1. Introduction
Antarctica, the southernmost landmass on Earth, covers an area of approximately 14 million km², making it the fifth largest continent in the world. It is subjected to extremely cold and dry conditions, with the lowest recorded temperature being -90°C. Approximately 98% of Antarctica is covered by ice and snow, with coastal temperatures typically ranging from 5°C to -35°C [
1]. Ice- and snow-free areas, present during the austral summer, are located around the continent’s coast. Most life forms in continental Antarctica are known to inhabit these ice-free and snow-free areas [
2].
The first record of fungi from this region was published around 1897-1899 by a Belgian expedition that collected
Sclerotium antarcticum from Danco Island near the Antarctic Peninsula [
3]. This expedition included the late Roald Amundsen (1872–1928), who was the first to reach the South Pole.
Syowa Station, the base for the Japanese Antarctic Research Expedition (JARE), is located on East Ongul Island (69° 1′ S, 39° 35′ E). The first report on fungi around Syowa Station was published in 1961 [
4], followed by three articles reporting 12 ascomycetes and four basidiomycetes [
5,
6,
7]. From the 1960s to 2013, no additional fungal species were reported by JARE near Syowa Station. Since 2013, fungal diversity has been reported from two sites: East Ongul Island, where Syowa Station is located, and the Skarvsnes ice-free area, about 60 km away from the station [
8,
9]. To date, these fungal surveys have reported a total of 77 fungal species, including 61 ascomycetes and 16 basidiomycetes [
10].
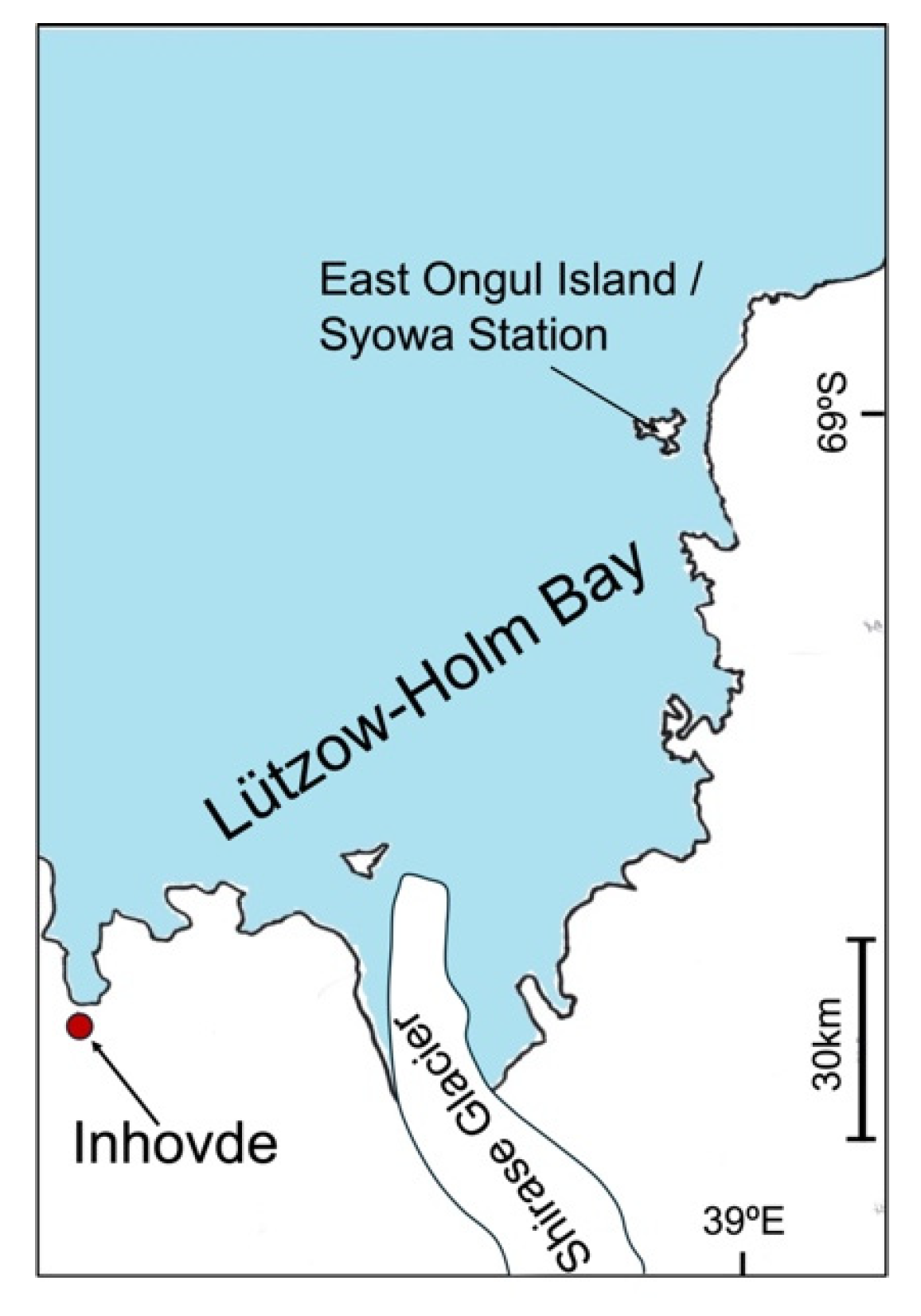
The Inhovde area (69°51′S, 37°06′W) is located between East Ongul Island and the Sør Rondane mountains. Despite previous fungal surveys near Syowa Station, the fungal diversity of the Inhovde area has been completely unexplored.
Fungi serve as decomposers of organic matter in ecosystems, supporting nutrient cycling. Therefore, understanding the diversity of fungi inhabiting Antarctica, which is experiencing rapid environmental changes, is crucial for research aimed at preserving the Antarctic environment [
11].
In this study, the diversity of fungi in the Inhovde area is reported for the first time, and the growth of these fungi at low temperatures is also investigated.
2. Materials and Methods
2.1. Sampling Site and Sample Collection
The Inhovde area (69°51′S, 37°06′W) is located in the Lützow Holm Bay, East Antarctica. This area is about 130 km from Syowa Station, beyond the Shirase Glacier (
Figure 1).
Ice and snow samples used in this study were collected from Inhovde area during the 56th JARE. The samples were stored at -20 °C immediately after collection until use.
2.2. Isolation of Fungal Strains
Each untreated sample (0.1 g) was plated directly on 5 × potato dextrose agar (PDA, Difco, Becton Dickinson, Tokyo, Japan) at pH8.0 containing 50 μg/mL chloramphenicol and incubated at 10 °C for up to 1 month. Fungal cultures were selected for further isolation and identification based on colony morphology and color, and fungi from colonies of different morphology and color were purified by repeated cultivation on fresh PDA at 10 °C.
2.3. Sequencing and Species Identification
DNA was extracted from fungal colonies using the NucleoSpin Microbial DNA kit (Takara Bio Inc., Shiga, Japan) following the manufacturer’s protocols. The extracted DNA was then amplified by polymerase chain reaction (PCR) using KOD-plus DNA polymerase (Toyobo, Osaka, Japan) with the primers ITS1F (5′-GTAACAAGGTTTCCGT) and NL4 (5′-GGTCCGTGTTTCAAGACGG). These primers target two DNA sequences commonly utilized in molecular fungal taxonomy: the ITS region and the D1/D2 domain of the LSU rDNA gene.
The PCR conditions were as follows: initial denaturation at 94 °C for 5 minutes; 35 cycles of 98 °C for 10 seconds (denaturation), 50-65 °C for 30 seconds (primer annealing), and 68 °C for 90 seconds (extension); followed by a final extension at 68 °C for 10 minutes. PCR was performed using an Eppendorf Master Cycler Nexus (Eppendorf Japan, Tokyo, Japan). The amplified DNA fragments were purified using Sephacryl S-400HR (Sigma-Aldrich, Tokyo, Japan). DNA sequencing was performed on an ABI Prism 3130xl sequencer (Applied Biosystems, Life Technologies, Tokyo, Japan), and the sequences were deposited in GenBank. The GenBank accession numbers for all sequences analyzed in this study are listed in
Supplementary Table S1. Fungal species identifications were conducted through BLAST analysis based on sequence homology.
The ITS region and D1/D2 domain sequences of
Cystobasidium sp. were aligned using CLUSTAL W (
http://clustalw.ddbj.nig.ac.jp/) and manually adjusted. The aligned sequences were used for phylogenetic reconstruction analysis by the neighbor-joining method in MEGA X [
12]. Bootstrap analysis with 1000 replicates was performed to determine the confidence of the tree nodes.
2.4. Determination of Growth Abilities of Fungal Strains in the Inhovde Area
Fungal growth tests at -3°C were conducted on PDA plates. Vitamin dependence of growth was tested in liquid medium in glass vials according to standard methods [
13] at 10°C. The amino acid requirement test was performed in liquid medium containing yeast nitrogen base without amino acids (YNB, Difco, Becton Dickinson, Tokyo, Japan) and 2% glucose at 10°C.
3. Results
3.1. Fungal Diversity of the Inhovde Area
In the current study, 148 fungal strains were isolated from ice and snow samples from the Inhovde area, of which 132 were successfully extracted and analyzed by DNA sequencing. Note that some strains were not identified to the species level in this study. When classification to the species level was not possible, the strains are presented at the genus or family level.
Based on the sequences of the ITS region and D1/D2 domain of the LSU rDNA gene, the strains were classified as basidiomycetes, belonging to six genera and 10 species: Cystobasidium ongulense, Cystobasidium sp., Glaciozyma watsonii, Leucosporidium yakuticum, Mrakia gelida, Mrakia robertii, Naganishia adeliensis, Naganishia albidosimilis, Naganishia friedmannii, and Vishniacozyma victoriae; and as ascomycetes, belonging to four genera and four species: Aotearoamyces sp., Leuconeurospora sp. (yeast-like), Pseudeurotium hygrophilum, and Symbiotaphrina microtheca(yeast-like). At the species level, the most frequently isolated fungi were V. victoriae (35.6%), G. watsonii (28.0%), and C. ongulense (15.2%). All species classified as basidiomycetes were in yeast form. Two of the four ascomycetous fungal species (Leuconeurospora sp. and Symbiotaphrina microtheca) exhibited yeast-like morphology.
Table 1.
Number of strains isolated from Inhovde area.
Table 1.
Number of strains isolated from Inhovde area.
| Speices |
Number of strains |
| Ascomycota |
|
|
Aotearoamyces sp. |
3 |
|
Leuconeurospora sp. |
1 |
| Pseudeurotium hygrophilum |
1 |
| Symbiotaphrina microtheca |
1 |
| Basidiomycota |
|
|
Cystobasidiumongulense
|
20 |
|
Cystobasidium sp. |
2 |
| Glaciozyma watsonii |
37 |
| Leucosporidium yakuticum |
1 |
| Mrakia gelida |
5 |
| Mrakia robertii |
9 |
| Naganishia adeliensis |
1 |
| Naganishia albidosimilis |
1 |
| Naganishia friedmannii |
4 |
| Vishniacozyma. victoriae |
46 |
| Total |
132 |
3.2. Growth Characteristics of Fungi in the Inhovde Area
In this study, one strain from each species was randomly selected. These selected strains were used as representative strains for each species in subsequent experiments.
To evaluate the growth characteristics of the selected fungal species at sub-zero temperatures, their ability to grow on PDA plates at -3 °C was examined. Additionally, their growth in vitamin-free and amino acid-free media at 10 °C was tested to assess their ability to thrive in oligotrophic environments. All 14 species were able to grow at -3 °C, as well as in vitamin-free and amino acid-free media (
Table 2).
4. Discussion
In the present study, 46 strains of
Vishniacozyma victoriae were isolated.
V. victoriae is a cosmopolitan yeast species. This yeast has been reported from various cold environments around the world including the Canadian High Arctic [
13]. In Antarctica, this yeast had also been identified from Victoria Land, Lichen Valley, Vestfold Hills, Davis Base, the Skarvsnes ice-free area and East Ongul Island [8,9,15,16,17].
In this study, 37 strains of Glaciozyma watsonii were isolated from the samples. G. watsonii was identified from meltwater stream sediment, glacial meltwater stream sediment and seawater [18-20]. The genus Glaciozyma is a fungus with a remarkable ability to adapt to cold temperatures, being able to grow even on PDA media frozen at -80°C (Tsuji unpublished data).
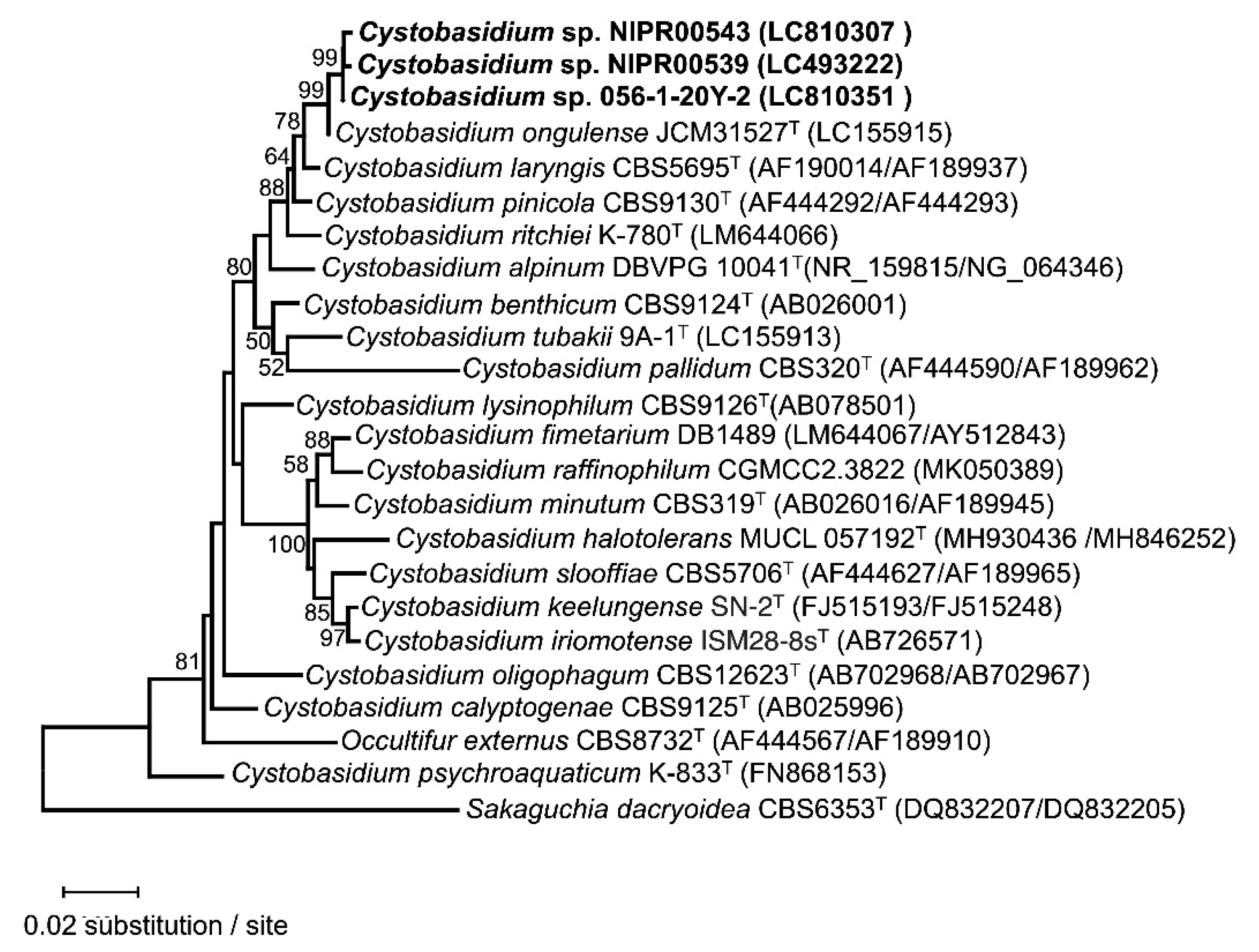
Cystobasidium ongulense strains were originally isolated from the soil of East Ongul Island, East Antarctica [21]. This yeast species has also been reported from Italian Alpus [22]. In the present study, 19 strains of Cysotobasidium ongulense were isolated from samples collected from Inhovde area. Three strains, Cystobasidium sp. NIPR00539, NIPR00543 and 056-1-20Y-2, were also closely related to C. ongulense but had two nucleotide substitutions in the ITS region and four in the D1/D2 region compared to the type strain, JCM 31527.
Phylogenetic analysis using sequences of the ITS and D1/D2 regions indicated that NIPR00539, NIPR00543 and 056-1-20Y-2 branched off from JCM 31527 (
Figure 2). This suggests that the three strains are new species closely related to
C. ongulense.
di Menna (1966) reported that approximately 24% of the culturable yeasts from Antarctic soil were Mrakia spp. [23] In this study, five Mrakia gelida and nine M. robertii strains were isolated from samples from the Inhovde area. M. gelida is one of the most commonly isolated yeasts from cold environments worldwide [24]. In Antarctica, this species has been isolated from soil at Scott Base and from lake sediment in the ice-free area of Skarvsnes, East Antarctica [8,9,22]. M. robertii was first reported from Mossell Lake, Antarctica, and the Italian Alps [26]. This yeast has also been reported from the Antarctic Peninsula and East Ongul Island. [9,26].
The genus Naganishia has been found in a variety of ecosystems, including marine waters, glaciers, soil, and Antarctica [27,28,29,30]. In this study, one strain of Naganishia adeliensis, one strain of N. albidosimilis, and four strains of N. friedmannii were isolated. There are also reports that N. friedmannii has a high biosurfactant production capacity in Antarctica. [31].
A strain of Leucosporidium yakuticum was obtained in this study. The original paper reported that this yeast was found in permafrost in Russia [32]. To the best of our knowledge, this is the second report of the fungus from Antarctica, following a report from the South Shetland Islands [16>], and the first from continental Antarctica.
Neighbor-join analysis of the ITS region and the LSU D1/D2 domain sequences of Cystobasidium sp. strains and closely related species. Cystobasidium sp. strains investigated in this study are highlighted in bold font. Sakaguchia dacryoidea CBS 6353 was designated as the outgroup. Bootstrap percentages of neighbor-join analysis over 50% from 1000 bootstrap replicates are shown from left on the branches. The scale bar represents 0.02 substitutions per nucleotide position.
The genus is one genus in a species, Aotearoamyces nothofagi, which is an ascomycetous fungus found in the forests of the Notochaetidae family in New Zealand.[34]. Aotearoamyces sp.cultured in this study showed low homology (94.9% in the ITS region and 97.3% in the D1/D2 region) with the type strain of A. nothofagi. This suggests that Aotearoamyces sp. isolated from the Inhovde region is a new species.
The genus Leuconeurospora is reported to have been isolated from King George Island, Antarctica [17]. To the best of our knowledge, this is the first report of this fungus from continental Antarctica.
Pseudeurotium hygrophilum has been reported to be isolated from sediment cores from Hope Bay, Antarctic Peninsula and from Lake Baikal, Russia [34,35]. In the present study, a strain of Pseudurotium hygrophilum was isolated from samples from the Inhovde area.Symbiotaphrina microtheca is also known as Sarea microtheca and Tromeropsis microtheca. Information on this fungus is limited but is probably the first report from continental Antarctica.
In this study, all representative strains indeed showed growth in vitamin-free media, amino acid-free media and at subzero temperatures. In general, continental Antarctica is characterized by an oligotrophic environment [21]. Considering this and the current observations, the fungi inhabiting the area may have acquired their growth characteristics to survive cold and oligotrophic environments such as that of the Inhovde area.
5. Conclusions
Of the 14 fungal species obtained in this study, the following six (two basidiomycetes and four conidiomycetes) were newly reported from the Syowa Station area: Cystobasidium sp., Leucosporidium yakuticum, Aotearoamyces sp., Leuconeurospora sp., Pseudeurotium hygrophilum, Symbiotaphrina microtheca. So far, 61 species of ascomycetous fungi and 16 species of basidiomycetous fungi have been reported from near the Showa Station area [36]. The results of the present study increased the total number of fungal species isolated from the Showa Station area to 83, of which 65 were ascomycetes and 18 were basidiomycetes. These results indicate that these fungi play an important role in nutrient cycling in the ecosystem of the Inhovde area at near-freezing temperatures.
In recent years, fungi inhabiting Antarctica have garnered attention as new microbial resources, due to their psychrophilic characteristics, including cold-active enzymes and secondary metabolites. The fungi isolated from the Inhovde area in this study also hold potential as new microbial resources that could contribute to achieving carbon neutrality.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Table S1: Species names, strain names and GenBank accession numbers of the ITS region and D1/Dr2 domain sequences of fungal isolates from Inhovde area.
Author Contributions
Conceived and designed the experiments: MT. Performed the laboratory experiments and analyses: MT. Analyzed the data: all authors. Wrote the manuscript: MT with input from all authors.
Funding
This work was supported by the NIPR Research Project (KP-404), a JSPS Grant-in-Aid for Scientific Research(B) to M. Tsuji (no. 23K28280), Institution for Fermentation, Osaka, for General Re-search Grant to M. Tsuji (no. G-2022-1-007). This work was also supported by “Strategic Research Projects “ grant from ROIS (Research Organization of Information and Systems) (no. 2022-SRP-01).
Data Availability Statement
The DNA sequences of Antarctic fungi obtained in this study have been deposited in the DNA Data Bank, and the list of accession numbers of the registered DNA sequences can be found in
Table S1.
Acknowledgments
The author would like to thank the National Institute of Polar Research, Japan, and The National Institute of Technology, Asahikawa College, Japan, for their support in obtaining these research results. We thank Ms. Mizuho Mori and Mr. Kenichi Watanabe for their technical support. We also thank Dr. Megumu Tsujimoto for providing samples.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ravindra, R.; Chaturvedi, A. Antarctica. In Encyclopedia of snow, ice and glaciers; Singh, V.P.; Singh, P.; Haritashya, U.K. Eds., Springer, Berlin, Germany, 2011; pp. 45–53.
- Onofri, S.; Zucconi, L.; Tosi, S. Continental Antarctic Fungi. IHW Verlag, München, Germany, 2007.
- Boomer, E.; Rousseau, M. Note préliminaire sur les champignons recueillis par l’Expedition Antarctique Belge. Bulletin de l’Académie Royale des Sciences de Belgique Classe des Sciences 1900, 8, 640–646. [Google Scholar]
- Tubaki, K. On some fungi isolated from the Antarctic materials. Biological results of the Japanese Antarctic Research Expedition, 1961, 14, 1- 9.
- Tubaki, K. Note on some fungi and yeasts from Antarctica. Antarctic Record (Tokyo), 1961, 11, 161–162. [Google Scholar]
- Soneda, M. On some yeasts from the Antarctic region. Biological results of the Japanese Antarctic Research Expedition,1961, 15, 3-10.
- Tubaki, K.; Asano, I. (1965). Additional species of fungi isolated from the Antarctic materials. JARE scientific reports. Series E: Biology, 1965, 27, 1–27. [Google Scholar]
- Tsuji, M.; Fujiu, S.; Xiao, N.; Hanada, Y.; Kudoh, S.; Kondo, H.; Tsuda, S.; Hoshino, T. Cold adaptation of fungi obtained from soil and lake sediment in the Skarvsnes ice-free area, Antarctic. FEMS Microbiol. Lett. 2013, 346, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, M. Genetic diversity of yeasts from East Ongul Island, East Antarctica and their extracellular enzymes secretion. Polar Biol. 2018, 41, 249–258. [Google Scholar] [CrossRef]
- Tsuji, M. A catalog of fungi recorded from the vicinity of Syowa Station. Mycoscience, 2018, 59, 319–324. [CrossRef]
- Welander, U. Microbial degradation of organic pollutants in soil in a cold climate. Soil Sediment Contam. 2005, 14, 281–291. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. ; Robert, V (2011) Methods for isolation, phenotypic characterization and maintenance of yeasts. In The yeast, a taxonomic study, 5th edn; Kurtzman, C.P.; Fell, J.W.; Boeckhout, T. Eds.; Elsevier, Amsterdam, Netherland, 2011; pp. 87-111. [CrossRef]
- Tsuji, M. Survey on fungi in Antarctica and High Arctic regions, and their impact on climate change. Climate, 2023, 11, 195. [CrossRef]
- Montes, M.J.; Belloch, C.; Galiana, M.; Garcia, M.D.; Andrés, C.; Ferrer, S.; Torres-Rodriguez, J.M.; Guinea, J. Polyphasic Taxonomy of a Novel Yeast Isolated from Antarctic Environment; Description of Cryptococcus victoriae sp. nov. Syst Appl Microbiol, 1999, 22, 97-105.
- Thomas-Hall, S.R.; Watson, K.; Scorzetti, G. (2002) Cryptococcus stzelliae sp. nov. and three novel strains of Cryptococcus victoriae, yeasts isolated from Antarctica soils. Int J Syst Evol Microbiol, 2002, 52, 2303-2308. [CrossRef]
- Vaz, A.B.M.; Rosa, L.H.; Vieira, M.L.A.; de Garcia, V.; Brandão, L.R.; Teixeira, L.C.R.S.; Moliné, M.; Libkind, D.; van Broock, M.; Rosa, C.A. The diversity, extracellular enzymatic activities and photoprotective compounds of yeasts isolated in Antarctica. Braz. J. Microbiol. 2011, 42, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Vishniac, H.S.; Klingler, J. (1986) Yeast in the Antarctic deserts. In Perspectives in microbial ecology, Mergusar F, Gantar M, Eds. Ljubljana, Slovene Society for Microbiology: 46-51.
- Vishniac, H.S.; Kurtzman, C.P. Cryptococcus antarcticus sp. nov. and Cryptococcus albidosimilis sp. nov., basidioblastomycetes from Antarctic soils. Int J Syst Evol Microbiol, 1992, 42, 547-553. [CrossRef]
- Turchetti, B.; Thomas-Hall, S.R.; Connell, L.B.; Branda, E.; Buzzini, P.; Theelen, B.; Boekhout, T. Psychrophilic yeasts from Antarctica and European glaciers: description of Glaciozyma gen. nov., Glaciozyma martinii sp. nov. and Glaciozyma watsonii sp. nov. Extremophiles, 2011, 15, 573-586. [CrossRef]
- Tsuji, M.; Tsujimoto, M.; Imura, S. (2017) Cystobasidium tubakii and Cystobasidium ongulense, new basidiomycetous yeast species isolated from East Ongul Island, East Antarctica. Mycoscience, 2017, 58, 103–107. [CrossRef]
- Turchetti, B.; Goretti, M.; Branda, E.; Diolaiuti, G.; D’Agata, C.; Smiraglia, C.; Onofri, A.; Buzzini, P. Influence of abiotic variables on culturable yeast diversity in two distinct Alpine glaciers. FEMS Microbiol Ecol, 2012, 86, 327-340. [CrossRef]
- di Menna, M.E. Yeasts in Antarctic soil. Antonie van Leeuwenhoek, 1966, 32, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Buzzini, P.; Branda, E.; Goretti, M.; Turchetti, B. Psychrophilic yeasts from worldwide glacial habitats: diversity, adaptation strategies and biotechnological potential. FEMS Microbiol Ecol, 2012, 82, 217-241. [CrossRef]
- Thomas-Hall, S.R.; Turchetti, B.; Buzzini, P.; Branda, E.; Boekhout, T.; Theelen, B.; Watson, K. Cold-adapted yeasts from Antarctica and Italian Alps-description of three novel species: Mrakia robertii sp. nov., Mrakia blollopis sp. nov. and Mrakiella niccombsii sp. nov. Extremophiles, 2010, 14, 47-59. [CrossRef]
- Ferreira, E.M.S.; de Sousa, F.M.P.; Rosa, L.H.; Pimenta, R.S. (2019) Taxonomy and richness of yeasts associated with angiosperms, bryophytes, and meltwater biofilms collected in the Antarctic Peninsula. Extremophiles, 2019, 23, 151-159. [CrossRef]
- Scorzetti, G.; Fell, J.W.; Fonseca, A.; Statzell-Tallman, A. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res, 2002, 2, 495–517. [CrossRef]
- Turchetti, B.; Buzzini, P.; Goretti, M.; Goretti, M.; Branda, E.; Diolaiuti, G.; D’Agata, C.; Smiraglia, C.; Vaughan-Martini, A. Psychrophilic yeasts in glacial environments of Alpine glaciers. FEMS Microbiol Ecol, 2008, 63, 73-83. [CrossRef]
- Vishniac, H.S. Cryptococcus Friedmannii, a new species of yeast from the Antarctic. Mycologia, 1985, 77, 149-153. [CrossRef]
- Vishniac, H.S.; Kurtzman, C.P. Cryptococcus antarcticus sp. nov. and Cryptococcus albidosimilis sp. nov., Basidioblastomycetes from Antarctic Soils. Int J Syst Bacteriol, 1992, 42, 547-553. [CrossRef]
- Chaves, F.D.S.; Brumano, L.P.; Franco Marcelino, P.R.; da Silva, S.S.; Sette, L.D.; Felipe, M.G.A. Biosurfactant production by Antarctic-derived yeasts in sugarcane straw hemicellulosic hydrolysate. Biomass Conv Bioref; 2023, 13, 5295-5305. [CrossRef]
- Golubev, W.I. New species of basidiomycetous yeasts, Rhodotorula creatinovora and R. yakutica, isolated from permafrost soils of Eastern-Siberian Arctic. Mikol Fitopatol, 1998, 32, 8-13.
- Quijada, L.; Johnston, P.R.; Cooper, J.A.Pfister, D.H. Overview of Phacidiales, including Aotearoamyces gen. nov. on Nothofagus. IMA Fungus, 2018, 9, 371-382. [CrossRef]
- Rosa, L.H.; Ogaki, M.B.; Lirio, J.M.; Vieira, R.; Coria, S.H.; Pinto, O.H.B.; Carvalho-Silva, M.; Convey, P.; Rosa, C.A.; Câmara, P.E.A.S. Fungal diversity in a sediment core from climate change impacted Boeckella Lake, Hope Bay, north-eastern Antarctic Peninsula assessed using metabarcoding. Extremophiles, 2022, 26, 16. [CrossRef]
- . Fedorova, M.D.; Kurakov, A.V. Microbiota of Bottom Sediments in the Coastal Zone of Lake Baikal. Contemp Probl Ecol, 2023, 16, 492-508. [CrossRef]
- Tsuji, M. (2019) An index of non-lichenized fungi recorded in the vicinity of Syowa Station, East Antarctica. In Fungi in Polar Regions, Tsuji M, Hoshino T. Eds., Oxford: CRC press, 1-16.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).