Submitted:
21 July 2024
Posted:
23 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Cell Culturing of Primary Colonocytes
2.2. CellTiter-Glo® Luminescent Cell Viability Assay
2.3. Mitochondrial Membrane Potential
2.4. Mitochondrial Bioenergetics
2.5. Lipidomics and Cell Exposure
2.6. Lipids Analysis in Colonocytes
2.7. Gene Expression Analysis
2.8. Statistical Analysis
3. Results
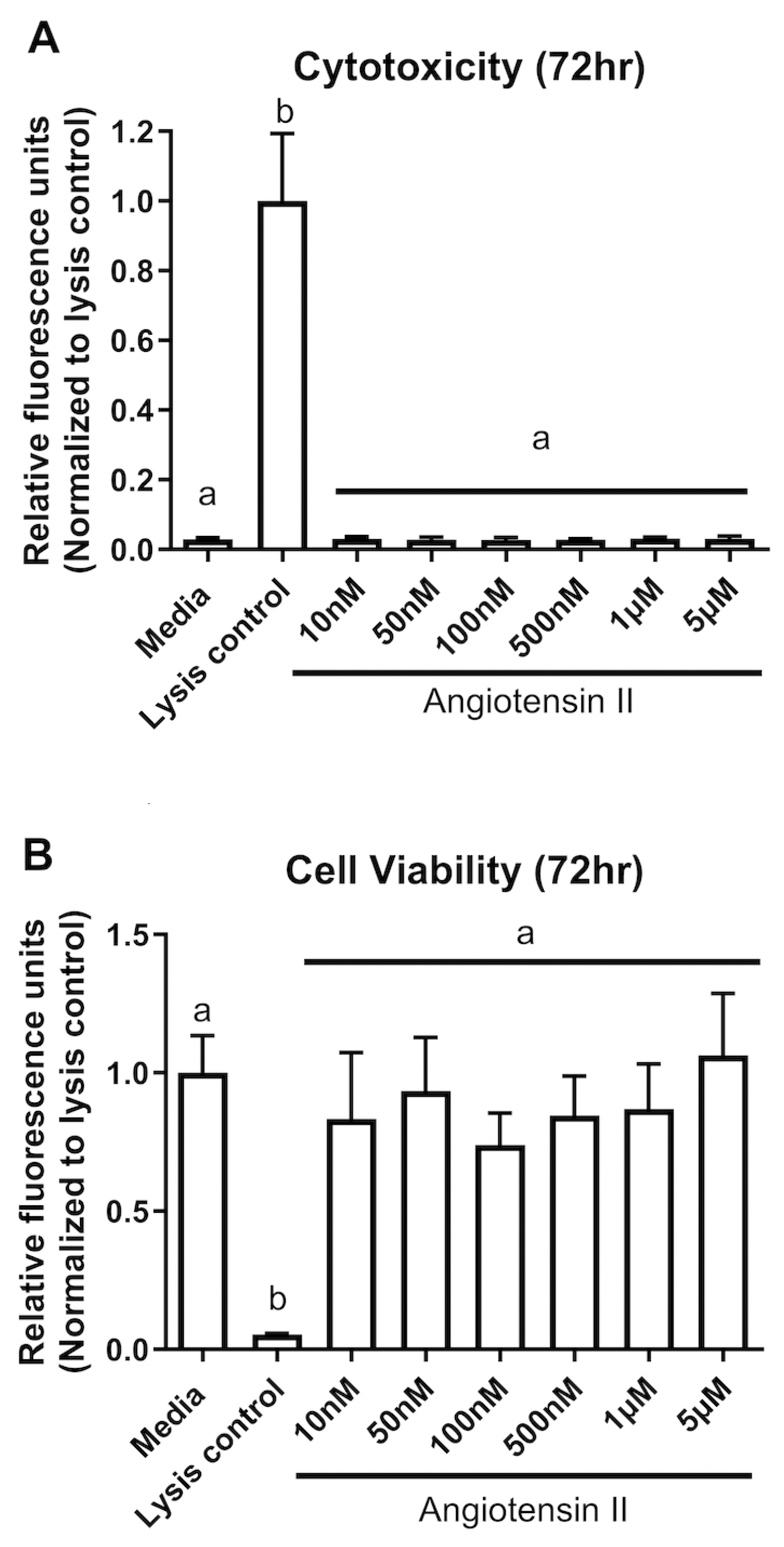
3.1. Cytotoxicity and cell viability
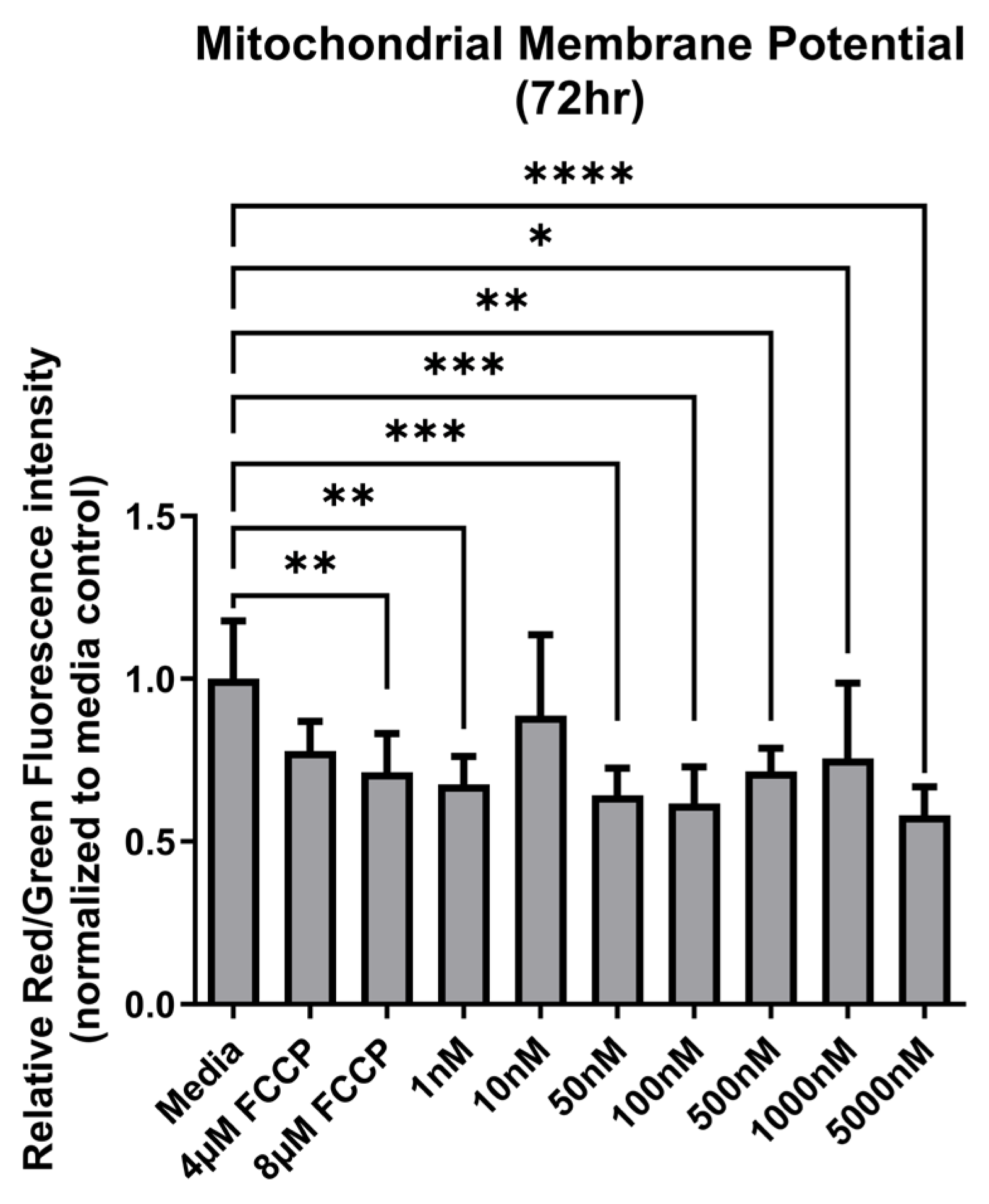
3.2. Mitochondrial Membrane Potential
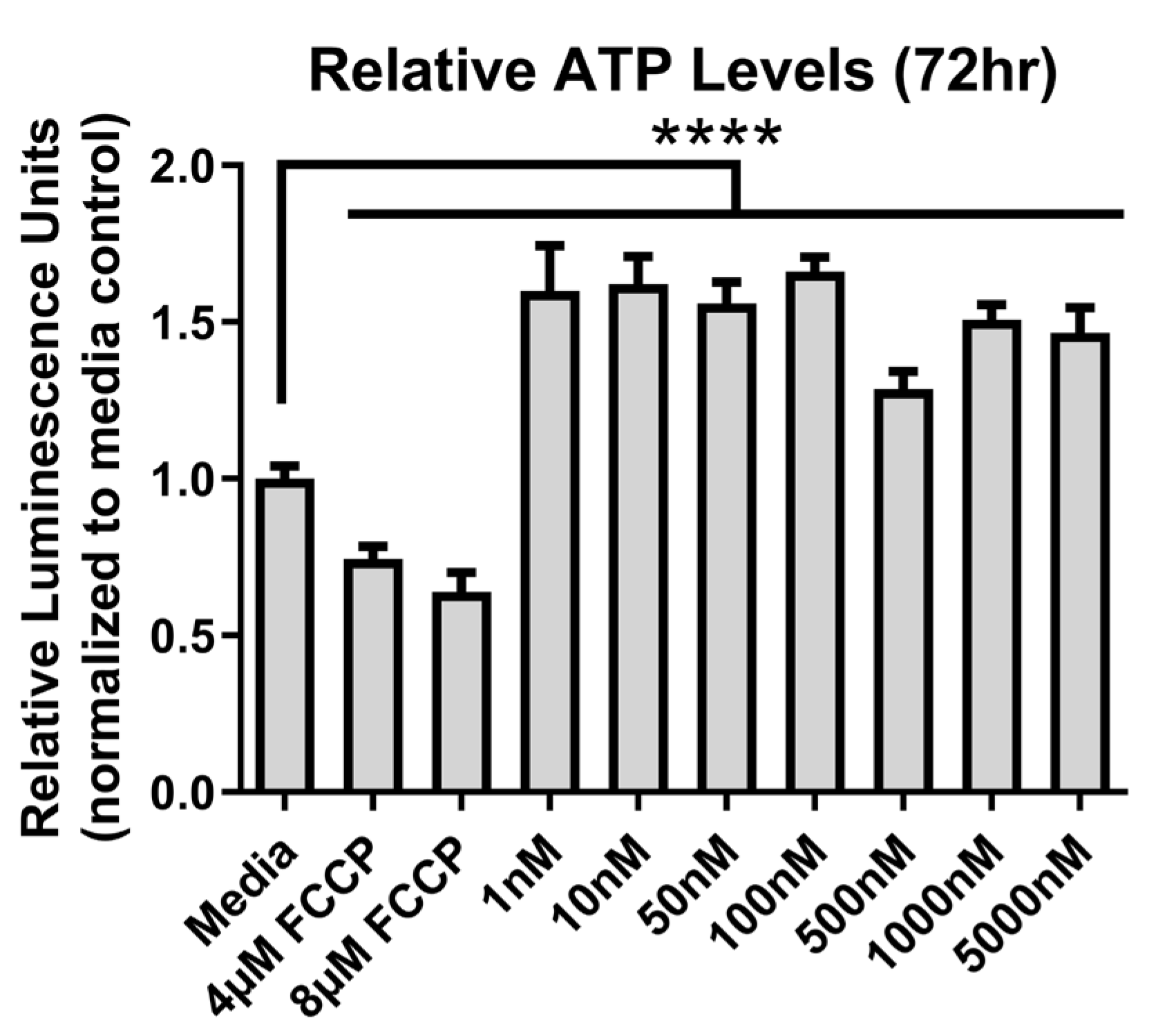
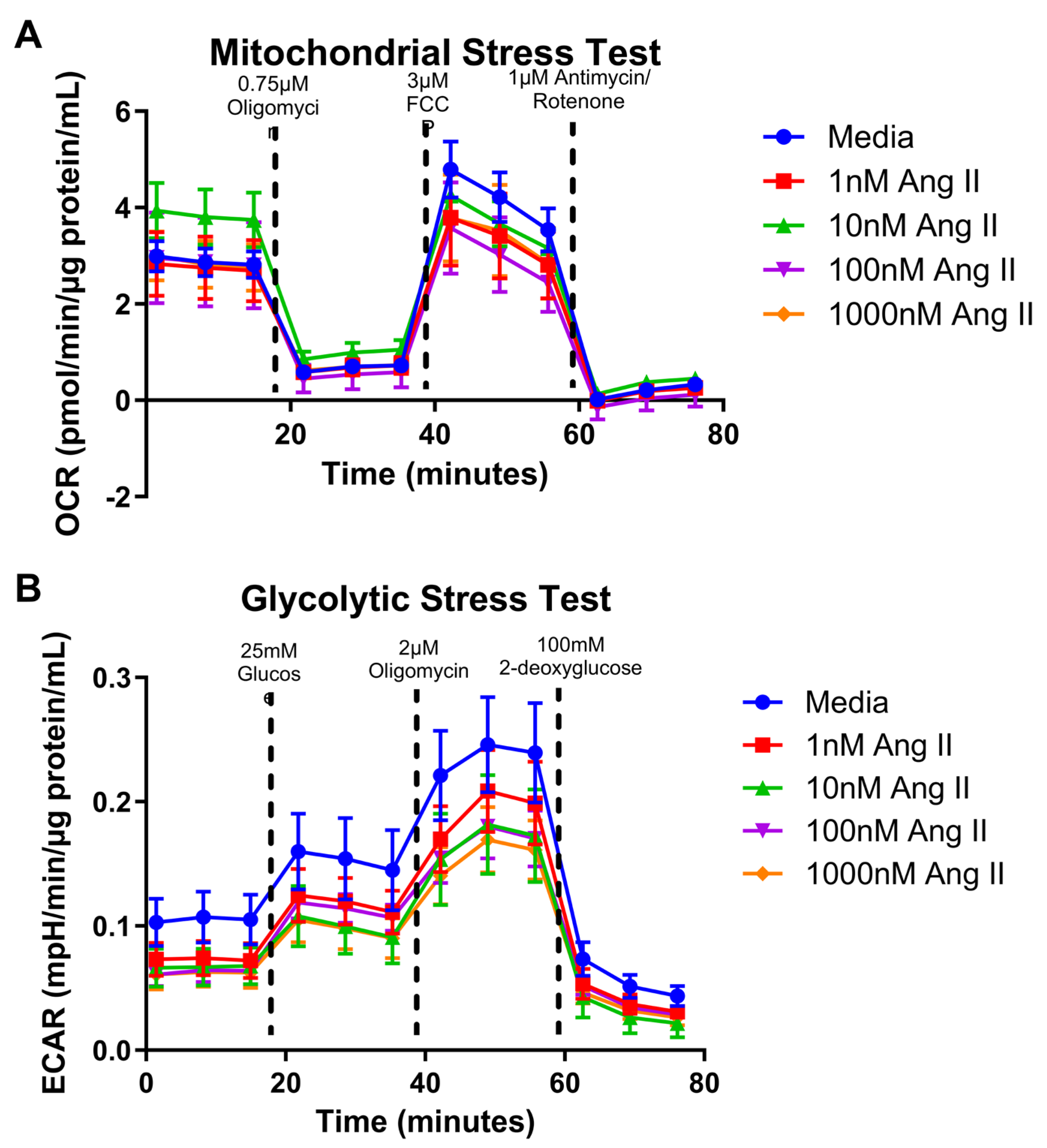
3.3. Mitochondrial Bioenergetics
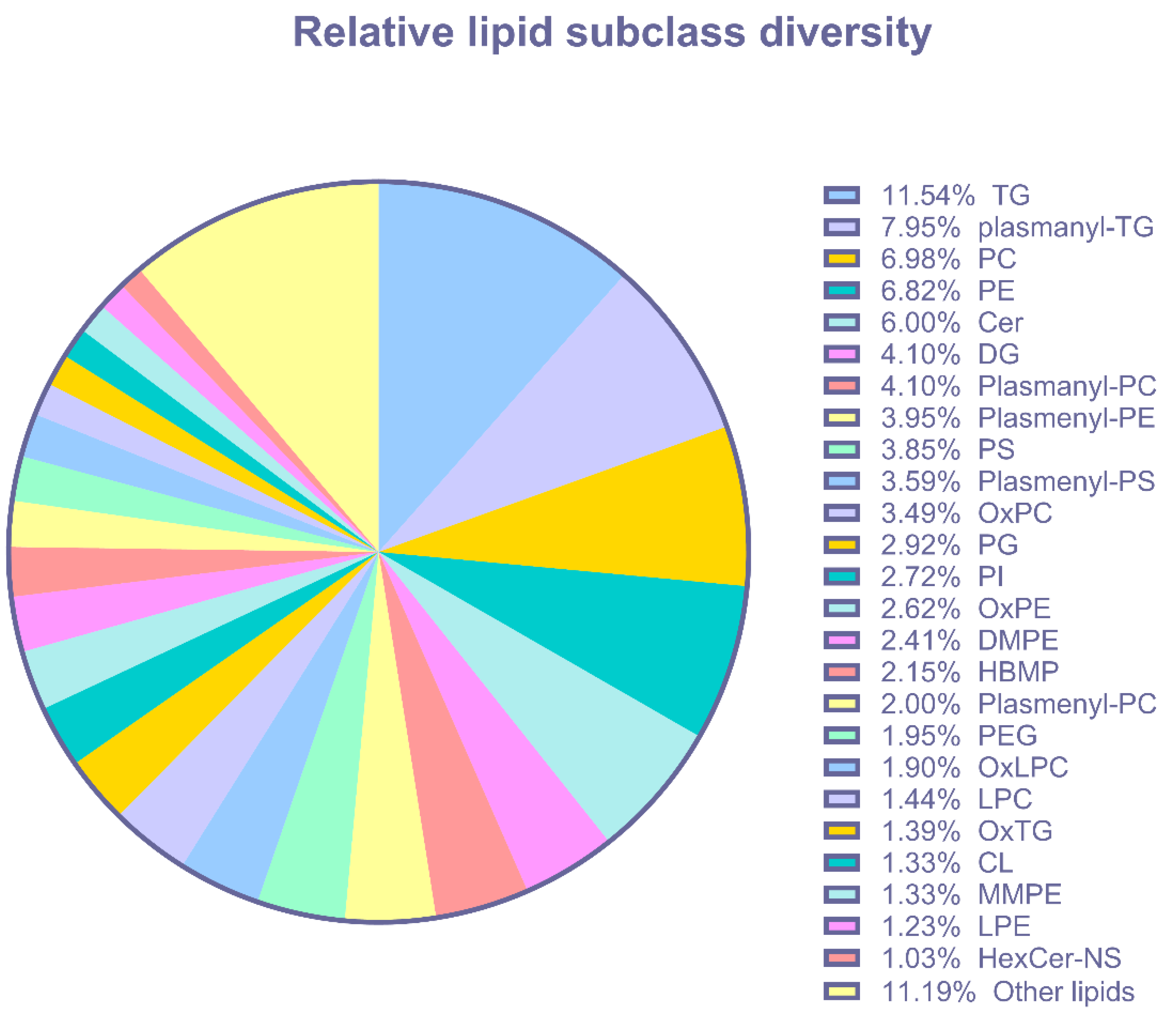
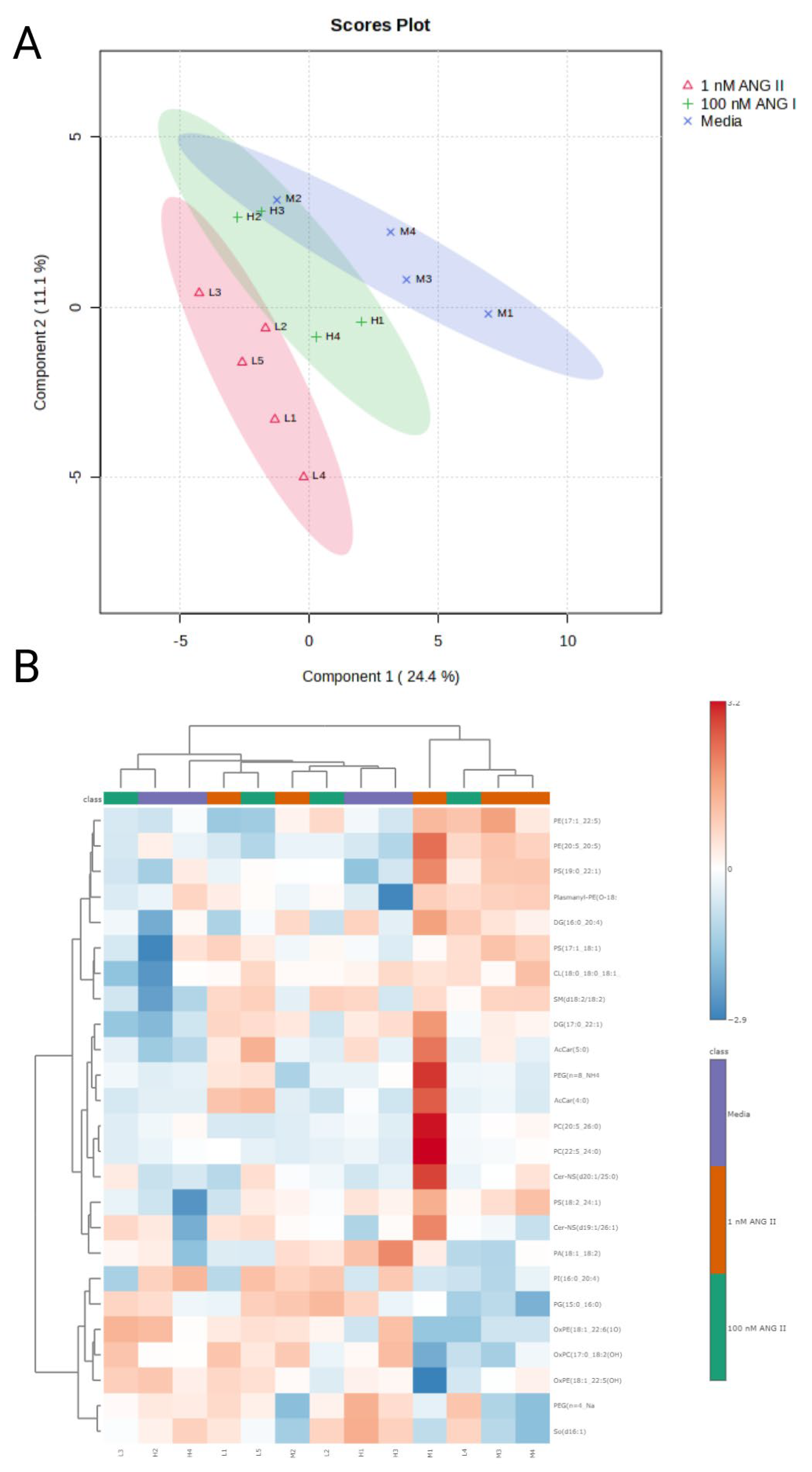
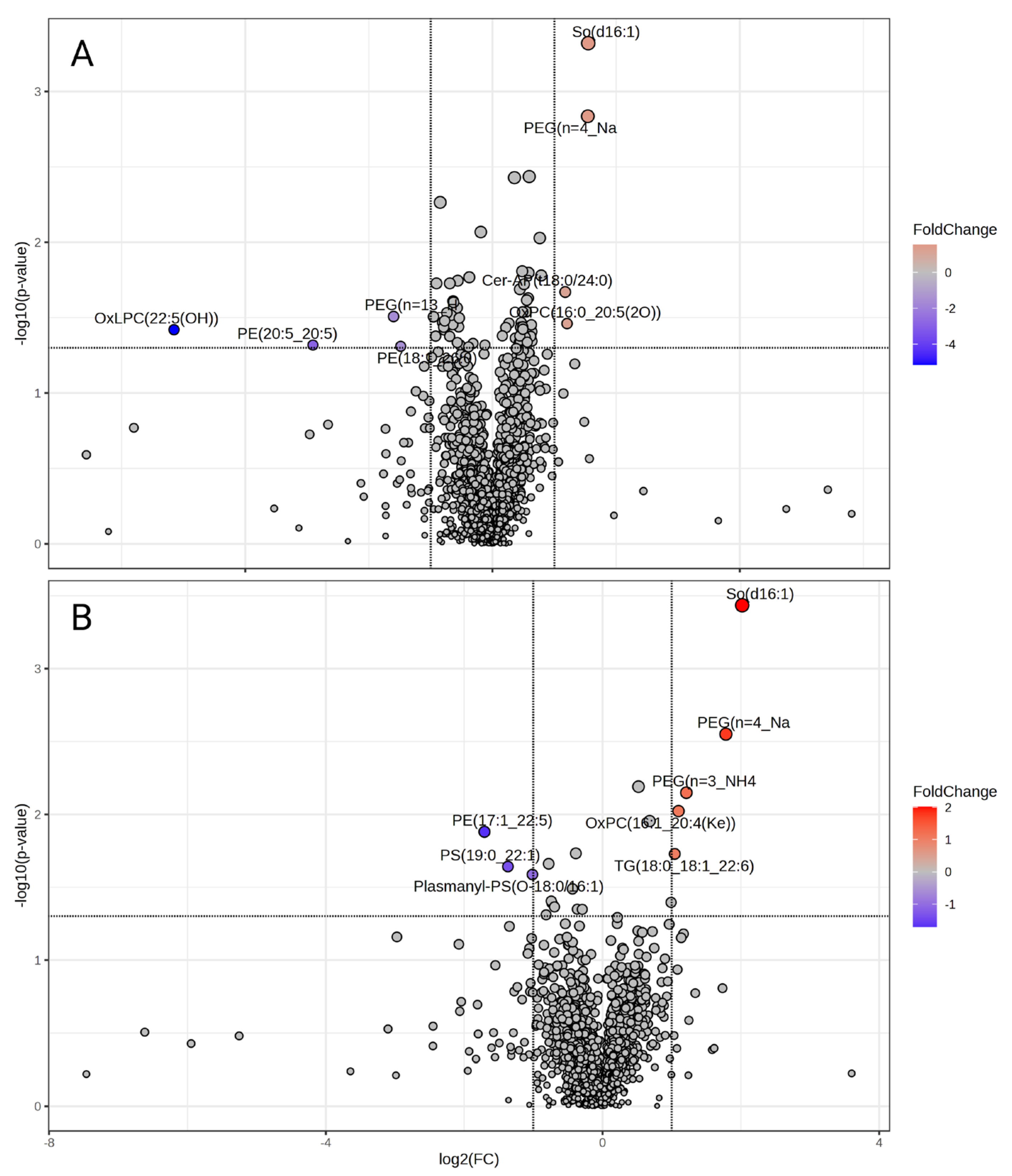
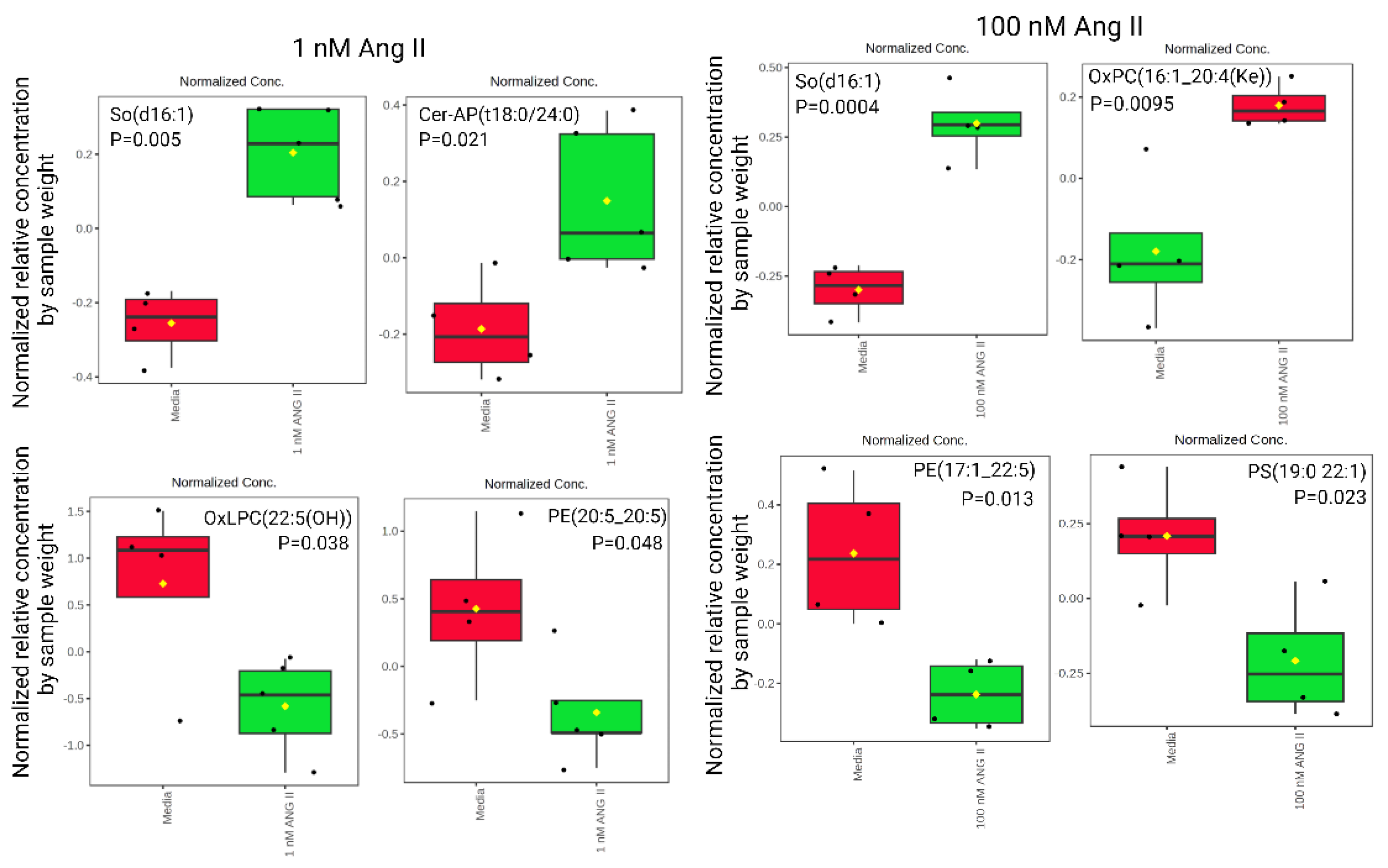
3.4. Lipid Abundance in Ang II Cultured Colonocytes
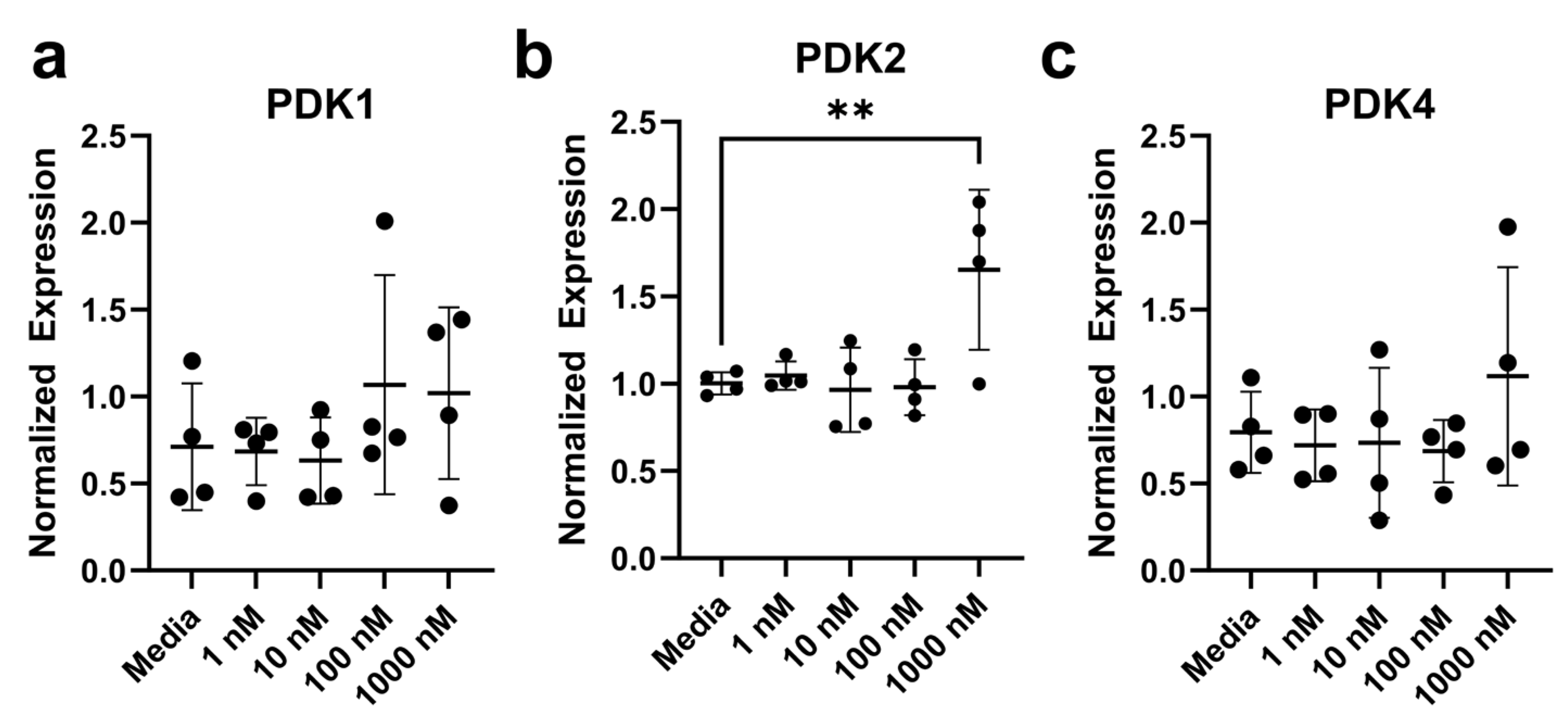
3.5. Expression Levels of Lipid and Metabolic-Related Genes in Colonocytes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Putnam, K.; et al. The renin-angiotensin system: A target of and contributor to dyslipidemias, altered glucose homeostasis, and hypertension of the metabolic syndrome. American Journal of Physiology-Heart and Circulatory Physiology, 2012, 302, H1219–H1230. [Google Scholar] [CrossRef] [PubMed]
- Villapol, S.; et al. The renin angiotensin system as a therapeutic target in traumatic brain injury. Neurotherapeutics, 2023, 20, 1565–1591. [Google Scholar] [CrossRef] [PubMed]
- Escobales; Nuñez, R.E.; Javadov, S. Mitochondrial angiotensin receptors and cardioprotective pathways. American Journal of Physiology-Heart and Circulatory Physiology 2019, 316, H1426–H1438. [Google Scholar] [CrossRef]
- Ait-Aissa, K.; et al. Protective role of short chain fatty acids against Angiotensin II-induced mitochondrial dysfunction in brain endothelial cells: Potential role of Heme Oxygenase 2. Physiology, 2023, 38, 5735216. [Google Scholar] [CrossRef]
- Kontaridis, M.I.; Chennappan, S. Mitochondria and the future of RASopathies: The emergence of bioenergetics. The Journal of Clinical Investigation 2022. [Google Scholar] [CrossRef]
- Kimura, S.; et al. Mitochondria-derived reactive oxygen species and vascular MAP kinases: Comparison of angiotensin II and diazoxide. Hypertension, 2005, 45, 438–444. [Google Scholar] [CrossRef]
- Larkin, J.E.; et al. Cardiac transcriptional response to acute and chronic angiotensin II treatments. Physiological genomics, 2004, 18, 152–166. [Google Scholar] [CrossRef] [PubMed]
- Steckelings, U.M.; et al. The angiotensin AT2 receptor: From a binding site to a novel therapeutic target. Pharmacological Reviews, 2022, 74, 1051–1135. [Google Scholar] [CrossRef]
- Abadir, P.M.; et al. Identification and characterization of a functional mitochondrial angiotensin system. Proceedings of the National Academy of Sciences, 2011, 108, 14849–14854. [Google Scholar] [CrossRef]
- Kawai, T.; et al. AT1 receptor signaling pathways in the cardiovascular system. Pharmacological research, 2017, 125, 4–13. [Google Scholar] [CrossRef]
- Valenzuela, R.; et al. Mitochondrial angiotensin receptors in dopaminergic neurons. Role in cell protection and aging-related vulnerability to neurodegeneration. Cell death & disease, 2016, 7, e2427–e2427. [Google Scholar]
- Goossens, G.; et al. Angiotensin II: A hormone that affects lipid metabolism in adipose tissue. International journal of obesity, 2007, 31, 382–384. [Google Scholar] [CrossRef] [PubMed]
- Fändriks, L. The angiotensin II type 2 receptor and the gastrointestinal tract. Journal of the renin-angiotensin-aldosterone system, 2010, 11, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; et al. Angiotensin II affects inflammation mechanisms via AMPK-related signalling pathways in HL-1 atrial myocytes. Scientific reports, 2017, 7, 10328. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; et al. Beneficial effects of astragaloside IV against angiotensin II-induced mitochondrial dysfunction in rat vascular smooth muscle cells. International journal of molecular medicine, 2015, 36, 1223–1232. [Google Scholar] [CrossRef] [PubMed]
- de Cavanagh, E.M.; et al. Angiotensin II, mitochondria, cytoskeletal, and extracellular matrix connections: An integrating viewpoint. American Journal of Physiology-Heart and Circulatory Physiology, 2009, 296, H550–H558. [Google Scholar] [CrossRef] [PubMed]
- Masi; Uliana, M.; Virdis, A. Angiotensin II and vascular damage in hypertension: Role of oxidative stress and sympathetic activation. Vascular pharmacology 2019, 115, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Dikalov, S.I.; Nazarewicz, R.R. Angiotensin II-induced production of mitochondrial reactive oxygen species: Potential mechanisms and relevance for cardiovascular disease. Antioxidants & redox signaling 2013, 19, 1085–1094. [Google Scholar]
- Johansson, B.; et al. Angiotensin II type 2 receptor-mediated duodenal mucosal alkaline secretion in the rat. American Journal of Physiology-Gastrointestinal and Liver Physiology, 2001, 280, G1254–G1260. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.P.; Debnam, E.S.; Leung, P.S. Involvement of an enterocyte renin–angiotensin system in the local control of SGLT1-dependent glucose uptake across the rat small intestinal brush border membrane. The Journal of physiology 2007, 584, 613–623. [Google Scholar] [CrossRef]
- Jin, X.-H.; et al. Regulation of jejunal sodium and water absorption by angiotensin subtype receptors. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology, 1998, 275, R515–R523. [Google Scholar] [CrossRef] [PubMed]
- Jose, P.A.; Raj, D. Gut microbiota in hypertension. Current opinion in nephrology and hypertension 2015, 24, 403. [Google Scholar] [CrossRef] [PubMed]
- Lam, Y.Y.; et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS ONE, 2012, 7, e34233. [Google Scholar] [CrossRef] [PubMed]
- Buttó, L.F.; Haller, D. Dysbiosis in intestinal inflammation: Cause or consequence. International Journal of Medical Microbiology 2016, 306, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Moschandrea, C.; et al. Mitochondrial dysfunction abrogates dietary lipid processing in enterocytes. Nature, 2024, 625, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Litvak; Byndloss, M.X.; Bäumler, A.J. Colonocyte metabolism shapes the gut microbiota. Science 2018, 362, eaat9076. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, C.L.; et al. Neurotoxicity assessment of triazole fungicides on mitochondrial oxidative respiration and lipids in differentiated human SH-SY5Y neuroblastoma cells. Neurotoxicology, 2020, 80, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.; et al. Neurotoxicity assessment of QoI strobilurin fungicides azoxystrobin and trifloxystrobin in human SH-SY5Y neuroblastoma cells: Insights from lipidomics and mitochondrial bioenergetics. Neurotoxicology, 2022, 91, 290–304. [Google Scholar] [CrossRef] [PubMed]
- Koelmel, J.P.; et al. Software tool for internal standard based normalization of lipids, and effect of data-processing strategies on resulting values. BMC bioinformatics, 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Souders, C.L.; et al. Interaction between Butyrate and Tumor Necrosis Factor α in Primary Rat Colonocytes. Biomolecules, 2023, 13, 258. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.; et al. A practical approach to RT-qPCR—Publishing data that conform to the MIQE guidelines. Methods, 2010, 50, S1–S5. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; et al. Biological effects of the benzotriazole ultraviolet stabilizers UV-234 and UV-320 in early-staged zebrafish (Danio rerio). Environmental pollution, 2019, 245, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Vajapey, R.; et al. The impact of age-related dysregulation of the angiotensin system on mitochondrial redox balance. Frontiers in physiology, 2014, 5, 439. [Google Scholar] [CrossRef] [PubMed]
- Friederich-Persson; Persson, P. Mitochondrial angiotensin II receptors regulate oxygen consumption in kidney mitochondria from healthy and type 1 diabetic rats. American Journal of Physiology-Renal Physiology, 2020, 318, F683–F688.
- Sechi, L.A.; et al. Characterization of angiotensin II receptor subtypes in rat heart. Circulation research, 1992, 71, 1482–1489. [Google Scholar] [CrossRef] [PubMed]
- Hallersund, P.; et al. Angiotensin II receptor expression and relation to Helicobacter pylori-infection in the stomach of the Mongolian gerbil. BMC gastroenterology, 2010, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, K.; et al. Localization of angiotensin II receptor and local renin-angiotensin system in human colonic mucosa. Gastroenterology, 2001, 5, A313–A314. [Google Scholar] [CrossRef]
- Casselbrant, A.; et al. Angiotensin II receptors are expressed and functional in human esophageal mucosa. American Journal of Physiology-Gastrointestinal and Liver Physiology, 2009, 297, G1019–G1027. [Google Scholar] [CrossRef]
- Hallersund, P.; et al. The expression of renin-angiotensin system components in the human gastric mucosa. Journal of the Renin-Angiotensin-Aldosterone System, 2011, 12, 54–64. [Google Scholar] [CrossRef]
- Spak, E.; et al. Changes in the mucosa of the Roux-limb after gastric bypass surgery. Histopathology, 2010, 57, 680–688. [Google Scholar] [CrossRef] [PubMed]
- Hagen, T.M.; et al. Mitochondrial decay in hepatocytes from old rats: Membrane potential declines, heterogeneity and oxidants increase. Proceedings of the National Academy of Sciences, 1997, 94, 3064–3069. [Google Scholar] [CrossRef] [PubMed]
- De Cavanagh, E.; et al. Angiotensin II blockade improves mitochondrial function in spontaneously hypertensive rats. Cell Mol Biol (Noisy-le-grand), 2005, 51, 573–578. [Google Scholar] [PubMed]
- Li, Y.; et al. Angiotensin II induces mitochondrial oxidative stress and mtDNA damage in osteoblasts by inhibiting SIRT1–FoxO3a–MnSOD pathway. Biochemical and biophysical research communications 2014, 455, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Ricci; Pastukh, V.; Schaffer, S.W. Involvement of the mitochondrial permeability transition pore in angiotensin II-mediated apoptosis. Experimental & Clinical Cardiology 2005, 10, 160. [Google Scholar]
- Go, Y.; et al. Inhibition of pyruvate dehydrogenase kinase 2 protects against hepatic steatosis through modulation of tricarboxylic acid cycle anaplerosis and ketogenesis. Diabetes, 2016, 65, 2876–2887. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; et al. Distinct structural mechanisms for inhibition of pyruvate dehydrogenase kinase isoforms by AZD7545, dichloroacetate, and radicicol. Structure, 2007, 15, 992–1004. [Google Scholar] [CrossRef] [PubMed]
- Woolbright, B.L.; Harris, R.A. PDK2: An Underappreciated Regulator of Liver Metabolism. Livers 2021, 1, 82–97. [Google Scholar] [CrossRef]
- Kang, H.-J.; et al. Pyruvate dehydrogenase kinase 1 and 2 deficiency reduces high-fat diet-induced hypertrophic obesity and inhibits the differentiation of preadipocytes into mature adipocytes. Experimental & Molecular Medicine, 2021, 53, 1390–1401. [Google Scholar]
- Sas, K.M.; et al. Renin-angiotensin system inhibition reverses the altered triacylglycerol metabolic network in diabetic kidney disease. Metabolomics, 2021, 17, 1–12. [Google Scholar] [CrossRef]
- Strazzullo, P. and F. Galletti, Impact of the renin-angiotensin system on lipid and carbohydrate metabolism. Current opinion in nephrology and hypertension, 2004, 13, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Seah, J.Y.H.; et al. Dietary fat and protein intake in relation to plasma sphingolipids as determined by a large-scale lipidomic analysis. Metabolites, 2021, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- Borodzicz-Jażdżyk, S.; et al. Sphingolipid metabolism and signaling in cardiovascular diseases. Frontiers in cardiovascular medicine, 2022, 9, 915961. [Google Scholar] [CrossRef] [PubMed]
- Cassilly, C.D.; Reynolds, T.B. PS, it’s complicated: The roles of phosphatidylserine and phosphatidylethanolamine in the pathogenesis of Candida albicans and other microbial pathogens. Journal of Fungi 2018, 4, 28. [Google Scholar] [CrossRef] [PubMed]
- Leventis, P.A.; Grinstein, S. The distribution and function of phosphatidylserine in cellular membranes. Annual review of biophysics 2010, 39, 407–427. [Google Scholar] [CrossRef] [PubMed]
- Vance, J.E.; Tasseva, G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids 2013, 1831, 543–554. [Google Scholar] [CrossRef]
- de Kloet, A.D.; Krause, E.G.; Woods, S.C. The renin angiotensin system and the metabolic syndrome. Physiology & behavior 2010, 100, 525–534. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




