Submitted:
21 July 2024
Posted:
23 July 2024
You are already at the latest version
Abstract
Keywords:
Introduction
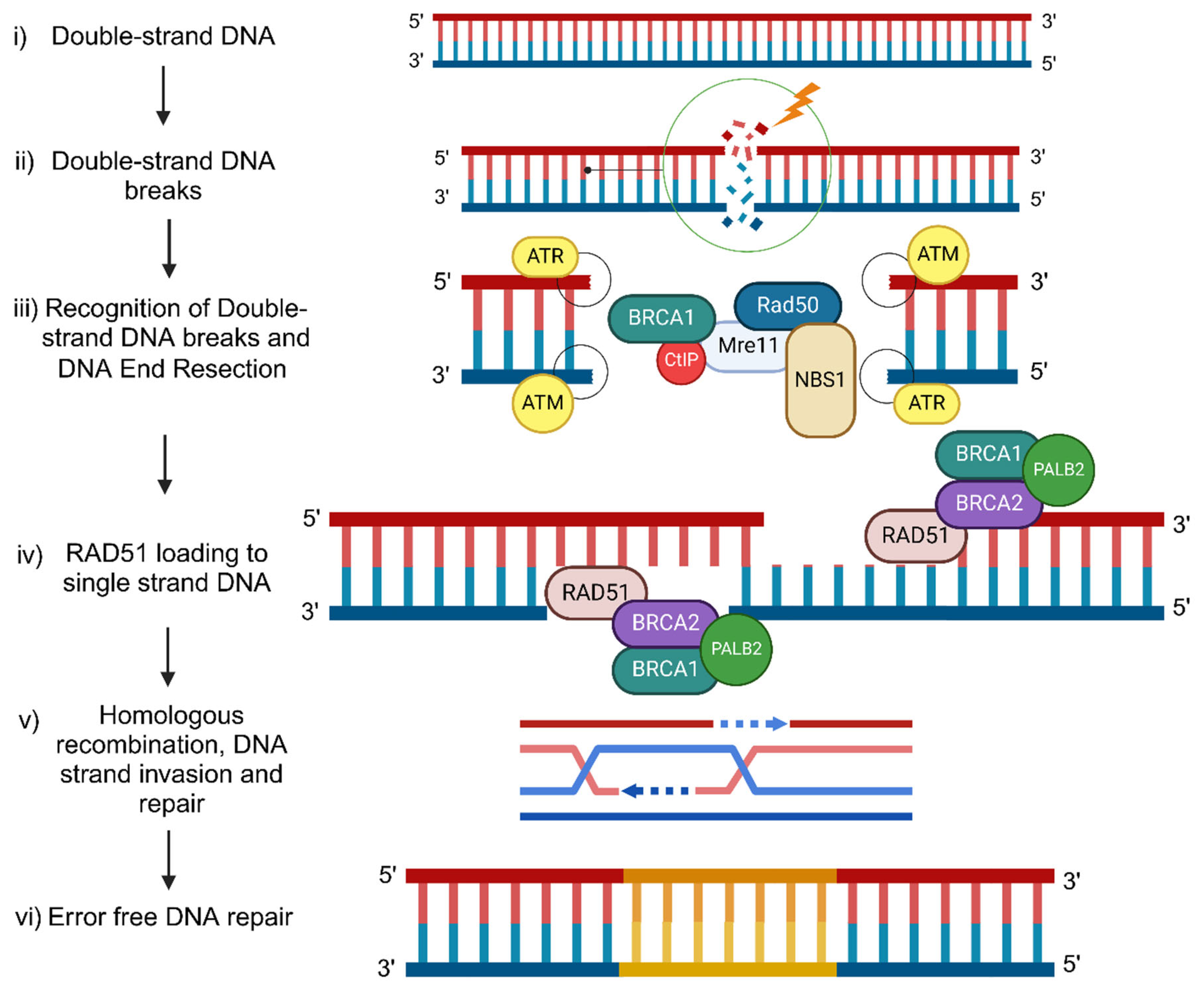
BRCA1/2 molecular mechanism of DNA damage response during Homologous Recombination
DSB: Double-Strand Breaks (DSB)
BRCA1/2-mutated cancers
BRCA1-like cancers
BRCA1/2 promoter methylation in cancers
BRCA1/2 promoter methylation in different cancer types
BrCa
OvCa
Prostate Cancer (PrCa)
Pancreatic Cancer (PaCa)
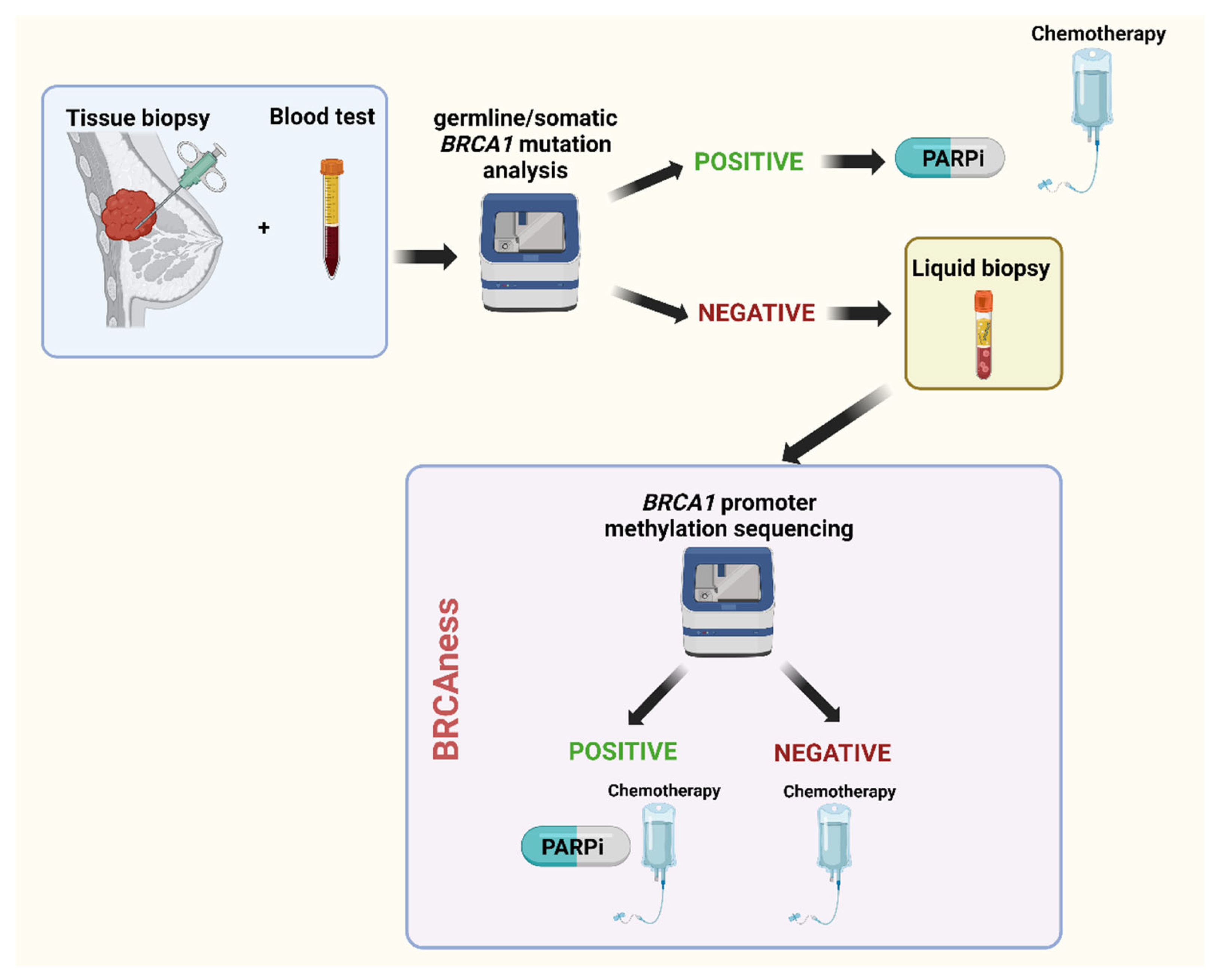
BRCA1/2 methylation in liquid biopsy as a predictive biomarker
BRCA1/2 methylation and treatment strategies
Conclusions
References
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer 2011, 12, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Prakash, R.; Zhang, Y.; Feng, W.; Jasin, M. Homologous Recombination and Human Health: The Roles of BRCA1, BRCA2, and Associated Proteins. Cold Spring Harb. Perspect. Biol. 2015, 7, a016600–a016600. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Wang, S.M. The BRCAness Landscape of Cancer. Cells 2022, 11, 3877. [Google Scholar] [CrossRef] [PubMed]
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.A.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.; Bennett, L.M.; Ding, W.; et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994, 266, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Black, D.M.; Solomon, E. The search for the familial breast/ovarian cancer gene. Trends Genet. 1993, 9, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Wooster, R.; Weber, B.L. Breast and Ovarian Cancer. New Engl. J. Med. 2003, 348, 2339–2347. [Google Scholar] [CrossRef] [PubMed]
- Pilarski, R. The Role of BRCA Testing in Hereditary Pancreatic and Prostate Cancer Families. Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Rebbeck, T.R.; Mitra, N.; Wan, F.; Sinilnikova, O.M.; Healey, S.; McGuffog, L.; Mazoyer, S.; Chenevix-Trench, G.; Easton, D.F.; Antoniou, A.C.; et al. Association of Type and Location ofBRCA1andBRCA2Mutations With Risk of Breast and Ovarian Cancer. JAMA 2015, 313, 1347–1361. [Google Scholar] [CrossRef] [PubMed]
- Kuchenbaecker, K.B.; Hopper, J.L.; Barnes, D.R.; Phillips, K.-A.; Mooij, T.M.; Roos-Blom, M.-J.; Jervis, S.; Van Leeuwen, F.E.; Milne, R.L.; Andrieu, N.; et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 2017, 317, 2402–2416. [Google Scholar] [CrossRef]
- Copson, E.R.; Maishman, T.C.; Tapper, W.J.; I Cutress, R.; Greville-Heygate, S.; Altman, D.G.; Eccles, B.; Gerty, S.; Durcan, L.T.; Jones, L.; et al. Germline BRCA mutation and outcome in young-onset breast cancer (POSH): a prospective cohort study. Lancet Oncol. 2018, 19, 169–180. [Google Scholar] [CrossRef]
- Goodwin, P.J.; Phillips, K.-A.; West, D.W.; Ennis, M.; Hopper, J.L.; John, E.M.; O'Malley, F.P.; Milne, R.L.; Andrulis, I.L.; Friedlander, M.L.; et al. Breast Cancer Prognosis in BRCA1 and BRCA2 Mutation Carriers: An International Prospective Breast Cancer Family Registry Population-Based Cohort Study. J. Clin. Oncol. 2012, 30, 19–26. [Google Scholar] [CrossRef]
- Schmidt, M.K.; Van Den Broek, A.J.; Tollenaar, R.A.; Smit, V.T.; Westenend, P.; Brinkhuis, M.; Oosterhuis, W.J.W.; Wesseling, J.; Janssen-Heijnen, M.L.; Jobsen, J.J.; et al. Breast Cancer Survival of BRCA1/BRCA2 Mutation Carriers in a Hospital-Based Cohort of Young Women. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef]
- Kurian, A.W.; Abrahamse, P.; Bondarenko, I.; Hamilton, A.S.; Deapen, D.; Gomez, S.L.; Morrow, M.; Berek, J.S.; Hofer, T.P.; Katz, S.J.; et al. Association of Genetic Testing Results With Mortality Among Women With Breast Cancer or Ovarian Cancer. JNCI J. Natl. Cancer Inst. 2021, 114, 245–253. [Google Scholar] [CrossRef]
- Bolton, K.L.; Chenevix-Trench, G.; Goh, C.; Sadetzki, S.; Ramus, S.J.; Karlan, B.Y.; Lambrechts, D.; Despierre, E.; Barrowdale, D.; McGuffog, L.; et al. Association Between BRCA1 and BRCA2 Mutations and Survival in Women With Invasive Epithelial Ovarian Cancer. JAMA 2012, 307, 382–390. [Google Scholar] [CrossRef]
- Dobrovic, A.; Simpfendorfer, D. Methylation of the BRCA1 gene in sporadic breast cancer. . 1997, 57, 3347–50. [Google Scholar]
- Catteau, A.; Harris, W.H.; Xu, C.-F.; Solomon, E. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene 1999, 18, 1957–1965. [Google Scholar] [CrossRef]
- Rabiau, N.; Thiam, M.O.; Satih, S.; Guy, L.; Kemeny, J.-L.; Boiteux, J.-P.; Fontana, L.; Bignon, Y.-J.; Bernard-Gallon, D. Methylation analysis of BRCA1, RASSF1, GSTP1 and EPHB2 promoters in prostate biopsies according to different degrees of malignancy. . 2009, 23, 387–91. [Google Scholar]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef]
- Herman, J.G.; Baylin, S.B. Gene Silencing in Cancer in Association with Promoter Hypermethylation. New Engl. J. Med. 2003, 349, 2042–2054. [Google Scholar] [CrossRef]
- Baylin, S.B. DNA methylation and gene silencing in cancer. Nat. Clin. Pr. Oncol. 2005, 2, S4–S11. [Google Scholar] [CrossRef]
- Tutt, A.N.; Garber, J.E.; Kaufman, B.; Viale, G.; Fumagalli, D.; Rastogi, P.; Gelber, R.D.; de Azambuja, E.; Fielding, A.; Balmaña, J.; et al. Adjuvant Olaparib for Patients with BRCA1- or BRCA2-Mutated Breast Cancer. New Engl. J. Med. 2021, 384, 2394–2405. [Google Scholar] [CrossRef]
- Arora, S.; Balasubramaniam, S.; Zhang, H.; Berman, T.; Narayan, P.; Suzman, D.; Bloomquist, E.; Tang, S.; Gong, Y.; Sridhara, R.; et al. FDA Approval Summary: Olaparib Monotherapy or in Combination with Bevacizumab for the Maintenance Treatment of Patients with Advanced Ovarian Cancer. Oncol. 2020, 26, e164–e172. [Google Scholar] [CrossRef]
- Tung, N.; Garber, J.E. PARP inhibition in breast cancer: progress made and future hopes. npj Breast Cancer 2022, 8, 1–5. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, S.; Cheng, S.; Yang, J.; Wang, Y. Clinical application of PARP inhibitors in ovarian cancer: from molecular mechanisms to the current status. J. Ovarian Res. 2023, 16, 1–15. [Google Scholar] [CrossRef]
- Mateo, J.; de Bono, J.S.; Fizazi, K.; Saad, F.; Shore, N.; Sandhu, S.; Chi, K.N.; Agarwal, N.; Olmos, D.; Thiery-Vuillemin, A.; et al. Olaparib for the Treatment of Patients With Metastatic Castration-Resistant Prostate Cancer and Alterations in BRCA1 and/or BRCA2 in the PROfound Trial. J. Clin. Oncol. 2024, 42, 571–583. [Google Scholar] [CrossRef]
- Fizazi, K.; Piulats, J.M.; Reaume, M.N.; Ostler, P.; McDermott, R.; Gingerich, J.R.; Pintus, E.; Sridhar, S.S.; Bambury, R.M.; Emmenegger, U.; et al. Rucaparib or Physician’s Choice in Metastatic Prostate Cancer. New Engl. J. Med. 2023, 388, 719–732. [Google Scholar] [CrossRef]
- Reiss, K.A.; Mick, R.; O'Hara, M.H.; Teitelbaum, U.; Karasic, T.B.; Schneider, C.; Cowden, S.; Southwell, T.; Romeo, J.; Izgur, N.; et al. Phase II Study of Maintenance Rucaparib in Patients With Platinum-Sensitive Advanced Pancreatic Cancer and a Pathogenic Germline or Somatic Variant in BRCA1, BRCA2, or PALB2. J. Clin. Oncol. 2021, 39, 2497–2505. [Google Scholar] [CrossRef]
- Li, S.; He, Y.; Li, C.; Liu, X.; Shen, Y.B.; Wu, Y.; Bai, N.; Li, Q.B. The association between the methylation frequency of BRCA1/2 gene promoter and occurrence and prognosis of breast carcinoma A meta-analysis. Medicine 2020, 99, e19345. [Google Scholar] [CrossRef]
- Kondrashova, O.; Topp, M.; Nesic, K.; Lieschke, E.; Ho, G.Y.; Harrell, M.I.; Zapparoli, G.V.; Hadley, A.; Holian, R.; Boehm, E.; et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Xu, Y.; Diao, L.; Chen, Y.; Liu, Y.; Wang, C.; Ouyang, T.; Li, J.; Wang, T.; Fan, Z.; Fan, T.; et al. Promoter methylation of BRCA1 in triple-negative breast cancer predicts sensitivity to adjuvant chemotherapy. Ann. Oncol. 2013, 24, 1498–1505. [Google Scholar] [CrossRef]
- Sahnane, N.; Carnevali, I.; Formenti, G.; Casarin, J.; Facchi, S.; Bombelli, R.; Di Lauro, E.; Memoli, D.; Salvati, A.; Rizzo, F.; et al. BRCA Methylation Testing Identifies a Subset of Ovarian Carcinomas without Germline Variants That Can Benefit from PARP Inhibitor. Int. J. Mol. Sci. 2020, 21, 9708. [Google Scholar] [CrossRef]
- Gorodetska, I.; Kozeretska, I.; Dubrovska, A. BRCA Genes: The Role in Genome Stability, Cancer Stemness and Therapy Resistance. J. Cancer 2019, 10, 2109–2127. [Google Scholar] [CrossRef]
- Moynahan, M.E.; Jasin, M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010, 11, 196–207. [Google Scholar] [CrossRef]
- Mandel, P. & Metais, P. [Not Available]. Comptes rendus des seances de la Societe de biologie et de ses filiales 142, 241-243 (1948).
- A Clapperton, J.; A Manke, I.; Lowery, D.M.; Ho, T.; Haire, L.F.; Yaffe, M.B.; Smerdon, S.J. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat. Struct. Mol. Biol. 2004, 11, 512–518. [Google Scholar] [CrossRef]
- Her, J.; Lee, N.S.; Kim, Y.; Kim, H. Factors forming the BRCA1-A complex orchestrate BRCA1 recruitment to the sites of DNA damage. Acta Biochim. et Biophys. Sin. 2016, 48, 658–664. [Google Scholar] [CrossRef]
- Zhao, F.; Kim, W.; Kloeber, J.A.; Lou, Z. DNA end resection and its role in DNA replication and DSB repair choice in mammalian cells. Exp. Mol. Med. 2020, 52, 1705–1714. [Google Scholar] [CrossRef]
- Zhao, W.; Steinfeld, J.B.; Liang, F.; Chen, X.; Maranon, D.G.; Ma, C.J.; Kwon, Y.; Rao, T.; Wang, W.; Sheng, C.; et al. BRCA1–BARD1 promotes RAD51-mediated homologous DNA pairing. Nature 2017, 550, 360–365. [Google Scholar] [CrossRef]
- Shah, J.B.; Pueschl, D.; Wubbenhorst, B.; Fan, M.; Pluta, J.; D’andrea, K.; Hubert, A.P.; Shilan, J.S.; Zhou, W.; Kraya, A.A.; et al. Analysis of matched primary and recurrent BRCA1/2 mutation-associated tumors identifies recurrence-specific drivers. Nat. Commun. 2022, 13, 1–19. [Google Scholar] [CrossRef]
- Fu, X.; Tan, W.; Song, Q.; Pei, H.; Li, J. BRCA1 and Breast Cancer: Molecular Mechanisms and Therapeutic Strategies. Front. Cell Dev. Biol. 2022, 10, 813457. [Google Scholar] [CrossRef]
- Petrucelli, N. , Daly, M. B. & Pal, T. in GeneReviews((R)) (eds M. P. Adam et al.) (1993).
- Casaubon, J. T. , Kashyap, S. P. in StatPearls ( 2024.
- Kalachand, R.D.; Stordal, B.; Madden, S.; Chandler, B.; Cunningham, J.; Goode, E.L.; Ruscito, I.; I Braicu, E.; Sehouli, J.; Ignatov, A.; et al. BRCA1Promoter Methylation and Clinical Outcomes in Ovarian Cancer: An Individual Patient Data Meta-Analysis. JNCI J. Natl. Cancer Inst. 2020, 112, 1190–1203. [Google Scholar] [CrossRef]
- Messina, C.; Cattrini, C.; Soldato, D.; Vallome, G.; Caffo, O.; Castro, E.; Olmos, D.; Boccardo, F.; Zanardi, E. BRCA Mutations in Prostate Cancer: Prognostic and Predictive Implications. J. Oncol. 2020, 2020, 1–7. [Google Scholar] [CrossRef]
- Lai, E.; Ziranu, P.; Spanu, D.; Dubois, M.; Pretta, A.; Tolu, S.; Camera, S.; Liscia, N.; Mariani, S.; Persano, M.; et al. BRCA-mutant pancreatic ductal adenocarcinoma. Br. J. Cancer 2021, 125, 1321–1332. [Google Scholar] [CrossRef]
- Miklikova, S.; Trnkova, L.; Plava, J.; Bohac, M.; Kuniakova, M.; Cihova, M. The Role of BRCA1/2-Mutated Tumor Microenvironment in Breast Cancer. Cancers 2021, 13, 575. [Google Scholar] [CrossRef]
- Hill, S.J.; Clark, A.P.; Silver, D.P.; Livingston, D.M. BRCA1 Pathway Function in Basal-Like Breast Cancer Cells. Mol. Cell. Biol. 2014, 34, 3828–3842. [Google Scholar] [CrossRef]
- Severson, T.M.; Peeters, J.; Majewski, I.; Michaut, M.; Bosma, A.; Schouten, P.C.; Chin, S.-F.; Pereira, B.; Goldgraben, M.A.; Bismeijer, T.; et al. BRCA1-like signature in triple negative breast cancer: Molecular and clinical characterization reveals subgroups with therapeutic potential. Mol. Oncol. 2015, 9, 1528–1538. [Google Scholar] [CrossRef]
- Aref-Eshghi, E.; McGee, J.D.; Pedro, V.P.; Kerkhof, J.; Stuart, A.; Ainsworth, P.J.; Lin, H.; Volodarsky, M.; McLachlin, C.M.; Sadikovic, B. Genetic and epigenetic profiling of BRCA1/2 in ovarian tumors reveals additive diagnostic yield and evidence of a genomic BRCA1/2 DNA methylation signature. J. Hum. Genet. 2020, 65, 865–873. [Google Scholar] [CrossRef]
- Nichols, C.A.; Gibson, W.J.; Brown, M.S.; Kosmicki, J.A.; Busanovich, J.P.; Wei, H.; Urbanski, L.M.; Curimjee, N.; Berger, A.C.; Gao, G.F.; et al. Loss of heterozygosity of essential genes represents a widespread class of potential cancer vulnerabilities. Nat. Commun. 2020, 11, 2517. [Google Scholar] [CrossRef]
- Maxwell, K.N.; Wubbenhorst, B.; Wenz, B.M.; De Sloover, D.; Pluta, J.; Emery, L.; Barrett, A.; Kraya, A.A.; Anastopoulos, I.N.; Yu, S.; et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Byrum, A.K.; Vindigni, A.; Mosammaparast, N. Defining and Modulating ‘BRCAness’. Trends Cell Biol. 2019, 29, 740–751. [Google Scholar] [CrossRef]
- Murai, J.; Pommier, Y. BRCAness, Homologous Recombination Deficiencies, and Synthetic Lethality. Cancer Res. 2023, 83, 1173–1174. [Google Scholar] [CrossRef] [PubMed]
- Schouten, P.C.; Marmé, F.; Aulmann, S.; Sinn, H.-P.; van Essen, H.F.; Ylstra, B.; Hauptmann, M.; Schneeweiss, A.; Linn, S.C. Breast Cancers with aBRCA1-like DNA Copy Number Profile Recur Less Often Than Expected after High-Dose Alkylating Chemotherapy. Clin. Cancer Res. 2015, 21, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.; Tutt, A.; Ashworth, A. Hallmarks of 'BRCAness' in sporadic cancers. Nat. Rev. Cancer 2004, 4, 814–819. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, S.; Brown, J.; Yamaguchi, K.; Hamanishi, J.; Yamanoi, K.; Takaya, H.; Kaneyasu, T.; Mori, S.; Mandai, M.; Matsumura, N. Utility of Homologous Recombination Deficiency Biomarkers Across Cancer Types. JCO Precis. Oncol. 2022, 6, e2200085. [Google Scholar] [CrossRef] [PubMed]
- Cruz, C.; Castroviejo-Bermejo, M.; Gutiérrez-Enríquez, S.; Llop-Guevara, A.; Ibrahim, Y.; Gris-Oliver, A.; Bonache, S.; Morancho, B.; Bruna, A.; Rueda, O.; et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann. Oncol. 2018, 29, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Bunting, S.F.; Callén, E.; Wong, N.; Chen, H.-T.; Polato, F.; Gunn, A.; Bothmer, A.; Feldhahn, N.; Fernandez-Capetillo, O.; Cao, L.; et al. 53BP1 Inhibits Homologous Recombination in Brca1-Deficient Cells by Blocking Resection of DNA Breaks. Cell 2010, 141, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Velazquez, C.; Orhan, E.; Tabet, I.; Fenou, L.; Orsetti, B.; Adélaïde, J.; Guille, A.; Thézénas, S.; Crapez, E.; Colombo, P.-E.; et al. BRCA1-methylated triple negative breast cancers previously exposed to neoadjuvant chemotherapy form RAD51 foci and respond poorly to olaparib. Front. Oncol. 2023, 13, 1125021. [Google Scholar] [CrossRef]
- Bouras, E.; Karakioulaki, M.; Bougioukas, K.I.; Aivaliotis, M.; Tzimagiorgis, G.; Chourdakis, M. Gene promoter methylation and cancer: An umbrella review. Gene 2019, 710, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulou, M.; Karaglani, M.; Balgkouranidou, I.; Biziota, E.; Koukaki, T.; Karamitrousis, E.; Nena, E.; Tsamardinos, I.; Kolios, G.; Lianidou, E.; et al. Circulating cell-free DNA in breast cancer: size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene 2019, 38, 3387–3401. [Google Scholar] [CrossRef]
- Panagopoulou, M.; Fanidis, D.; Aidinis, V.; Chatzaki, E. ENPP2 Methylation in Health and Cancer. Int. J. Mol. Sci. 2021, 22, 11958. [Google Scholar] [CrossRef]
- Panagopoulou, M.; Esteller, M.; Chatzaki, E. Circulating Cell-Free DNA in Breast Cancer: Searching for Hidden Information towards Precision Medicine. Cancers 2021, 13, 728. [Google Scholar] [CrossRef] [PubMed]
- Ibragimova, I. & Cairns, P. Assays for hypermethylation of the BRCA1 gene promoter in tumor cells to predict sensitivity to PARP-inhibitor therapy. [CrossRef]
- Birgisdottir, V.; Stefansson, O.A.; Bodvarsdottir, S.K.; Hilmarsdottir, H.; Jonasson, J.G.; Eyfjord, J.E. Epigenetic silencing and deletion of the BRCA1 gene in sporadic breast cancer. Breast Cancer Res. 2006, 8, R38–R38. [Google Scholar] [CrossRef] [PubMed]
- Glodzik, D.; Bosch, A.; Hartman, J.; Aine, M.; Vallon-Christersson, J.; Reuterswärd, C.; Karlsson, A.; Mitra, S.; Niméus, E.; Holm, K.; et al. Comprehensive molecular comparison of BRCA1 hypermethylated and BRCA1 mutated triple negative breast cancers. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Brianese, R.C.; Nakamura, K.D.d.M.; Almeida, F.G.d.S.R.d.; Ramalho, R.F.; Barros, B.D.d.F.; e Ferreira, E.N.; Formiga, M.N.d.C.; de Andrade, V.P.; de Lima, V.C.C.; Carraro, D.M. BRCA1 deficiency is a recurrent event in early-onset triple-negative breast cancer: a comprehensive analysis of germline mutations and somatic promoter methylation. Breast Cancer Res. Treat. 2018, 167, 803–814. [Google Scholar] [CrossRef] [PubMed]
- A Stefansson, O.; Hilmarsdottir, H.; Olafsdottir, K.; Tryggvadottir, L.; Sverrisdottir, A.; Johannsson, O.T.; Jonasson, J.G.; E Eyfjord, J.; Sigurdsson, S. BRCA1 Promoter Methylation Status in 1031 Primary Breast Cancers Predicts Favorable Outcomes Following Chemotherapy. JNCI Cancer Spectr. 2020, 4, pkz100. [Google Scholar] [CrossRef] [PubMed]
- Lønning, P.E.; Nikolaienko, O.; Pan, K.; Kurian, A.W.; Eikesdal, H.P.; Pettinger, M.; Anderson, G.L.; Prentice, R.L.; Chlebowski, R.T.; Knappskog, S. Constitutional BRCA1 Methylation and Risk of Incident Triple-Negative Breast Cancer and High-grade Serous Ovarian Cancer. JAMA Oncol. 2022, 8, 1579–1587. [Google Scholar] [CrossRef] [PubMed]
- Stefansson, O.A.; Jonasson, J.G.; Olafsdottir, K.; Hilmarsdottir, H.; Olafsdottir, G.; Esteller, M.; Johannsson, O.T.; Eyfjord, J.E. CpG island hypermethylation ofBRCA1and loss of pRb as co-occurring events in basal/triple-negative breast cancer. Epigenetics 2011, 6, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.L.; Nguyen, T.T.; Doan, V.T.H.; Vo, L.T.T. Methylation Profiles of BRCA1, RASSF1A and GSTP1 in Vietnamese Women with Breast Cancer. 19, 1887. [Google Scholar] [CrossRef]
- Parrella, P.; Poeta, M.L.; Gallo, A.P.; Prencipe, M.; Scintu, M.; Apicella, A.; Rossiello, R.; Liguoro, G.; Seripa, D.; Gravina, C.; et al. Nonrandom Distribution of Aberrant Promoter Methylation of Cancer-Related Genes in Sporadic Breast Tumors. Clin. Cancer Res. 2004, 10, 5349–5354. [Google Scholar] [CrossRef] [PubMed]
- Kontorovich, T.; Cohen, Y.; Nir, U.; Friedman, E. Promoter methylation patterns of ATM, ATR, BRCA1, BRCA2 and P53 as putative cancer risk modifiers in Jewish BRCA1/BRCA2 mutation carriers. Breast Cancer Res. Treat. 2008, 116, 195–200. [Google Scholar] [CrossRef]
- Sahnane, N.; Rivera, D.; Libera, L.; Carnevali, I.; Banelli, B.; Facchi, S.; Gismondi, V.; Paudice, M.; Cirmena, G.; Vellone, V.G.; et al. Pyrosequencing Assay for BRCA1 Methylation Analysis. J. Mol. Diagn. 2023, 25, 217–226. [Google Scholar] [CrossRef]
- Kawachi, A.; Yamashita, S.; Okochi-Takada, E.; Hirakawa, A.; Tsuda, H.; Shimomura, A.; Kojima, Y.; Yonemori, K.; Fujiwara, Y.; Kinoshita, T.; et al. BRCA1 promoter methylation in breast cancer patients is associated with response to olaparib/eribulin combination therapy. Breast Cancer Res. Treat. 2020, 181, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Geissler, F.; Nesic, K.; Kondrashova, O.; Dobrovic, A.; Swisher, E.M.; Scott, C.L.; Wakefield, M.J. The role of aberrant DNA methylation in cancer initiation and clinical impacts. Ther. Adv. Med Oncol. 2024, 16. [Google Scholar] [CrossRef] [PubMed]
- Lønning, P.E.; Nikolaienko, O.; Pan, K.; Kurian, A.W.; Eikesdal, H.P.; Pettinger, M.; Anderson, G.L.; Prentice, R.L.; Chlebowski, R.T.; Knappskog, S. Constitutional BRCA1 Methylation and Risk of Incident Triple-Negative Breast Cancer and High-grade Serous Ovarian Cancer. JAMA Oncol. 2022, 8, 1579–1587. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, J.; Xu, Y.; Li, Z.; Wen, X.; Yao, L.; Xie, Y.; Deng, D. BRCA1promoter methylation associated with poor survival in Chinese patients with sporadic breast cancer. Cancer Sci. 2009, 100, 1663–1667. [Google Scholar] [CrossRef] [PubMed]
- Blanc-Durand, F.; Tang, R.; Pommier, M.; Nashvi, M.; Cotteret, S.; Genestie, C.; Le Formal, A.; Pautier, P.; Michels, J.; Kfoury, M.; et al. Clinical Relevance of BRCA1 Promoter Methylation Testing in Patients with Ovarian Cancer. Clin. Cancer Res. 2023, 29, 3124–3129. [Google Scholar] [CrossRef] [PubMed]
- Pradjatmo, H.; Dasuki, D.; Anwar, M.; Mubarika, S.; Harijadi, H. Methylation Status and Immunohistochemistry of BRCA1 in Epithelial Ovarian Cancer. Asian Pac. J. Cancer Prev. 2014, 15, 9479–9485. [Google Scholar] [CrossRef] [PubMed]
- Ruscito, I.; Dimitrova, D.; Vasconcelos, I.; Gellhaus, K.; Schwachula, T.; Bellati, F.; Zeillinger, R.; Benedetti-Panici, P.; Vergote, I.; Mahner, S.; et al. BRCA1 gene promoter methylation status in high-grade serous ovarian cancer patients – A study of the tumour Bank ovarian cancer (TOC) and ovarian cancer diagnosis consortium (OVCAD). Eur. J. Cancer 2014, 50, 2090–2098. [Google Scholar] [CrossRef] [PubMed]
- Bednarz, N.; Eltze, E.; Semjonow, A.; Rink, M.; Andreas, A.; Mulder, L.; Hannemann, J.; Fisch, M.; Pantel, K.; Weier, H.-U.G.; et al. BRCA1 Loss Preexisting in Small Subpopulations of Prostate Cancer Is Associated with Advanced Disease and Metastatic Spread to Lymph Nodes and Peripheral Blood. Clin. Cancer Res. 2010, 16, 3340–3348. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Porter, N.; Borges, M.; Gauthier, C.; Ferguson, L.; Huang, B.; Nanda, N.; He, J.; Laheru, D.; Hruban, R.H.; et al. Examination of ATM, BRCA1, and BRCA2 promoter methylation in patients with pancreatic cancer. Pancreatology 2021, 21, 938–941. [Google Scholar] [CrossRef]
- Peng, D.-F.; Kanai, Y.; Sawada, M.; Ushijima, S.; Hiraoka, N.; Kitazawa, S.; Hirohashi, S. DNA methylation of multiple tumor-related genes in association with overexpression of DNA methyltransferase 1 (DNMT1) during multistage carcinogenesis of the pancreas. Carcinog. 2006, 27, 1160–1168. [Google Scholar] [CrossRef]
- Abdallah, R.; Zhao, S.; Garinet, S.; Hormigos, K.; Le Corre, D.; Cros, J.; Toralla, K.P.; Bats, A.S.; Augustin, J.; Bachet, J.-B.; et al. BRCA1 and RAD51C promotor methylation in human resectable pancreatic adenocarcinoma. Clin. Res. Hepatol. Gastroenterol. 2022, 46, 101880. [Google Scholar] [CrossRef] [PubMed]
- Zheng-Lin, B.; Rainone, M.; Varghese, A.M.; Yu, K.H.; Park, W.; Berger, M.; Mehine, M.; Chou, J.; Capanu, M.; Mandelker, D.; et al. Methylation Analyses Reveal Promoter Hypermethylation as a Rare Cause of “Second Hit” in Germline BRCA1-Associated Pancreatic Ductal Adenocarcinoma. Mol. Diagn. Ther. 2022, 26, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, F.; Xu, R.; Zhang, S.; Peng, X.; Feng, Y.; Wang, J.; Lu, C. Promoter methylation of BRCA1 in the prognosis of breast cancer: a meta-analysis. Breast Cancer Res. Treat. 2013, 142, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Azzollini, J.; Pesenti, C.; Pizzamiglio, S.; Fontana, L.; Guarino, C.; Peissel, B.; Plebani, M.; Tabano, S.; Sirchia, S.M.; Colapietro, P.; et al. Constitutive BRCA1 Promoter Hypermethylation Can Be a Predisposing Event in Isolated Early-Onset Breast Cancer. Cancers 2019, 11, 58. [Google Scholar] [CrossRef] [PubMed]
- de Ruijter, T.C.; Veeck, J.; de Hoon, J.P.J.; van Engeland, M.; Tjan-Heijnen, V.C. Characteristics of triple-negative breast cancer. J. Cancer Res. Clin. Oncol. 2010, 137, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Almansour, N.M. Triple-Negative Breast Cancer: A Brief Review About Epidemiology, Risk Factors, Signaling Pathways, Treatment and Role of Artificial Intelligence. Front. Mol. Biosci. 2022, 9, 836417. [Google Scholar] [CrossRef]
- Bednarz-Knoll, N.; Eltze, E.; Semjonow, A.; Brandt, B. BRCAness in prostate cancer. Oncotarget 2019, 10, 2421–2422. [Google Scholar] [CrossRef] [PubMed]
- Swisher, E.M.; Lin, K.K.; Oza, A.M.; Scott, C.L.; Giordano, H.; Sun, J.; Konecny, G.E.; Coleman, R.L.; Tinker, A.V.; O’Malley, D.M.; et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017, 18, 75–87. [Google Scholar] [CrossRef]
- Elazezy, M.; Prieske, K.; Kluwe, L.; Oliveira-Ferrer, L.; Peine, S.; Müller, V.; Woelber, L.; Schmalfeldt, B.; Pantel, K.; Joosse, S.A. BRCA1 promoter hypermethylation on circulating tumor DNA correlates with improved survival of patients with ovarian cancer. Mol. Oncol. 2021, 15, 3615–3625. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Huang, Y.; Chen, C.; Zhang, X.; Xing, Y.; Gu, Y.; Zhang, M.; Cai, L.; Xu, S.; et al. Intratumor heterogeneity of breast cancer detected by epialleles shows association with hypoxic microenvironment. Theranostics 2021, 11, 4403–4420. [Google Scholar] [CrossRef]
- Ashour, M.; Shafik, H.E. Frequency of germline mutations in BRCA1 and BRCA2 in ovarian cancer patients and their effect on treatment outcome. Cancer Manag. Res. 2019, ume 11, 6275–6284. [Google Scholar] [CrossRef]
- Bowtell, D.D.L. The genesis and evolution of high-grade serous ovarian cancer. Nat. Rev. Cancer 2010, 10, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Nesic, K.; et al. BRCA1 secondary splice-site mutations drive exon-skipping and PARP inhibitor resistance. medRxiv : the preprint server for health sciences. [CrossRef]
- Litwin, M.S.; Tan, H.-J. The Diagnosis and Treatment of Prostate Cancer. JAMA 2017, 317, 2532–2542. [Google Scholar] [CrossRef] [PubMed]
- Rajwa, P.; Quhal, F.; Pradere, B.; Gandaglia, G.; Ploussard, G.; Leapman, M.S.; Gore, J.L.; Paradysz, A.; Tilki, D.; Merseburger, A.S.; et al. Prostate cancer risk, screening and management in patients with germline BRCA1/2 mutations. Nat. Rev. Urol. 2023, 20, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Omari, A.; Nastały, P.; Stoupiec, S.; Bałabas, A.; Dąbrowska, M.; Bielińska, B.; Huss, S.; Pantel, K.; Semjonow, A.; Eltze, E.; et al. Somatic aberrations of BRCA1 gene are associated with ALDH1, EGFR, and tumor progression in prostate cancer. Int. J. Cancer 2018, 144, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Bilici, A. Prognostic factors related with survival in patients with pancreatic adenocarcinoma. World J. Gastroenterol. 2014, 20, 10802–12. [Google Scholar] [CrossRef] [PubMed]
- Lal, G.; Liu, G.; Schmocker, B.; Kaurah, P.; Ozcelik, H.; A Narod, S.; Redston, M.; Gallinger, S. Inherited predisposition to pancreatic adenocarcinoma: role of family history and germ-line p16, BRCA1, and BRCA2 mutations. . 2000, 60, 409–16. [Google Scholar] [PubMed]
- Iqbal, J.; Ragone, A.; Lubinski, J.; Lynch, H.T.; Moller, P.; Ghadirian, P.; Foulkes, W.D.; Armel, S.; Eisen, A.; Neuhausen, S.L.; et al. The incidence of pancreatic cancer in BRCA1 and BRCA2 mutation carriers. Br. J. Cancer 2012, 107, 2005–2009. [Google Scholar] [CrossRef] [PubMed]
- Golan, T.; Sella, T.; O'Reilly, E.M.; Katz, M.H.G.; Epelbaum, R.; Kelsen, D.P.; Borgida, A.; Maynard, H.; Kindler, H.; Friedmen, E.; et al. Overall survival and clinical characteristics of BRCA mutation carriers with stage I/II pancreatic cancer. Br. J. Cancer 2017, 116, 697–702. [Google Scholar] [CrossRef]
- Tivey, A.; Church, M.; Rothwell, D.; Dive, C.; Cook, N. Circulating tumour DNA — looking beyond the blood. Nat. Rev. Clin. Oncol. 2022, 19, 600–612. [Google Scholar] [CrossRef]
- Panagopoulou, M.; Karaglani, M.; Balgkouranidou, I.; Pantazi, C.; Kolios, G.; Kakolyris, S.; Chatzaki, E. Circulating cell-free DNA release in vitro: kinetics, size profiling, and cancer-related gene methylation. J. Cell. Physiol. 2019, 234, 14079–14089. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulou, M.; Drosouni, A.; Fanidis, D.; Karaglani, M.; Balgkouranidou, I.; Xenidis, N.; Aidinis, V.; Chatzaki, E. ENPP2 Promoter Methylation Correlates with Decreased Gene Expression in Breast Cancer: Implementation as a Liquid Biopsy Biomarker. Int. J. Mol. Sci. 2022, 23, 3717. [Google Scholar] [CrossRef] [PubMed]
- Papadakis, V.M.; Cheimonidi, C.; Panagopoulou, M.; Karaglani, M.; Apalaki, P.; Katsara, K.; Kenanakis, G.; Theodosiou, T.; Constantinidis, T.C.; Stratigi, K.; et al. Label-Free Human Disease Characterization through Circulating Cell-Free DNA Analysis Using Raman Spectroscopy. Int. J. Mol. Sci. 2023, 24, 12384. [Google Scholar] [CrossRef] [PubMed]
- Nikanjam, M.; Kato, S.; Kurzrock, R. Liquid biopsy: current technology and clinical applications. J. Hematol. Oncol. 2022, 15, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Heidrich, I.; Ačkar, L.; Mossahebi Mohammadi, P.; Pantel, K. Liquid biopsies: Potential and challenges. Int. J. Cancer 2021, 148, 528–545. [Google Scholar] [CrossRef] [PubMed]
- S, S.K.; Swamy, S.N.; Premalatha, C.S.; Pallavi, V.R.; Gawari, R. Aberrant Promoter Hypermethylation of RASSF1a and BRCA1 in Circulating Cell-Free Tumor DNA Serves as a Biomarker of Ovarian Carcinoma. Asian Pac. J. Cancer Prev. 2019, 20, 3001–3005. [Google Scholar] [CrossRef] [PubMed]
- de Caceres, I.I.; Battagli, C.; Esteller, M.; Herman, J.G.; Dulaimi, E.; Edelson, M.I.; Bergman, C.; Ehya, H.; Eisenberg, B.L.; Cairns, P. Tumor Cell-Specific BRCA1 and RASSF1A Hypermethylation in Serum, Plasma, and Peritoneal Fluid from Ovarian Cancer Patients. Cancer Res. 2004, 64, 6476–6481. [Google Scholar] [CrossRef] [PubMed]
- Melnikov, A.; Scholtens, D.; Godwin, A.; Levenson, V. Differential Methylation Profile of Ovarian Cancer in Tissues and Plasma. J. Mol. Diagn. 2009, 11, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Cristall, K.; Bidard, F.-C.; Pierga, J.-Y.; Rauh, M.J.; Popova, T.; Sebbag, C.; Lantz, O.; Stern, M.-H.; Mueller, C.R. A DNA methylation-based liquid biopsy for triple-negative breast cancer. npj Precis. Oncol. 2021, 5, 1–13. [Google Scholar] [CrossRef]
- Liu, L.; Sun, L.; Li, C.; Li, X.; Zhang, Y.; Yu, Y.; Xia, W. Quantitative detection of methylation of FHIT and BRCA1 promoters in the serum of ductal breast cancer patients. Bio-Medical Mater. Eng. 2015, 26, S2217–S2222. [Google Scholar] [CrossRef]
- Sturgeon, S.R.; Balasubramanian, R.; Schairer, C.; Muss, H.B.; Ziegler, R.G.; Arcaro, K.F. Detection of promoter methylation of tumor suppressor genes in serum DNA of breast cancer cases and benign breast disease controls. Epigenetics 2012, 7, 1258–1267. [Google Scholar] [CrossRef] [PubMed]
- de Ruijter, T.C.; van der Heide, F.; Smits, K.M.; Aarts, M.J.; van Engeland, M.; Heijnen, V.C.G. Prognostic DNA methylation markers for hormone receptor breast cancer: a systematic review. Breast Cancer Res. 2020, 22, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yen, J.; Chen, S.; Jenkins, C.; Overstreet, B.; Fu, Y.; Zhao, J.; Jiang, T.; Drusbosky, L.; Pettitt, S.; Dorschner, M.; et al. Abstract 6603: BRCA1 promoter methylation in sporadic breast cancer patients detected by liquid biopsy. Cancer Res. 2023, 83, 6603–6603. [Google Scholar] [CrossRef]
- Koukaki, T.; Balgkouranidou, I.; Biziota, E.; Karayiannakis, A.; Bolanaki, H.; Karamitrousis, E.; Zarogoulidis, P.; Deftereos, S.; Charalampidis, C.; Ioannidis, A.; et al. Prognostic significance of BRCA1 and BRCA2 methylation status in circulating cell-free DNA of Pancreatic Cancer patients. J. Cancer 2024, 15, 2573–2579. [Google Scholar] [CrossRef] [PubMed]
- Vasseur, A.; Kiavue, N.; Bidard, F.-C.; Pierga, J.-Y.; Cabel, L. Clinical utility of circulating tumor cells: an update. Mol. Oncol. 2021, 15, 1647–1666. [Google Scholar] [CrossRef] [PubMed]
- Stordal, B.; Timms, K.; Farrelly, A.; Gallagher, D.; Busschots, S.; Renaud, M.; Thery, J.; Williams, D.; Potter, J.; Tran, T.; et al. BRCA1/2 mutation analysis in 41 ovarian cell lines reveals only one functionally deleterious BRCA1 mutation. Mol. Oncol. 2013, 7, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Cortesi, L.; Rugo, H.S.; Jackisch, C. An Overview of PARP Inhibitors for the Treatment of Breast Cancer. Target. Oncol. 2021, 16, 255–282. [Google Scholar] [CrossRef]
- Moschetta, M.; George, A.; Kaye, S.B.; Banerjee, S. BRCA somatic mutations and epigenetic BRCA modifications in serous ovarian cancer. Ann. Oncol. 2016, 27, 1449–1455. [Google Scholar] [CrossRef] [PubMed]
- Jacot, W.; Thezenas, S.; Senal, R.; Viglianti, C.; Laberenne, A.-C.; Lopez-Crapez, E.; Bibeau, F.; Bleuse, J.-P.; Romieu, G.; Lamy, P.-J. BRCA1 promoter hypermethylation, 53BP1 protein expression and PARP-1 activity as biomarkers of DNA repair deficit in breast cancer. BMC Cancer 2013, 13, 523–523. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, L.; Huang, B.; Li, X.; Yang, L.; Hu, X.; Jiang, Y.; Shao, Z.; Wang, Z. Efficacy and mechanism of the combination of PARP and CDK4/6 inhibitors in the treatment of triple-negative breast cancer. J. Exp. Clin. Cancer Res. 2021, 40, 1–18. [Google Scholar] [CrossRef]
- Gelmon, K.A.; Tischkowitz, M.; Mackay, H.; Swenerton, K.; Robidoux, A.; Tonkin, K.; Hirte, H.; Huntsman, D.; Clemons, M.; Gilks, B.; et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011, 12, 852–861. [Google Scholar] [CrossRef] [PubMed]
- Veeck, J.; Ropero, S.; Setien, F.; Gonzalez-Suarez, E.; Osorio, A.; Benitez, J.; Herman, J.G.; Esteller, M. BRCA1 CpG Island Hypermethylation Predicts Sensitivity to Poly(Adenosine Diphosphate)- Ribose Polymerase Inhibitors. J. Clin. Oncol. 2010, 28, e563–e564. [Google Scholar] [CrossRef] [PubMed]
- Kondrashova, O.; Topp, M.; Nesic, K.; Lieschke, E.; Ho, G.Y.; Harrell, M.I.; Zapparoli, G.V.; Hadley, A.; Holian, R.; Boehm, E.; et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Dimitrova, D.; Ruscito, I.; Olek, S.; Richter, R.; Hellwag, A.; Türbachova, I.; Woopen, H.; Baron, U.; Braicu, E.I.; Sehouli, J. Germline mutations of BRCA1 gene exon 11 are not associated with platinum response neither with survival advantage in patients with primary ovarian cancer: understanding the clinical importance of one of the biggest human exons. A study of the Tumor Bank Ovarian Cancer (TOC) Consortium. Tumor Biol. 2016, 37, 12329–12337. [Google Scholar] [CrossRef]
- Drost, R.; Dhillon, K.K.; Van Der Gulden, H.; Van Der Heijden, I.; Brandsma, I.; Cruz, C.; Chondronasiou, D.; Castroviejo-Bermejo, M.; Boon, U.; Schut, E.; et al. BRCA1185delAG tumors may acquire therapy resistance through expression of RING-less BRCAJ. Clin. Investig. 2016, 126, 2903–2918. [Google Scholar] [CrossRef] [PubMed]
- Jacot, W.; Lopez-Crapez, E.; Mollevi, C.; Boissière-Michot, F.; Simony-Lafontaine, J.; Ho-Pun-Cheung, A.; Chartron, E.; Theillet, C.; Lemoine, A.; Saffroy, R.; et al. BRCA1 Promoter Hypermethylation is Associated with Good Prognosis and Chemosensitivity in Triple-Negative Breast Cancer. Cancers 2020, 12, 828. [Google Scholar] [CrossRef] [PubMed]
- Buisson, R.; Dion-Côté, A.-M.; Coulombe, Y.; Launay, H.; Cai, H.; Stasiak, A.Z.; Stasiak, A.; Xia, B.; Masson, J.-Y. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 2010, 17, 1247–1254. [Google Scholar] [CrossRef] [PubMed]
- Annunziata, C.M.; O'Shaughnessy, J. Poly (ADP-Ribose) Polymerase as a Novel Therapeutic Target in Cancer. Clin. Cancer Res. 2010, 16, 4517–4526. [Google Scholar] [CrossRef] [PubMed]
- Drew, Y.; Mulligan, E.A.; Vong, W.-T.; Thomas, H.D.; Kahn, S.; Kyle, S.; Mukhopadhyay, A.; Los, G.; Hostomsky, Z.; Plummer, E.R.; et al. Therapeutic Potential of Poly(ADP-ribose) Polymerase Inhibitor AG014699 in Human Cancers With Mutated or Methylated BRCA1 or BRCA2. JNCI J. Natl. Cancer Inst. 2010, 103, 334–346. [Google Scholar] [CrossRef]
- Vos, S.; Moelans, C.B.; van Diest, P.J. BRCA promoter methylation in sporadic versus BRCA germline mutation-related breast cancers. Breast Cancer Res. 2017, 19, 64. [Google Scholar] [CrossRef]
- Guo, M.; Peng, Y.; Gao, A.; Du, C.; Herman, J.G. Epigenetic heterogeneity in cancer. Biomark. Res. 2019, 7, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Prieske, K.; Prieske, S.; Joosse, S.A.; Trillsch, F.; Grimm, D.; Burandt, E.; Mahner, S.; Schmalfeldt, B.; Milde-Langosch, K.; Oliveira-Ferrer, L.; et al. Loss of BRCA1 promotor hypermethylation in recurrent high-grade ovarian cancer. Oncotarget 2017, 8, 83063–83074. [Google Scholar] [CrossRef] [PubMed]
- Harvey-Jones, E.; Raghunandan, M.; Robbez-Masson, L.; Thanussuyah, A.; Liccardo, R.; Yablonovitch, A.; Cai, M.; Drusbosky, L.; Dorschner, M.; Pardo, L.M.; et al. Abstract 6094: Longitudinal analysis of PARP inhibitor and platinum resistance in BRCA1/2m breast cancer using liquid biopsy. Cancer Res. 2023, 83, 6094–6094. [Google Scholar] [CrossRef]
- Jayaram, A.; Wingate, A.; Wetterskog, D.; Wheeler, G.; Sternberg, C.; Jones, R.; Berruti, A.; Lefresne, F.; Lahaye, M.; Thomas, S.; et al. Plasma tumor gene conversions after one cycle abiraterone acetate for metastatic castration-resistant prostate cancer: a biomarker analysis of a multicenter international trial. Ann. Oncol. 2021, 32, 726–735. [Google Scholar] [CrossRef]
| Cancer type | Biomaterial | BRCA1 methylation (%) | Correlation | Reference |
|---|---|---|---|---|
| BrCa | Tissue | 9.1 | Diagnosis at a young age | Birgisdottir et al [65] |
| 3.0 | Improved Survival after chemotherapy | Stefansson et al [68] | ||
| 12.4 | Incidence of TNBC | Lonning et al [77] | ||
| 26.0 | Worse Survival | Chen et al [78] | ||
| 24.1 | Improved Survival after chemotherapy | Glodzik et al [66] | ||
| TNBC | Tissue | 20.6 | Improved Survival | Brianese et al [67] |
| OvCa | Tissue | 16.3 | Young age, Advanced stage, Improved Survival | Kalachand et al [43] |
| 19 (high methylation) 14 (low methylation) |
High methylation with GIS and PARPi treatment option | Durand et al [79] | ||
| 89.9 | None | Pradjatmo et al [80] | ||
| 5.2 | Partially BRCAness prediction | Aref-Eshghi et al [49] | ||
| 19.3 | None | Sahnane et al | ||
| HGSOC | Tissue | 14.8 | Young age | Ruscito et al [81] |
| PrCa | Tissue | 100.0 | None | Rabiau et al [17] |
| 0.0 | None | Bednarz et al [82] | ||
| PaCa | blood lymphocytes | 0.3 | None | Zhou et al [83] |
| Tissue | 60.3 | Poorer tumor differentiation, protein expression levels |
Peng et al. [84] | |
| PaCa | Tissue | 0.0 | None | Abdalah et al. [85] |
| PaCa | Tissue & Blood lymphocytes | 3.6 | None | Zhen-Lin et al. [86] |
140. Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





