1. Introduction
This study was undertaken in response to an article published in JDD Sept 20231, mainly related to the in vitro gene expression analysis segment. In that study, a 3-dimensional (3D) full-thickness human skin model (EpiDermFT, MatTek Corporation, Ashland, MA) was utilized with prior irradiation to simulate photodamaged skin. Topical application of the test product, a topical firming and toning body lotion (FTB – SkinMedica®, Allergan Aesthetics, an AbbVie Company, Irvine, CA) and multiple comparator products were applied, including TransFORM Body Treatment with TriHex Technology® (ATF – Alastin Skincare®, a Galderma company Fort Worth, Texas, USA). The expression of various genes related to extracellular matrix (ECM), angiogenesis, autophagy, etc.) was then measured after 24 hours of incubation with the selected products using RT-PCR.
The results showed only the tested agent—FTB—upregulated all the reported genes, with a remarkable nonresponse or downregulation of genes across for all the other 8 competitors.
When one product produces an isolated and unique response in a multitude of selected genes among a host of comparable products, this sets off alarm bells. Thus, we undertook this study to assess the repeatability of the reported findings and further investigate gene upregulation at more than one time point to observe gene transcription effects. We then further validated these results by examining protein expression in an appropriate ex vivo model as the translation sequence to the gene response.
2. Materials and Methods
Ex Vivo Model and Treatments
All experiments were conducted by 3D Genomics (Carlsbad, CA) as an independent laboratory using an established ex vivo model2. Initial skin culture and histology preparation involved photodamaged skin derived from patients undergoing facelift procedures (study approved under Veritas Institutional Review Board—study ID # 3192). Discarded skin was received within 2 h of removal from patients. All skin processing was conducted under BSL2 laboratory conditions. The skin was washed in PBS and defatted if necessary. Any visible hairs were shaved using a scalpel. The skin was then cut into ~5mm x 5mm square pieces and placed into transwells suspended in 6 well plates. About 2.0ml of Skin Media (DMEM/F12 Media, Adenine (50 uM), CaCl2 (1.88 mM), T3 Tri-iodothyronine (0.02 nM), Insulin-Transferrin-Selenium-Ethanolamine (ITS -X), Antibiotic-Antimycotic / Penicillin/Streptomycin 1%, 2% Heat Inactivated FBS, Glutagro 1%, and Gentamicin (0.01 mg/ml) was added to each well, and about 200ul to 300ul was added to each transwell to surround the skin sample while maintaining an air-exposed epidermal surface. Media was changed daily. The skin in the transwell cultures was kept under standard conditions in the 37˚C 5% CO2 incubator for about 72 hours before initiating treatment. The study was conducted in 2 parts, including tissue staining and gene expression assessments. These studies aimed to replicate the in vitro parts from Makino et al. 2023 JDD using their product (FTB) compared to Alastin body product (ATF- TransFORM).
Treatments were added each day. About 100ul to 500ul of each compound formulation (TransFORM and FTB) was placed on the surface of a sterile petri plate. Skin was retrieved from the transwell culture plate using sharp forceps to pick up the skin at the edge with minimal forceps compression. The skin was gently swiped over the formulation to completely cover the epidermis and then returned to the transwell culture plate. The treatment with above process was repeated daily for seven days. One set was left untreated as the baseline comparison. A total of 18 skin samples were processed, all from one individual. Twelve skin samples were used for the immunohistochemistry arm and 6 for the gene expression arm.
Gene Expression Analysis
Skin samples (untreated and treated with TransFORM and FTB; n=2) were collected on days 1 (to compare to the Makino et al. 2023 study) and 3 (as an additional assessment). Upon removal from treatment, the tissue samples were washed in PBS and immediately stored in RNALater at -80C until all 18 samples were collected. RNA was prepared from each skin sample using the Qiagen RNEasy protocol. The mRNA was quantitated on the bioanalyzer.
Sybr green qPCR oligos were purchased from Integrated DNA Technologies for 12 genes (
Table 1), with the oligo sequences indicated by the human gene names. The Takara OneStep RT-PCR kit was used to determine the Ct values of the 18 RNA samples x 11 genes in a single 384-well plate for all reactions. HPRT1 was used as the housekeeping gene. The Ct method was used to quantify the gene expression after normalizing to the housekeeping gene.
Immunofluorescence
The treated skin samples were retrieved after 3 days and 7 days of treatment. There was a total of 6 skin samples (2 each for untreated, TransFORM, and FTB) for each time point. The retrieved skin samples were washed twice in PBS and then fixed in 10% neutral buffered formalin (“NBF”) for 24 hours at 4˚C. The skin was then washed in PBS and stored in 70% ethanol. After all the samples were collected, they were paraffin-embedded and sectioned for immunostaining. Slides were stained with an anti-tropoelastin antibody (Elastin Products Co.). The primary antibody was detected with a fluor-conjugated secondary (anti-rabbit 647) (Jackson Immunoresearch). Sections were counterstained with DAPI (ThermoFisher) and imaged on a Zeiss Axio Observer running Zeiss Zen software.
3. Results
RT-PCR Differences
Makino et al. used a 3D full-thickness skin model treated with UV light. The control was treated with H2O, FTB and TransFORM were applied topically to the other samples (15ul) for 24 hrs., and RNA was harvested. The data were normalized to GAPDH. Our study used our established ex vivo model. The treatments were applied as described above. RNA was harvested on days 1 and 3, and the data was normalized to HPRT.
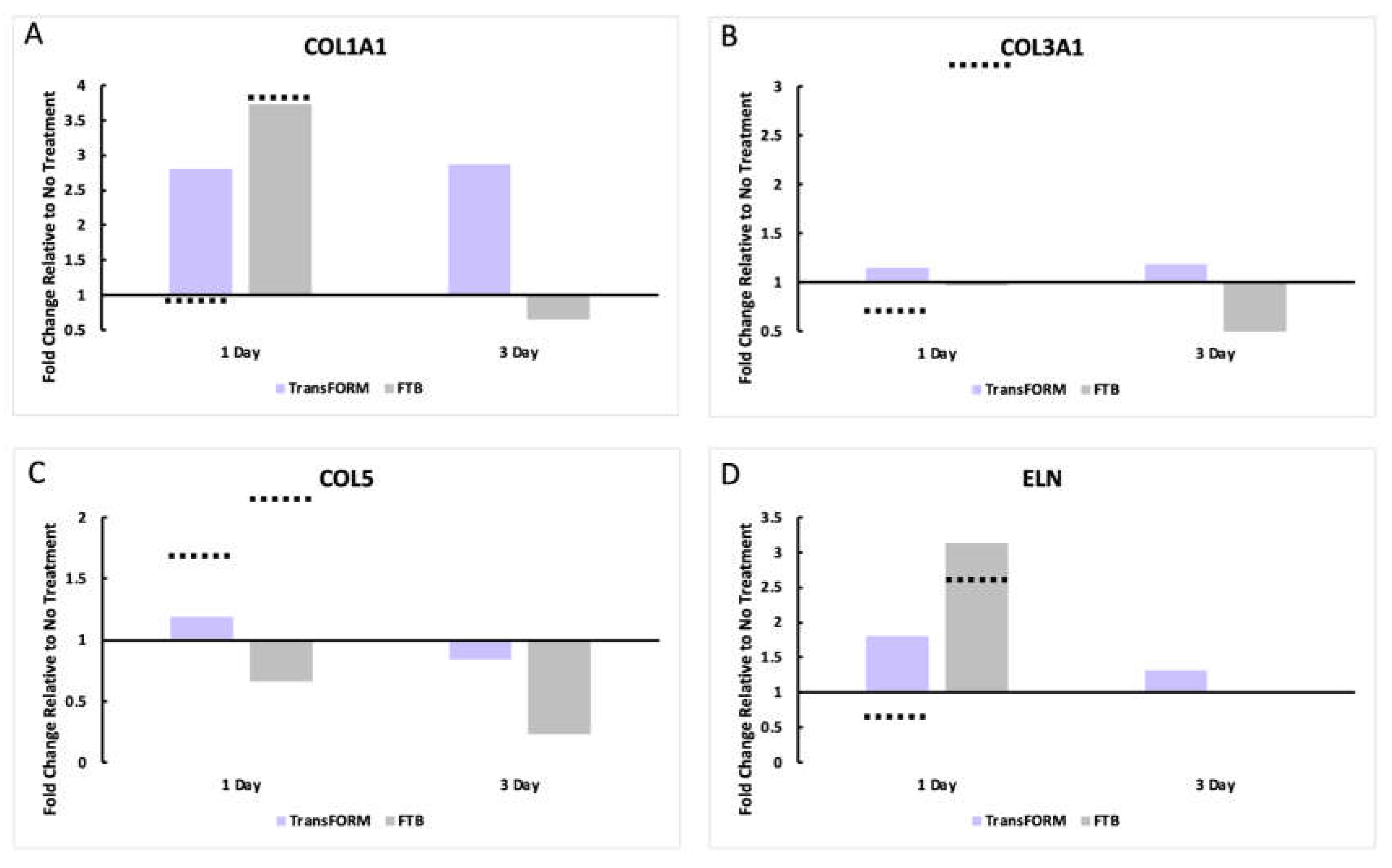
ECM Gene Expression Analysis and Comparison
At 24 hrs., COL1A1 expression on the models treated with FTB was very similar between our study and the previous report (~3.75-fold greater than control). However, after TransFORM treatment, we found a ~2.8-fold increase, and Makino et al. reported that COL1A1 expression did not change with TransFORM treatment. At day 3, we discovered that COL1A1 expression was maintained after TransFORM and diminished to baseline after FTB (
Figure 1A).
The ex vivo skin model was established, and the skin was left untreated or treated with TransFORM or FTB for 24 hrs. and 3 days. Gene expression was analyzed by RT-PCR to assess (A) COL1A1 (collagen I), (B) COL3A1 (collage III), (C) COL5 (collagen V), and (D) ELN (elastin) to evaluate ECM-related genes. The data are presented as the fold-change relative to the untreated sample. The dashed lines represent the approximate expression values presented by Makino et al. as a comparison.COL3A1 expression at 24 hours after treatment with FTB and TransFORM was not different from that of the untreated group in the current study. However, Makino et al. reported a >3-fold upregulation with FTB and a downregulation with TransFORM. At day 3, we observed a slight increase with TransFORM and an evident downregulation with FTB (Figure 1B).
Makino et al. reported an upregulation of >1.5-fold and >2-fold for TransFORM and FTB, respectively, at 24 hours for COL5. Our data revealed a 1.2-fold upregulation with TransFORM and a downregulation with FTB. At day 3, both treatments showed reduced gene expression compared to the control, and the downregulation with FTB was greater than at 24 hrs. (
Figure 1C).
At 24 hrs., TransFORM and FTB upregulated ELN (1.8-fold and 3.1-fold, respectively). In contrast, Makino et al. demonstrated a lower level of upregulation with FTB and a downregulation with TransFORM. After 3 days, ELN was still upregulated (1.3-fold) with TransFORM whereas, was downregulated by FTB and was similar to control (
Figure 1D).
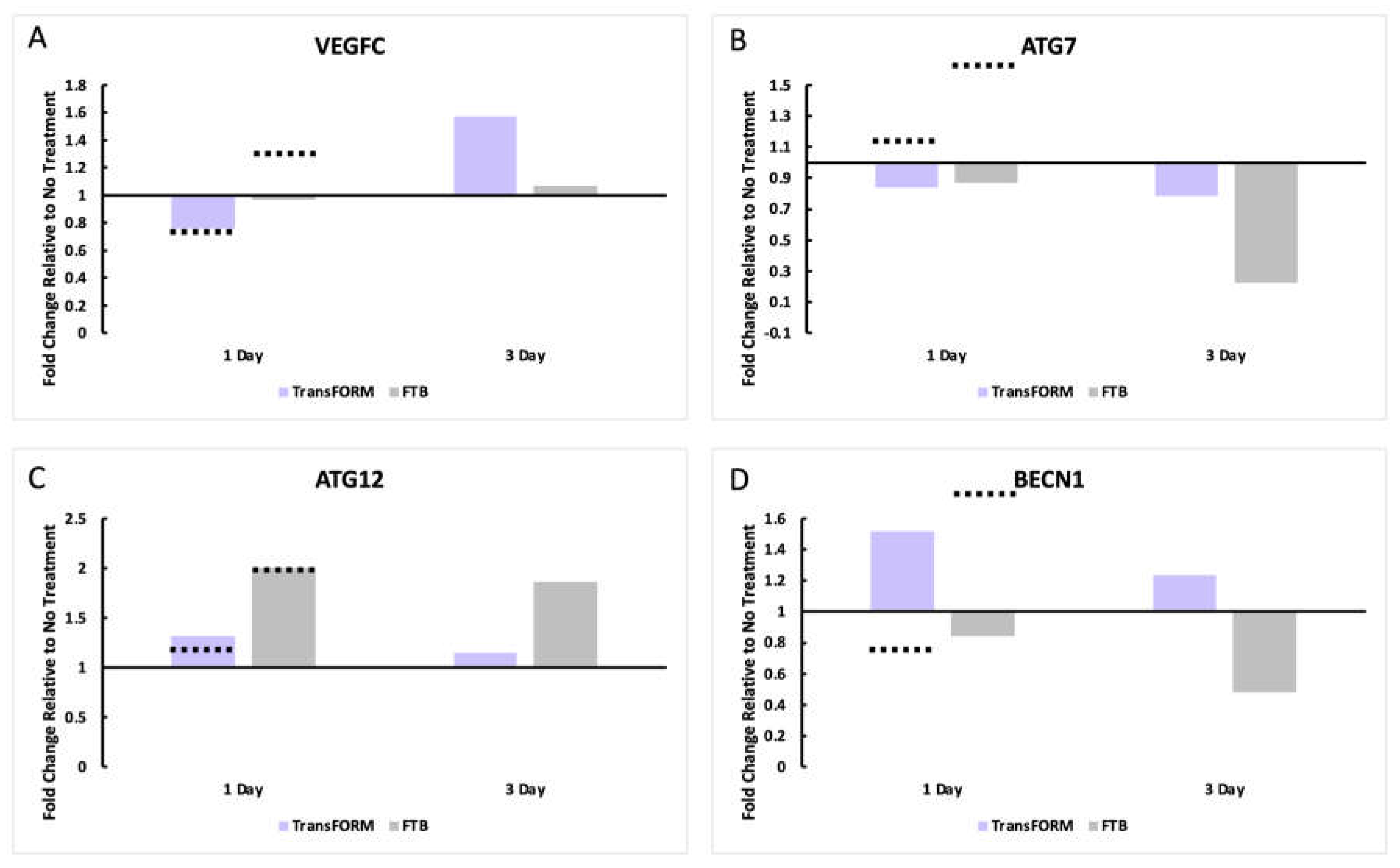
Lymphatic Vessel and Autophagy Gene Expression Analysis and Comparison
At 24 hrs., VEGFC expression was downregulated with TransFORM in our study and by Makino et al. at a similar level. With FTB, Makino et al. reported an upregulation (>1.25-fold), and our results demonstrated no difference compared to the control. At day 3, TransFORM upregulated VEGFC (1.6-fold), while FTB remained unchanged compared to the control (
Figure 2A).
Makino et al. reported that at 24 hrs., both TransFORM and FTB upregulated ATG7 and ATG12, but the upregulation by FTB was greater for both genes (
Figure 2B,C). In contrast, ATG7 was slightly downregulated for both TransFORM and FTB (
Figure 2B). While ATG12 was upregulated by both, mirroring the Makino et al. findings (
Figure 2C). At day 3, the downregulation was potentiated for both treatments for ATG7, and the levels for ATG12 were similar for both treatments, compared to the 24 hrs. results (
Figure 2B,C).
At 24 hrs., Makino et al. reported an upregulation of BECN1 with FTB and a downregulated with TransFORM. Our study revealed the opposite trend. After 3 days, BECN1 remained upregulated with TransFORM and was further downregulated with FTB (
Figure 2D).
The ex vivo skin model was established, and the skin was left untreated or treated with TransFORM or FTB for 24 hrs. and 3 days. Gene expression was analyzed by RT-PCR to assess (A) VEGFC (vascular endothelial growth factor C) to evaluate a lymphatic vessel gene and (B) ATG7 (Autophagy Related 7), (C) ATG12 (Autophagy Related 12), and (D) BECN1 (Beclin1) to evaluate autophagy-related genes. The data are presented as the fold-change relative to the untreated sample. The dashed lines represent the approximate expression values presented by Makino et al. as a comparison.
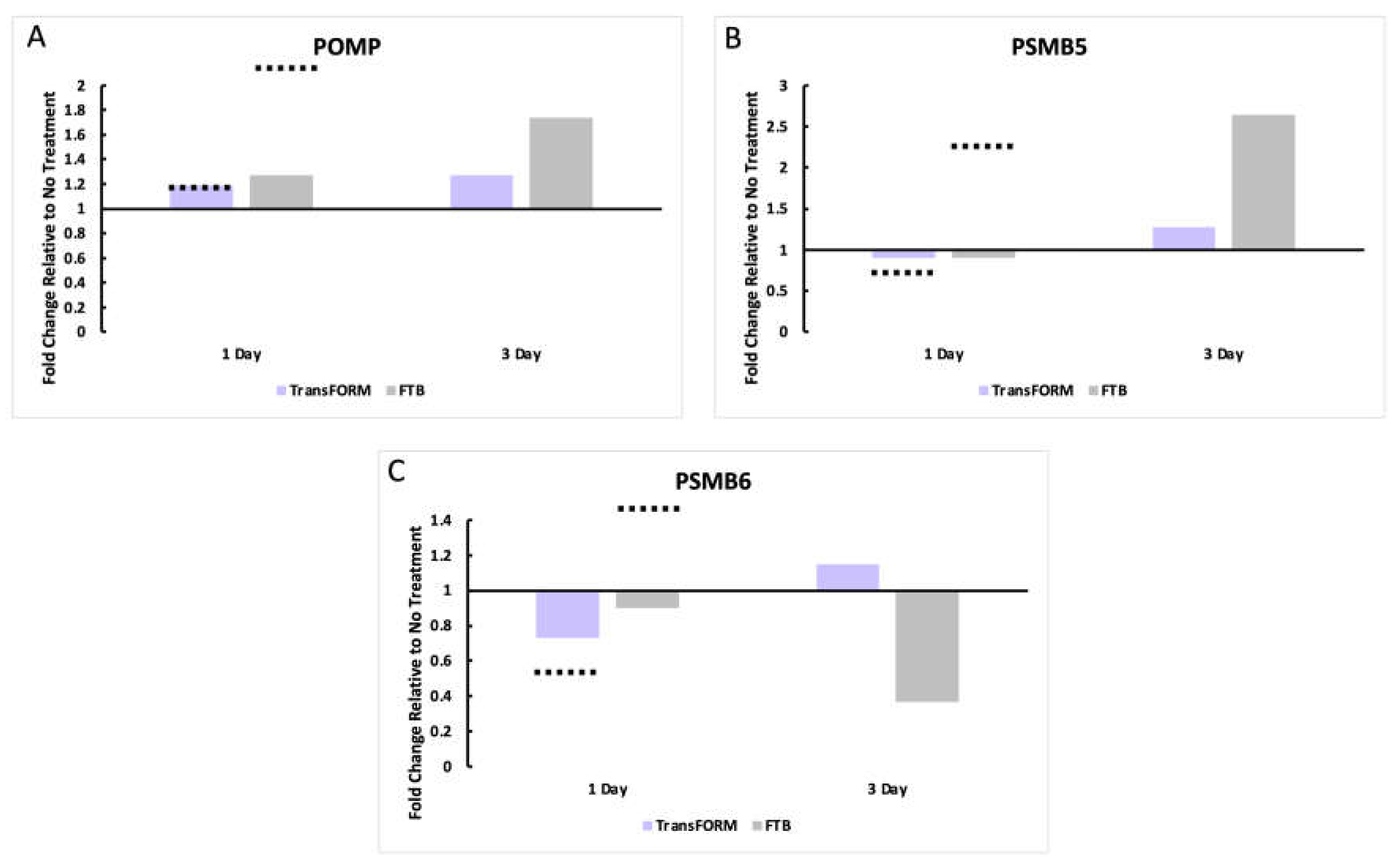
Proteosome Gene Expression Analysis and Comparison
After 24 hrs. with TransFORM treatment, the upregulation of POMP observed by Makino et al. and our group was similar. In contrast, with FTB, we found an upregulation of 1.3-fold, and Makino et al. reported an increase of >2.0-fold. After 3 days, the upregulation of POMP by both treatments we observed at 24 hrs. was potentiated (
Figure 3A).
In our study, PSMB5 was unchanged with TransFORM and FTB after 24 hrs. In contrast, Makino et al. demonstrated a downregulation with TransFORM and an upregulation with FTB (>2-fold). After 3 days, both treatments increased the expression of PSMB5 (
Figure 3B). PSMB6 was downregulated by TransFORM in both our study and as reported by Makino et al. at 24 hrs. However, with FTB, we found no change compared to the control, and Makino et al. demonstrated an upregulation (>1.5-fold). After 3 days, PSMB6 was slightly upregulated with TransFORM and exponentially downregulated with FTB (
Figure 3C).
The ex vivo skin model was established, and the skin was left untreated or treated with TransFORM or FTB for 24 hrs. and 3 days. Gene expression was analyzed by RT-PCR to assess (A) POMP (Proteasome Maturation Protein), (B) PSMB5 (Proteasome 20S Subunit Beta 5), and (C) PSMB6 (Proteasome 20S Subunit Beta 6) to evaluate proteasome-related genes. The data are presented as the fold-change relative to the untreated sample. The dashed lines represent the approximate expression values presented by Makino et al. as a comparison.
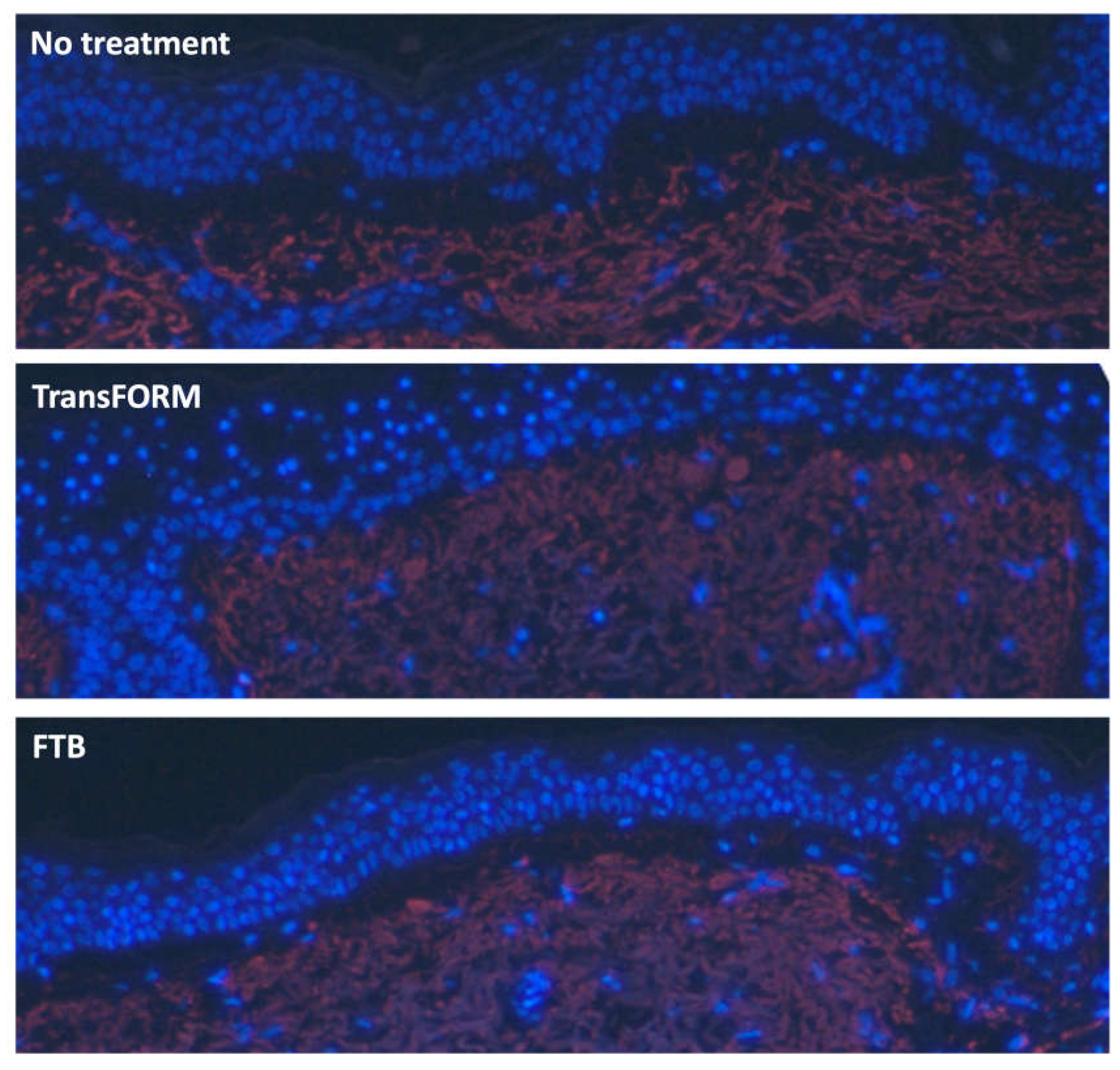
Ex vivo Translational Results
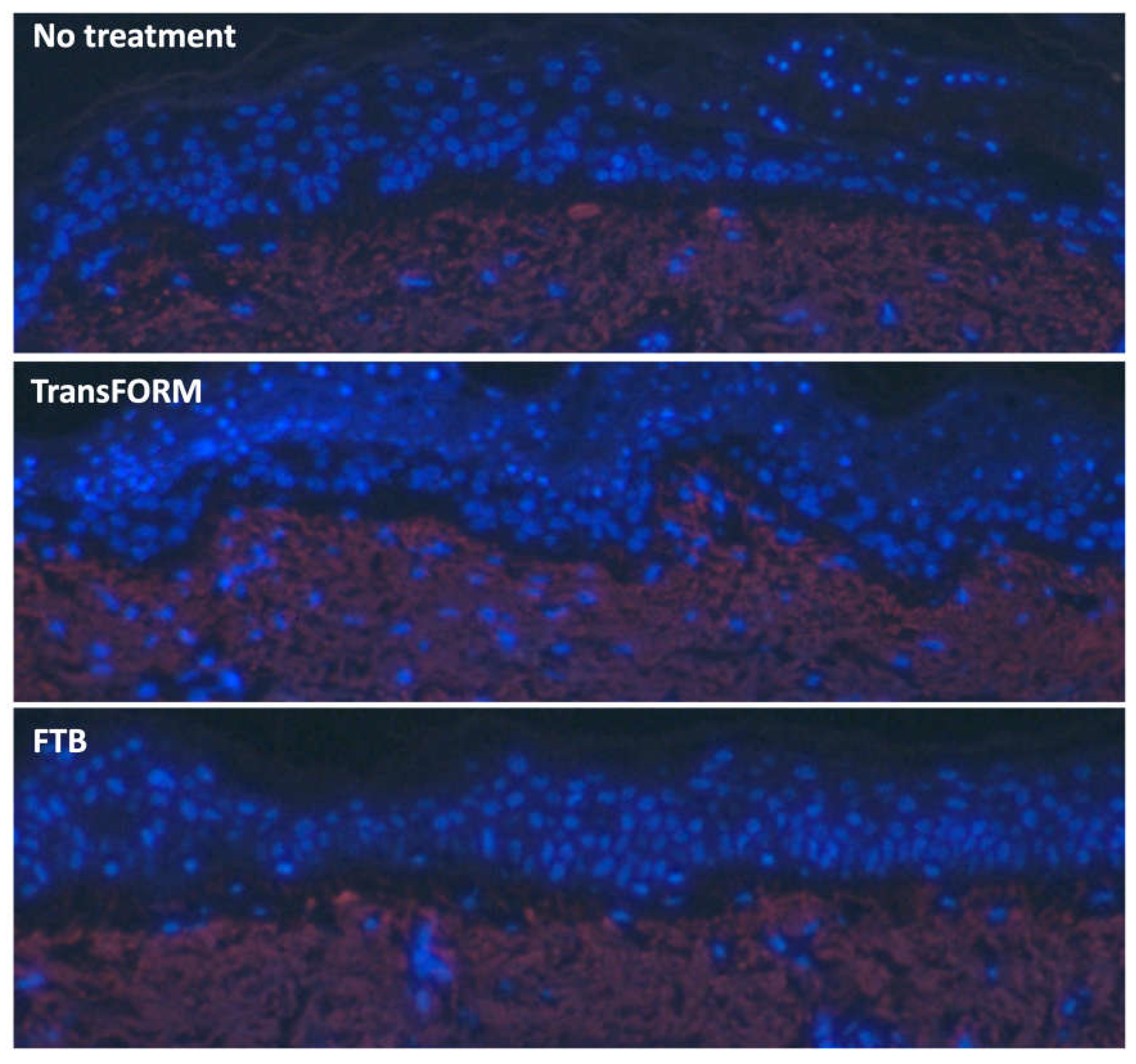
On day 3, both TransFORM and FTB show enhanced elastin in the papillary dermis. However, TransFORM shows elastin fibers extending into the dermal-epidermal junction (DEJ). The gap in the DEJ in the nontreated is similar to the gap after treatment with FTB (
Figure 4). Whereas sample treated with TrasFORM is showing a more intact and tighter DEJ.
The ex vivo skin model was established, and the skin was left untreated or treated with TransFORM or FTB for 3 days. The tissue was processed for immunostaining to assess tropoelastin expression (red). The tissue was counter-stained with DAPI (blue) to detect the nuclei.
On day 7, both TransFORM and FTB showed enhanced elastin staining in the papillary dermis, slightly denser than on day 3. For FTB, the gap in the DEJ remained. For TransFORM, elastin fibers were in the DEJ as on Day 3, and there was an enhanced cellular accumulation in the dermis, likely fibroblast cellular proliferative response. In addition, very significant DEJ undulation was apparent only in the TransFORM group, indicating healthy rejuvenation of the area (
Figure 5).
The ex vivo skin model was established, and the skin was left untreated or treated with TransFORM or FTB for 7 days. The tissue was processed for immunostaining to assess tropoelastin expression (red). The tissue was counter-stained with DAPI (blue) to detect the nuclei.
4. Discussion
Gene transcription studies in ex vivo models need to be carefully designed to not favor one comparator over another. Considerations include, among others; has the study agent been optimized to suit that particular model?; have sufficient time points been selected to account for differing modes of action of different formulations?; are the absorption parameters of the model adequate to test all comparators?
In an effort to avoid these pitfalls, we selected an ex vivo model that already comprises of photodamaged skin (face-lift patients), obviating the need to use acute UV stress exposure in non-sun-damaged (abdominal skin) models. The acute UV light exposure model differs from chronic sun damage and exposes formulations to unique stressors not necessarily relatable to normal physiological situations.
It is imperative to re-examine the model and design of the study when only one agent out of a host of other comparators appears to succeed in upregulating multiple genes. This would suggest parameters within the design that favor that particular test agent. This study bears out this suggestion, and we have demonstrated that almost all reported results differed when examined at multiple time points in a more physiologically matched ex vivo model.
In this study, the gene expression studies were selected or ‘superselected’ in accordance with those chosen by authors of the previous paper1. We excluded many other possible gene differentiators that may have favored TransFORM over FTB and merely recapitulated those reported in the previous study. It is apparent that by measuring the gene expressions at 2 time points, day 1 and day 3, a more comprehensive picture is obtained. It is also evident that the 3-day time point dramatically alters the comparison picture with TransFORM in cases of collagen, elastin, VEGFC, and many autophagic genes, outperforming FTB in most cases. This was not reflected in the previous study, and thus, false conclusions were drawn. This situation of single time points is not new, and we have had to previously dispute similar single time points used as comparators with similar incorrect conclusions drawn from those studies2.
To further elucidate the activity of the formulation, longer-term ex vivo histological analyses were carried out to determine the translational component of the gene upregulation. Tropoelastin staining was carried out to assess stimulation of this protein, an important component of a body product expected to aid in skin tightening. It is evident that although both products stimulated elastin, TransFORM appears to promote elastin to inhabit the DEJ at Day 3, filling this gap far more efficiently than FTB. On day 7, the pattern was maintained with increased rete peg folding and an increased fibroblast presence (DAPI stain) in the dermis, representing a more regenerative ECM milieu.
In the photodamaged control model, the DEJ demonstrates a flattened and irregular morphology characterized by the retractions of the rete pegs or epidermal undulations, indicative of aged skin. Conversely, in the skin ex vivo model treated with TransFORM, alterations at the DEJ level became notably accentuated by day 7, with the undulations showing early signs of becoming more prominent. The rete pegs are essential for enhancing the mechanical characteristics of the skin and preserving homeostasis (3). This observation coincides with a discernible increase in DAPI and tropoelastin staining, indicative of heightened cellular regeneration and proliferation processes.
Contrary to the observed trends in the skin ex vivo model treated with TransFORM, the skin ex vivo model treated with FTB presents a departure from these changes. Instead, after FTB treatment, the tissue exhibited a flattened DEJ morphology, accompanied by a thinning of the epidermal layer and a notable reduction in DAPI staining intensity by day 7.
5. Conclusions
It behooves us as scientists to methodically question results that appear to be significantly slanted in one direction. In this study, we repeated gene expression analysis in previously selected genes and followed this with an ex vivo analysis of histological changes comparing 2 formulations. It is apparent that by adding extra examination points and re-examining the data, the first study published is questionable in its conclusions. In the current model, TransFORM body product outperformed FTB in almost all parameters measured, both from a gene expression standpoint and based on the histological changes observed in an appropriate ex vivo model.
Author Contributions
Conceptualization, ADW. and MEZ.; methodology,ADW and MEZ.; validation,MEZ.; formal analysis, MEZ.; data curation ADW and MEZ.; writing—original draft preparation, ADW, MEZ and FS.; writing—review and editing FS..; supervision ADW.; project administration, ADW and FS.; funding acquisition ADW and FS. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Alastin Skincare, a Galderma company.
Conflicts of Interest
All authors are employees of Galderma. However these studies were carried out by 3D Genomics (Carlsbad CA), and independent company.
References
- Makino, E.T.; Jiang, L.I.; Acevedo, S.F.; et al. Development of a Topical Formulation for Improving Body Skin Quality. Journal of drugs in dermatology 2023, 22, 887–897. [Google Scholar] [PubMed]
- Widgerow, A.D.; Ziegler, M.E.; Garruto, J.A.; Shafiq, F. Antioxidants with proven efficacy and elastin-conserving vitamin C-A new approach to free radical defense. J Cosmet Dermatol. 2023, 22, 3320–3328. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Sun, L.; Liu, Z.; Li, M.; Cao, Y.; Han, L.; Wang, J.; Wu, X.; Sang, S. Rete ridges: Morphogenesis, function, regulation, and reconstruction. Acta Biomater. 2023, 155, 19–34. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).