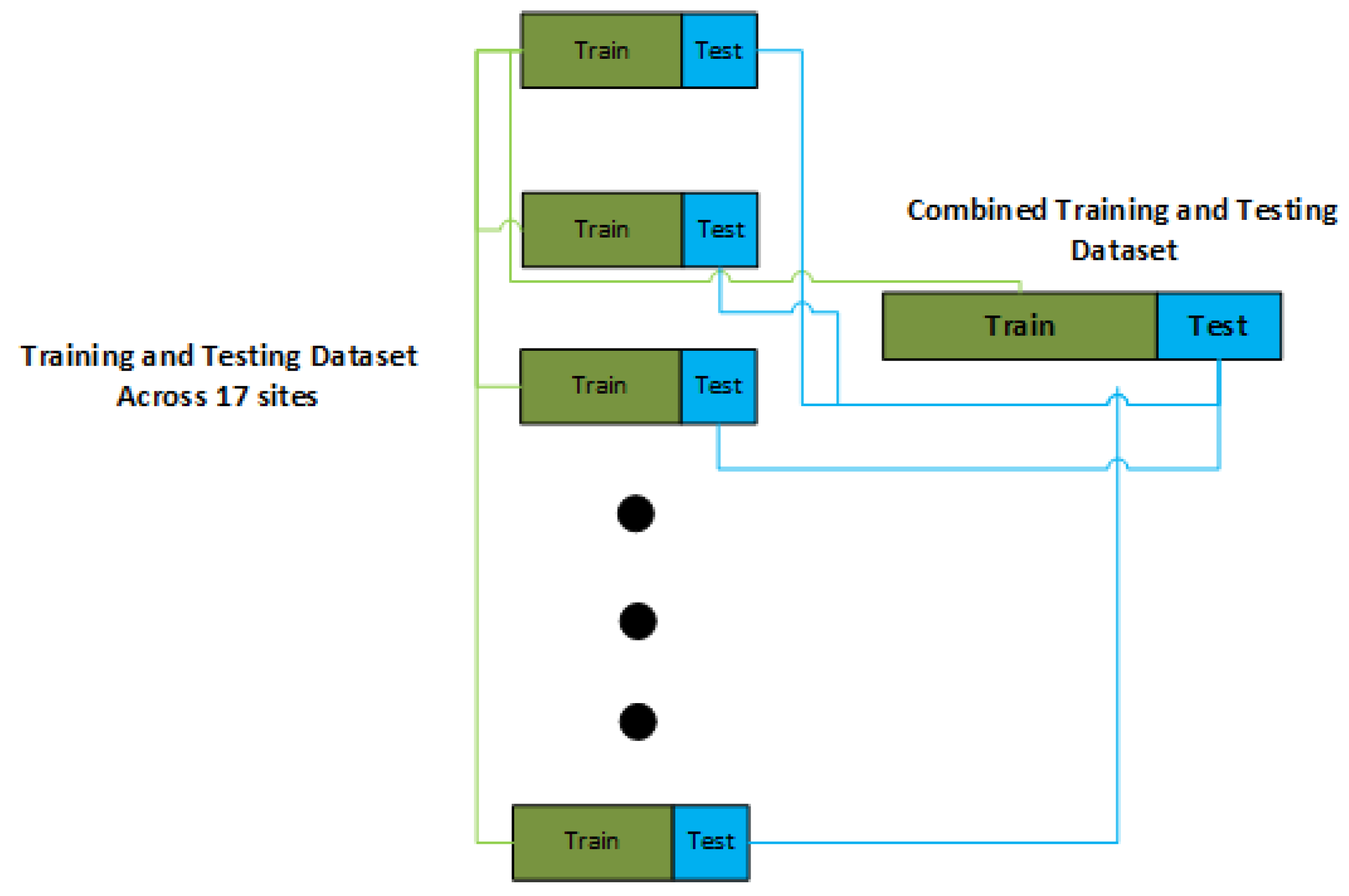
2.2. Stratified Training and Testing Split
The imbalance nature of dataset as explained and shown in the previous section necessitates us to use a stratified based partition of dataset for training and testing as shown in the
Figure 2. Therefore , henceforth in the rest of study we use a stratified based split of dataset into the training and testing so that every site gets its representation in the training and testing .
2.3. Methodology
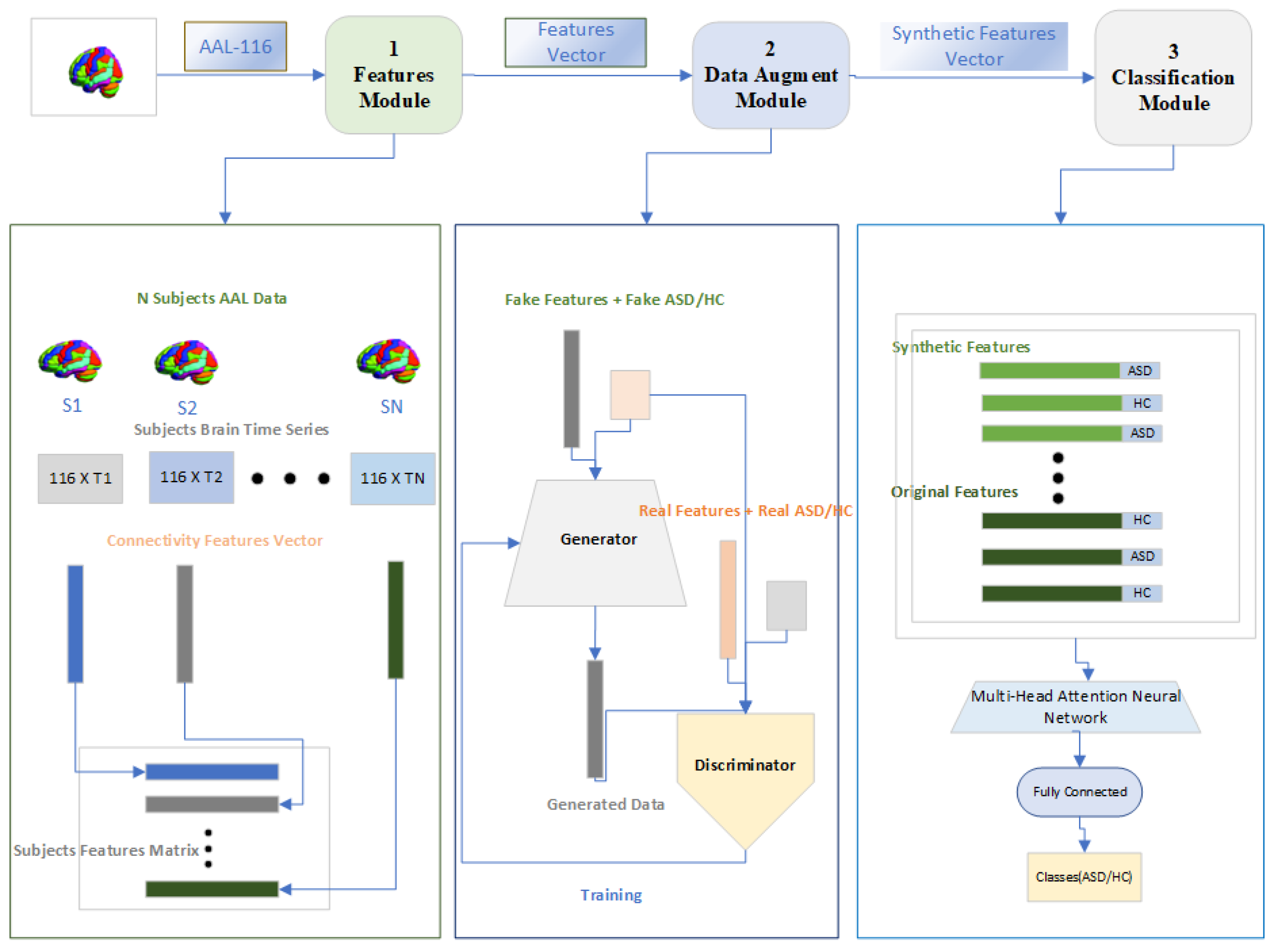
In the
Figure 3 We now explain our proposed method for the classification of ASD. The proposed model consists of three modules which we call as Features Module , Data Augment Module and a Classification Module. The general flow of the proposed approach is as follows , Firstly , the connectivity features will be calculated for both class of subjects . Secondly, The connectivity features will be used to train a Generative Adversarial Network to generate synthetic features and class per subjects so that the problem of over-fitting while training the DL model be reduced . Finally , A classification module will be used for the classification of subjects to ASD and HC using the Multi-Head attention module.
In the following subsections , We briefly describe each the three modules that were shown in the
Figure 3.
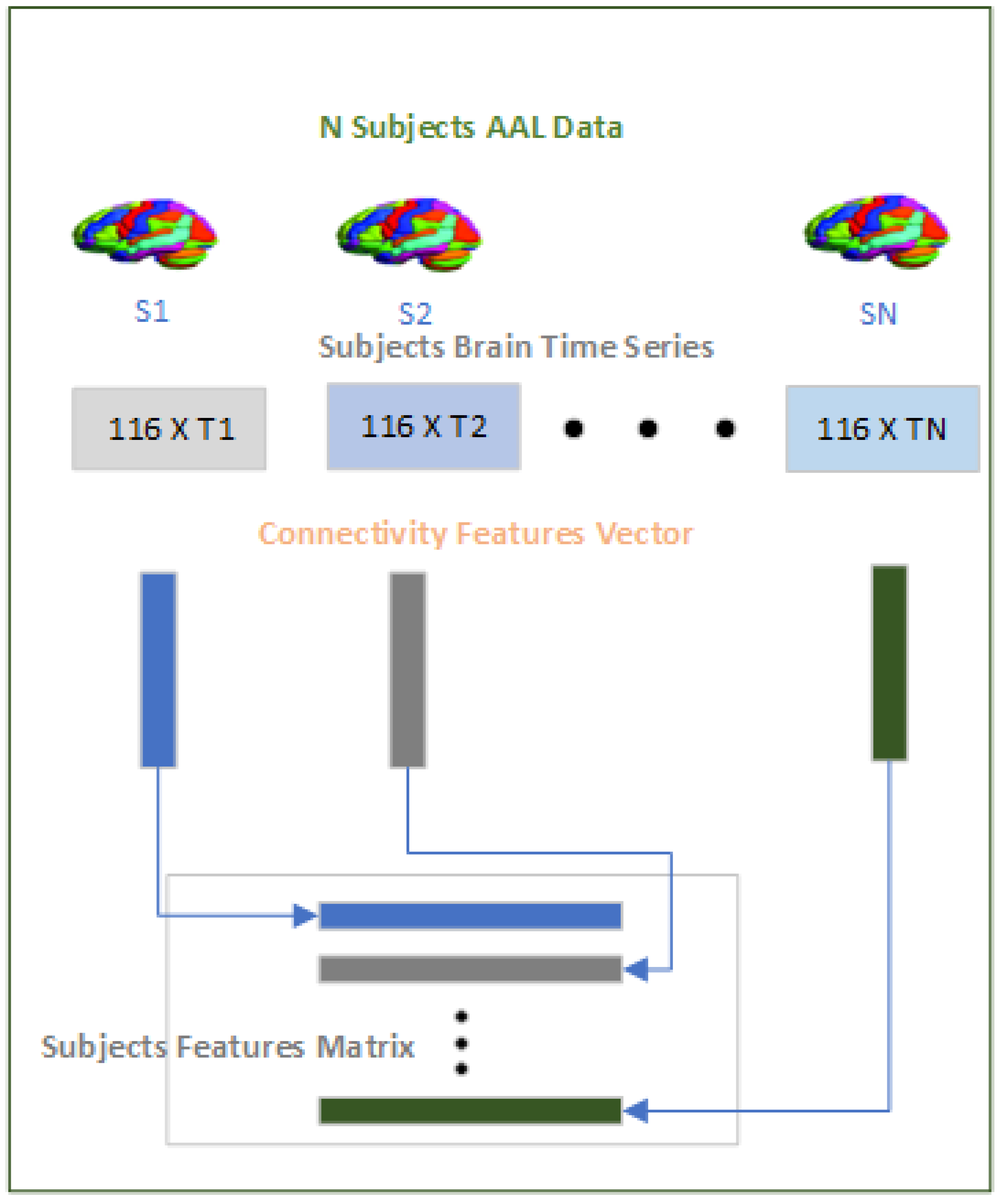
2.3.1. Features Module
The purpose of the features module as shown in
Figure 4 is to convert the time series data on the brain regions across subjects to a fixed length dimension that can be processed by the Data Augment Module and subsequently by the Classification Module. The time series data on the 116 brain regions vary sometimes within site and also across sites and tackling the varying time series data is a significant challenge because taking a fixed size window for the time series across subjects not only violates the inherent structure of the pre-processed fMRI but also limit generalization over the subjects. To tackle this challenge we have used a Pearson’s correlation
"r" as given in Eq
1 to measure the functional connectivity between any two brain regions
"x" and
"y". Since we are using the AAL-116 template in the study where each subject has 116 brain regions so the correlation will result in a symmetric 116x116 matrix from which we extracted the upper diagonal "6670" values excluding the diagonal because the diagonal is always 1 as the correlation between the two same brain regions will always be 1. Therefore for each of the subjects whether ASD or HC we will have a "6670" dimensional connectivity features vector which will be used subsequently in the rest of the modules.
The motivations of using the functional connectivity features is deeply rooted in how the brain actually performs a task as it has been shown with significant evidence that brain regions when doing a task work in pairs and functional connectivity has proved to be associated with cognition and motor control in human beings [
29,
30].
2.3.2. Data Augment Module
The purpose of the Data Augment Module is to generate synthetic 6670 dimensional features across ASD and HC because the size of dataset corresponding to the features dimension is very less leading to the over-fitting of the DL models. We have used Generative Adversarial Networks (GAN) [
31] based approach to generated synthetic features across subjects and since we want not only features but also want to condition the features across the subjects so we conditioned our GANs based on the subject condition that is either belonging to ASD or HC. The objective function of the Conditional GAN (CGAN) as explained in [
32] is given in Eq
2 . Where
"G" and
"D" are the Generator and Discriminator ,
"x" is the features vector ,
"y" is the class of the subject and
"z" is the random vector . The main purpose of the GAN minmax approach is to fool the discriminator over time by the generator by generating subjects as close to the real data. .
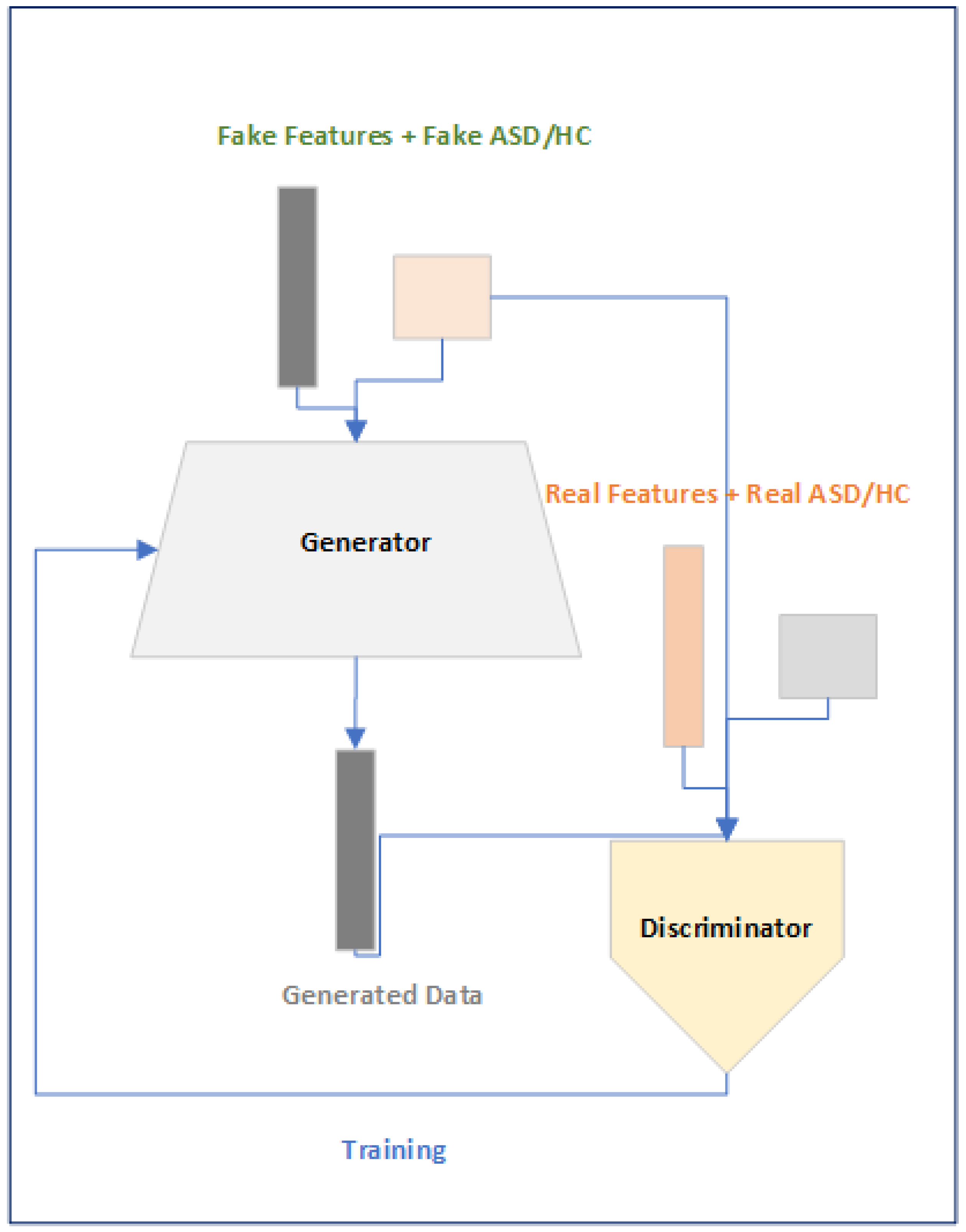
Figure 5.
Data Augment Module.
Figure 5.
Data Augment Module.
The motivations of using the CGAN based approach is to overcome the problem of using a fixed size subject time series data and to minimize the problem of over-fitting that is faced when training the DL model.
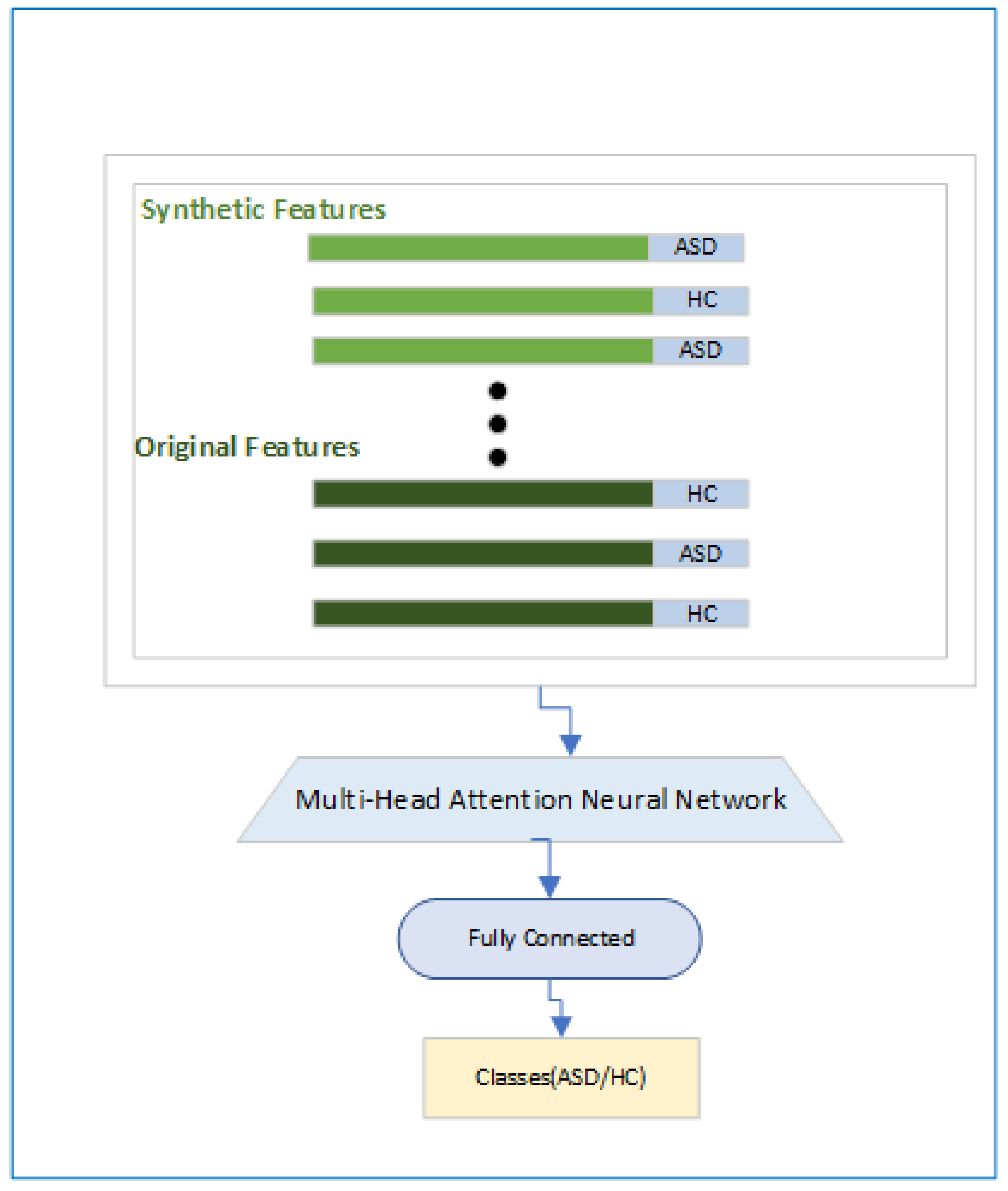
2.3.3. Classification Module
In the Classification Module in
Figure 6 we have shown that how the final objective of the classification will be achieved. Here first we will have features and associated labels for the original and synthetic ASD and HC subjects and then a Multi-Head attention [
33] based module will be used so that focus is on the relevant features followed by a fully connected layer and a classification head.The Multi-Head attention based module is designed using the Eq
3 where
"Q",
"K" and
"V" are called the query, key and value and
"d" is normalization constant.
2.3.4. Algorithm
In the following section we explain the proposed approach using an algorithm as written in the Algorithm 1 so that the reader has a clear mind map of the step-wise working of the proposed approach.
|
Algorithm 1: Proposed GAN Powered Multi-Head Attention Approach’s Algorithm |
- 1:
Input ← D , Output ← {} , ganEpochs, synthEpochs , classEpochs - 2:
X,Y ← D (X ← Features Matrix , Y = ASD/HC) - 3:
, , , ← Train-Test-Split(X,Y) - 4:
← {}, ← {} - 5:
(Features Module): - 6:
for, , , in (,,,) do - 7:
=, = - 8:
← PearsonCorr() - 9:
← PearsonCorr() - 10:
← UpperTria() - 11:
← UpperTria() - 12:
← U { , } - 13:
← U { , } - 14:
end for - 15:
(Data Augment Module): - 16:
← {} - 17:
for epoch in ganEpochs do - 18:
, ← Random() - 19:
, ← Generator(,) - 20:
, ← Subset() - 21:
Discriminator(,) - 22:
Discriminator(,) - 23:
end for - 24:
for epoch in synthEpochs do - 25:
, ← Generator() - 26:
← U { , } - 27:
end for - 28:
(Classification Module): - 29:
for epoch in classEpochs do - 30:
← MultiHead(,) - 31:
← FC1() - 32:
← FC2() - 33:
← FC3() - 34:
← Class() - 35:
end for - 36:
Output ← () |
The proposed algorithm works by taking an input as the subject data and associated label and output the accuracy of the model on the testing dataset. Algorithm is divided into the three sub modules which we cal as Features Module , Data Augment Module and the Classification Module. The function "Train-Test-Split" as described in the line 3 is used to partition the dataset into the training and testing dataset by considering stratification. The function "PearsonCorr" is the correlation coefficient as mentioned in Eq
1 and the function "UpperTria" is used to extract the unique values from the symmetric 116 x 116 connectivity matrix. The function "Generator" and "Discriminator" are the neural networks used to train the CGAN in the adversarial manner. The function "MultiHead" , "FC’s" and "Class" are the multi-head based attention layer , fully connected layers and the classification layer of the Classification Module.
3. Results
In the following three subsections , we now describe experimental settings and the types of experiments that we performed in the current study.
3.1. Experimental Settings
We used Denoising Autoencoder to reconstruct data features and map the generated cGAN based features to validate generated subjects data distribution with the original subjects features data.
3.1.1. Denoising Autoencoder
Denoising Autoencoder has 5 encoder layers of size 4000,3000,2000,1000,500 and 5 decoder layers of size 1000,2000,3000,4000,6670 ,"Batch Normalization" and "Relu" activation are used after each of the encoder and decoder layers. "Learning Rate" was set to 0.00001, "Epochs" set to 5000 , "Batch Size" set to 64 and the "Adam" optimization was used for the training of training while a random "Gaussian" noise was added to the input to make the decoder part of the model more robust.
3.1.2. CGAN
The Generator has a total of 5 layers, noise dimension for random vector generation was set to 500 and the condition dimension for ASD and HC was also set to 500 as our purpose is to train the conditional GAN. Before the first layer we used and embedding layer for converting the two categories to a combined noise and condition layer resulting in 1000 dimension. After the embedding layer , the layers have sizes of 2000,3000,4000,5000 and 6670 , each layer was followed by Spectral Normalization layer, Batch Normalization layer and a leakyRelu activation of 0.1. The Discriminator has 5 layers of sizes 4000,2000,1000,500 and 1 where each layer was followed by spectral normalization and a LeakyRelu of 0.1. Before the first layer of the Discriminator the Embedding layer was used to convert the ASD/HC categories to 500 conditional dimension and then the first input layer has input size of Features size that is 6670 and a conditional dimension size of 500. The learning rate for the Discriminator and Generator was set to 0.00001 , Optimization was set to Adam , Batch Size to 64 and the total Epochs were set to 5000 for training of the conditional GAN.
3.1.3. Classification
The Classification module consisted of a Multi-Head attention block , which has input of size 6670 , followed by Multi-Head Attention layer with 10 heads , A skip layer from input to the out of Multi-Head Attention Layer and a Normalization layer followed by a three feed forward layers of size 3000,2000,1000,1 containing skip connections from previous layer and a normalization layer . We set Epochs to 5000 , Batch Size to 64 , Learning Rate to 0.0001 and Optimizer to Adam for the training of the classification module.
3.2. Experiments
There are two kinds of experiments that we conducted for the current work , An overall accuracy comparison where the whole dataset was first divided into the training and testing split using stratification and then the model got trained on the combined training data and then tested on the combined testing dataset. A site wise comparison where we used the largest site NYU for the training , trained the model on this site and then tested on the remaining sites but excluding the site CMU because it contained too few subjects to compare.
3.2.1. Overall Comparison
In the first experiment we compared the results of the proposed approach with the state-of-the-art approaches as shown in
Table 2. It can be seen that our proposed approach accuracy , precision and recall have outperformed the other approaches which validates the effectiveness of the proposed approach on the whole data.
3.2.2. Site Wise Comparison
In the second comparison we used the "NYU" training total dataset as the training dataset and tested the trained model on the rest of the 15 sites excluding the "CMU" site as done in [
21] . We have shown the results of the proposed approach with the state-of-the-art approaches and it can be seen that our proposed approach has outperformed the other approaches in 10 out of the 15 sites validating the effectiveness of the proposed approach.
Table 3.
Site wise dataset comparison with the State-of-the-Art(%).
Table 3.
Site wise dataset comparison with the State-of-the-Art(%).
| |
Site |
Size |
MHSA[21] |
MVES[22] |
ASD-GANNet |
| 1 |
CALTECH |
37 |
64.60 |
71.40 |
71.60 |
| 2 |
KKI |
39 |
79.60 |
75.00 |
75.40 |
| 3 |
LEUVEN |
61 |
70.40 |
72.10 |
73.20 |
| 4 |
MaxMun |
42 |
66.40 |
61.50 |
66.90 |
| 5 |
OUSH |
23 |
66.00 |
- |
60.00 |
| 6 |
Olin |
25 |
76.00 |
73.50 |
76.00 |
| 7 |
Pitt |
45 |
65.80 |
74.50 |
73.00 |
| 8 |
SBL |
26 |
89.30 |
64.30 |
65.00 |
| 9 |
SDSU |
33 |
75.70 |
79.30 |
79.60 |
| 10 |
Stanford |
36 |
77.50 |
69.20 |
70.00 |
| 11 |
Trinity |
44 |
73.10 |
73.30 |
74.00 |
| 12 |
UCLA |
75 |
69.30 |
77.60 |
78.50 |
| 13 |
USM |
113 |
76.90 |
87.30 |
88.10 |
| 14 |
UM |
61 |
74.90 |
75.70 |
75.90 |
| 15 |
Yale |
48 |
75.10 |
80.00 |
81.10 |
4. Discussion
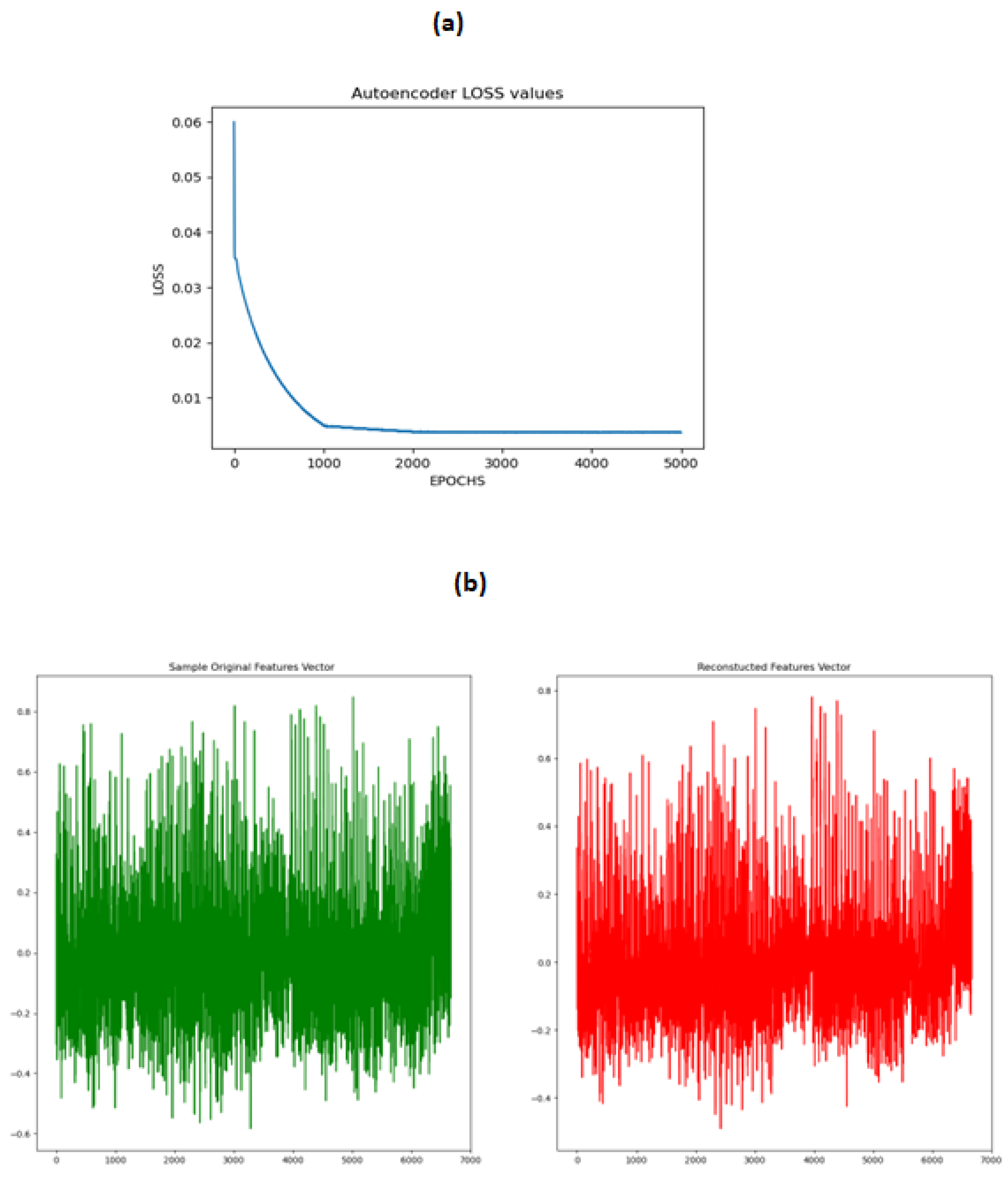
Generating the synthetic subject’s connectivity features using CGAN is not a trivial task because we have no idea in advance about the goodness of the synthetic subject’s features set. To tackle this issue we sampled a small subset of the training data and trained a Denoising Autoencoder on this dataset . The objective was that the after training the Autoencoder on a small subset of subjects, during the CGAN training stage the generated subject will be fed to the trained Autoencoder and then the reconstructed subject using the decoder of the Autoencoder will be compared to the original generated subject using the mean squared error to decide if the generated subject lies in the latent space of the previously trained subjects. This step is crucial because even after the minimization of the loss of the Generator and the Discriminator of the CGAN , we should know how close the generated subject is to the distribution of the original subjects. The Autoencoder loss and the comparison of the original and the reconstructed features are shown in
Figure 7 .
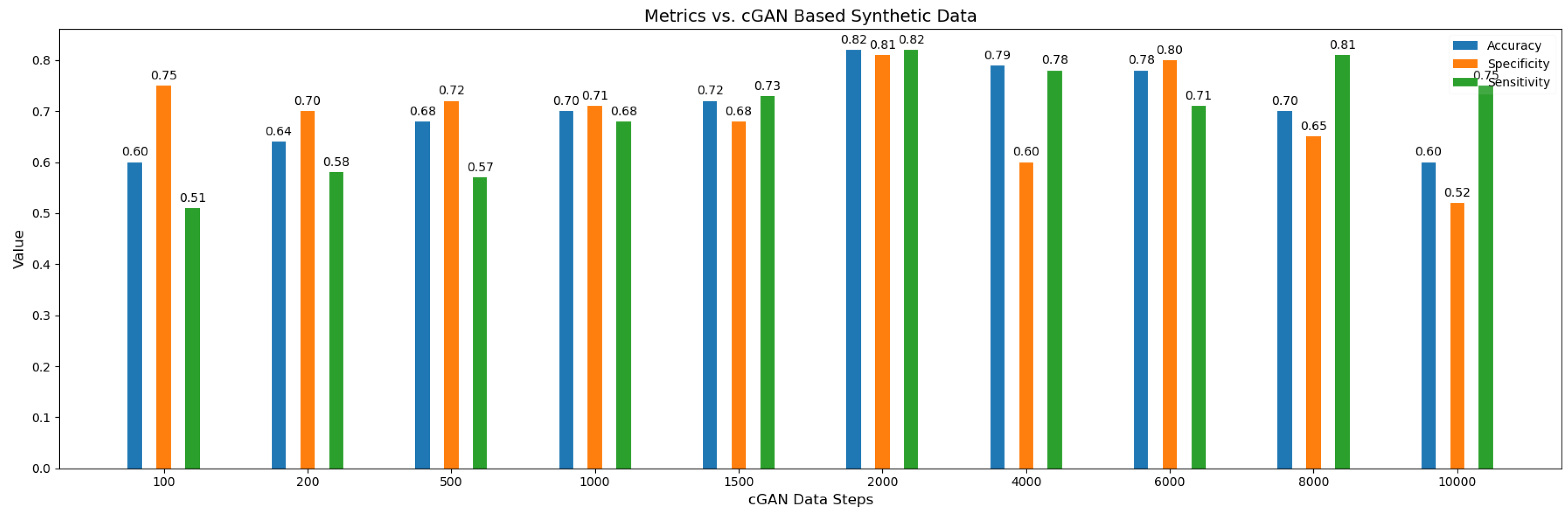
We have also done experimentation with varying number of synthetic subjects as shown in
Figure 8 , It can be seen the best metrics result when the number of the synthetic subjects are 2000 . There appears to be trend in the values of the accuracy across the synthetic subjects as when the number of synthetic subjects are lower then the values of accuracy is low but it gradually increases until peaking at when the number of subjects are 2000 which shows the over-fitting issue gets resolved gradually by the model when attaining sufficient number of subjects. On the other hand too many synthetic subjects is also not a favorable scenario for the model because in that case model will learn more and more the distribution of synthetic data instead of the original data and in a way will forget about the true distribution resulting in lower accuracy.
4.1. Ablation Studies
We have also compared the performance of our model by taking out the essential components out of our proposed approach to analyze how the proposed model behaves in scenarios when those components are present or absent from the model. For the purpose of comparison , we did two kinds of experiments where first we only used the GAN component named as "GAN Only" in which the down stream task of classification was done using a simple neural network consisting of 7 fully connected layers of 4000,3000,2000,1000,500,50,1 where each layer was followed by "BatchNorm" , "ReLU" and "Dropout" of 10%. In the second experiment we only used the "Muti-Head" attention layer named as "Multi-Head Only" and did not use GAN for synthetic data generation.
For both kinds of experiments the results of cross validated accuracy on the whole dataset and on the site wise are displayed in
Table 4 and
Table 5 respectively. It is obvious from both the tables that the accuracy results suffer significantly in both the comparison , which proves that both the GAN and Multi-Head attention component are essential for the proposed approach .
5. Conclusions
We proposed ASD-GANNet , a GAN based data augmentation technique for the classification of ASD on the fMRI pre-processed ABIDE dataset. Instead of directly generating the subjects we tackled the problem of varying time points across subjects by generating the fixed length connectivity features . Our proposed framework is promising as we not only achieved good performance but also compared our results with the state-of-the-art methods in literature to prove the effectiveness of the proposed approach. The major limitation of the proposed approach is that we were not able to generate the original time varying like synthetic data using GANs . In the future work we will explore how the subjects belonging to ASD or HC could be generated instead of the fixed length correlation features and how the synthetic subjects distributions align with the overall original data distribution and the site wise distribution of the ASD and HC subjects.
Author Contributions
N.A.K conceived the idea , did the experiments , did the detailed analysis of the research work and prepared the manuscript. S.X repeatedly analyzed the results , did critical analysis , reviewed the draft and gave valuable suggestions.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 61772426).
Data Availability Statement
ABIDE pre-processed fMRI dataset is a publicly available dataset maintained at
http://preprocessed-connectomes-project.org/abide/. To download the dataset users can go to the Github tab of the mentioned url and under the ”abide” repo can run the script named ”download_abide_preproc.py” to download all the pre-processed fMRI dataset of ASD.
Acknowledgments
We are grateful to China National Natural Science Foundation , China Scholarships Council , Data Mining Laboratory of Northwestern Polytechnical University , China for giving us administrative and moral support to pursue the proposed research works.
Conflicts of Interest
”The authors declare no conflicts of interest”.
Abbreviations
The following abbreviations are used in this manuscript:
| ABIDE |
Autism Brain Imaging Data Exchange |
| ASD |
Autism Spectrum Disorder |
| CGAN |
Conditional Generative Adversarial Networks |
| fMRI |
Functional Magnetic Resonance Imaging |
| GAN |
Generative Adversarial Networks |
| WGAN |
Wasserstein Generative Adversarial Networks |
| rs-fMRI |
Resting State Functional Magnetic Resonance Imaging |
References
- American Psychiatric Association, D.; Association, A.P.; others. Diagnostic and statistical manual of mental disorders: DSM-5; Vol. 5, American psychiatric association Washington, DC, 2013.
- Lord, C.; Elsabbagh, M.; Baird, G.; Veenstra-Vanderweele, J. Autism spectrum disorder. The lancet 2018, 392, 508–520. [Google Scholar]
- Hirota, T.; King, B.H. Autism spectrum disorder: A review. Jama 2023, 329, 157–168. [Google Scholar]
- Gabbay-Dizdar, N.; Ilan, M.; Meiri, G.; Faroy, M.; Michaelovski, A.; Flusser, H.; Menashe, I.; Koller, J.; Zachor, D.A.; Dinstein, I. Early diagnosis of autism in the community is associated with marked improvement in social symptoms within 1–2 years. Autism 2022, 26, 1353–1363. [Google Scholar]
- Iyama-Kurtycz, T.; Iyama-Kurtycz, T. Obstacles to diagnosing autism spectrum disorder and myths about autism. Diagnosing and Caring for the Child with Autism Spectrum Disorder: A Practical Guide for the Primary Care Provider 2020, pp. 9–17.
- McCarty, P.; Frye, R.E. Early detection and diagnosis of autism spectrum disorder: Why is it so difficult? Seminars in Pediatric Neurology. Elsevier, 2020, Vol. 35, p. 100831.
- Soares, J.M.; Magalhães, R.; Moreira, P.S.; Sousa, A.; Ganz, E.; Sampaio, A.; Alves, V.; Marques, P.; Sousa, N. A Hitchhiker’s guide to functional magnetic resonance imaging. Frontiers in neuroscience 2016, 10, 218898. [Google Scholar]
- Chow, M.S.; Wu, S.L.; Webb, S.E.; Gluskin, K.; Yew, D. Functional magnetic resonance imaging and the brain: A brief review. World journal of radiology 2017, 9, 5. [Google Scholar]
- Lajoie, I.; Nugent, S.; Debacker, C.; Dyson, K.; Tancredi, F.B.; Badhwar, A.; Belleville, S.; Deschaintre, Y.; Bellec, P.; Doyon, J.; others. Application of calibrated fMRI in Alzheimer’s disease. NeuroImage: Clinical 2017, 15, 348–358. [Google Scholar]
- Yap, K.H.; Manan, H.A.; Sharip, S. Heterogeneity in brain functional changes of cognitive processing in ADHD across age: a systematic review of task-based fMRI studies. Behavioural Brain Research 2021, 397, 112888. [Google Scholar]
- Santana, C.P.; de Carvalho, E.A.; Rodrigues, I.D.; Bastos, G.S.; de Souza, A.D.; de Brito, L.L. rs-fMRI and machine learning for ASD diagnosis: A systematic review and meta-analysis. Scientific reports 2022, 12, 6030. [Google Scholar]
- Guo, L.; Bai, G.; Zhang, H.; Lu, D.; Zheng, J.; Xu, G. Cognitive functioning in temporal lobe epilepsy: a BOLD-fMRI study. Molecular neurobiology 2017, 54, 8361–8369. [Google Scholar]
- Casteràs, D.P.; Cano, M.; Camprodon, J.; Loo, C.; Palao, D.; Soriano-Mas, C.; Cardoner, N. A multimetric systematic review of fMRI findings in patients with MDD receiving ECT. Eur Neuropsychopharmacol 2021, 44, S47. [Google Scholar] [CrossRef]
- Wolters, A.F.; van de Weijer, S.C.; Leentjens, A.F.; Duits, A.A.; Jacobs, H.I.; Kuijf, M.L. “Resting-state fMRI in Parkinson’s disease patients with cognitive impairment: A meta-analysis”: Answer to Wang and colleagues. Parkinsonism & Related Disorders 2019, 66, 253–254. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Communications of the ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Nazir, M.; Shakil, S.; Khurshid, K. Role of deep learning in brain tumor detection and classification (2015 to 2020): A review. Computerized medical imaging and graphics 2021, 91, 101940. [Google Scholar]
- Lavecchia, A. Deep learning in drug discovery: opportunities, challenges and future prospects. Drug discovery today 2019, 24, 2017–2032. [Google Scholar] [CrossRef]
- Lauriola, I.; Lavelli, A.; Aiolli, F. An introduction to deep learning in natural language processing: Models, techniques, and tools. Neurocomputing 2022, 470, 443–456. [Google Scholar]
- Yang, J.; Hu, M.; Hu, Y.; Zhang, Z.; Zhong, J. Diagnosis of Autism Spectrum Disorder (ASD) Using Recursive Feature Elimination–Graph Neural Network (RFE–GNN) and Phenotypic Feature Extractor (PFE). Sensors 2023, 23, 9647. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Feng, F.; Ye, S.; Mao, Y.; Chen, X.; Li, Y.; Ning, M.; Zhang, M. Multi-head self-attention mechanism-based global feature learning model for ASD diagnosis. Biomedical Signal Processing and Control 2024, 91, 106090. [Google Scholar]
- Kang, L.; Chen, J.; Huang, J.; Jiang, J. Autism spectrum disorder recognition based on multi-view ensemble learning with multi-site fMRI. Cognitive Neurodynamics 2023, 17, 345–355. [Google Scholar] [CrossRef]
- Zhang, F.; Wei, Y.; Liu, J.; Wang, Y.; Xi, W.; Pan, Y. Identification of Autism spectrum disorder based on a novel feature selection method and Variational Autoencoder. Computers in Biology and Medicine 2022, 148, 105854. [Google Scholar] [CrossRef]
- Wang, N.; Yao, D.; Ma, L.; Liu, M. Multi-site clustering and nested feature extraction for identifying autism spectrum disorder with resting-state fMRI. Medical image analysis 2022, 75, 102279. [Google Scholar]
- Cao, M.; Yang, M.; Qin, C.; Zhu, X.; Chen, Y.; Wang, J.; Liu, T. Using DeepGCN to identify the autism spectrum disorder from multi-site resting-state data. Biomedical Signal Processing and Control 2021, 70, 103015. [Google Scholar]
- Di Martino, A.; Yan, C.G.; Li, Q.; Denio, E.; Castellanos, F.X.; Alaerts, K.; Anderson, J.S.; Assaf, M.; Bookheimer, S.Y.; Dapretto, M.; others. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular psychiatry 2014, 19, 659–667. [Google Scholar]
- Craddock, C.; Benhajali, Y.; Chu, C.; Chouinard, F.; Evans, A.; Jakab, A.; Khundrakpam, B.S.; Lewis, J.D.; Li, Q.; Milham, M.; others. The neuro bureau preprocessing initiative: open sharing of preprocessed neuroimaging data and derivatives. Frontiers in Neuroinformatics 2013, 7, 5. [Google Scholar]
- Korhonen, O.; Saarimäki, H.; Glerean, E.; Sams, M.; Saramäki, J. Consistency of regions of interest as nodes of fMRI functional brain networks. Network Neuroscience 2017, 1, 254–274. [Google Scholar]
- Lips, M.A.; Wijngaarden, M.A.; van der Grond, J.; van Buchem, M.A.; de Groot, G.H.; Rombouts, S.A.; Pijl, H.; Veer, I.M. Resting-state functional connectivity of brain regions involved in cognitive control, motivation, and reward is enhanced in obese females. The American journal of clinical nutrition 2014, 100, 524–531. [Google Scholar]
- Bijsterbosch, J.D.; Woolrich, M.W.; Glasser, M.F.; Robinson, E.C.; Beckmann, C.F.; Van Essen, D.C.; Harrison, S.J.; Smith, S.M. The relationship between spatial configuration and functional connectivity of brain regions. elife 2018, 7, e32992. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Communications of the ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Mirza, M.; Osindero, S. Conditional generative adversarial nets. arXiv preprint arXiv:1411.1784, arXiv:1411.1784 2014.
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Advances in neural information processing systems 2017, 30. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).