Submitted:
22 July 2024
Posted:
24 July 2024
You are already at the latest version
Abstract
Keywords:
I. Introduction
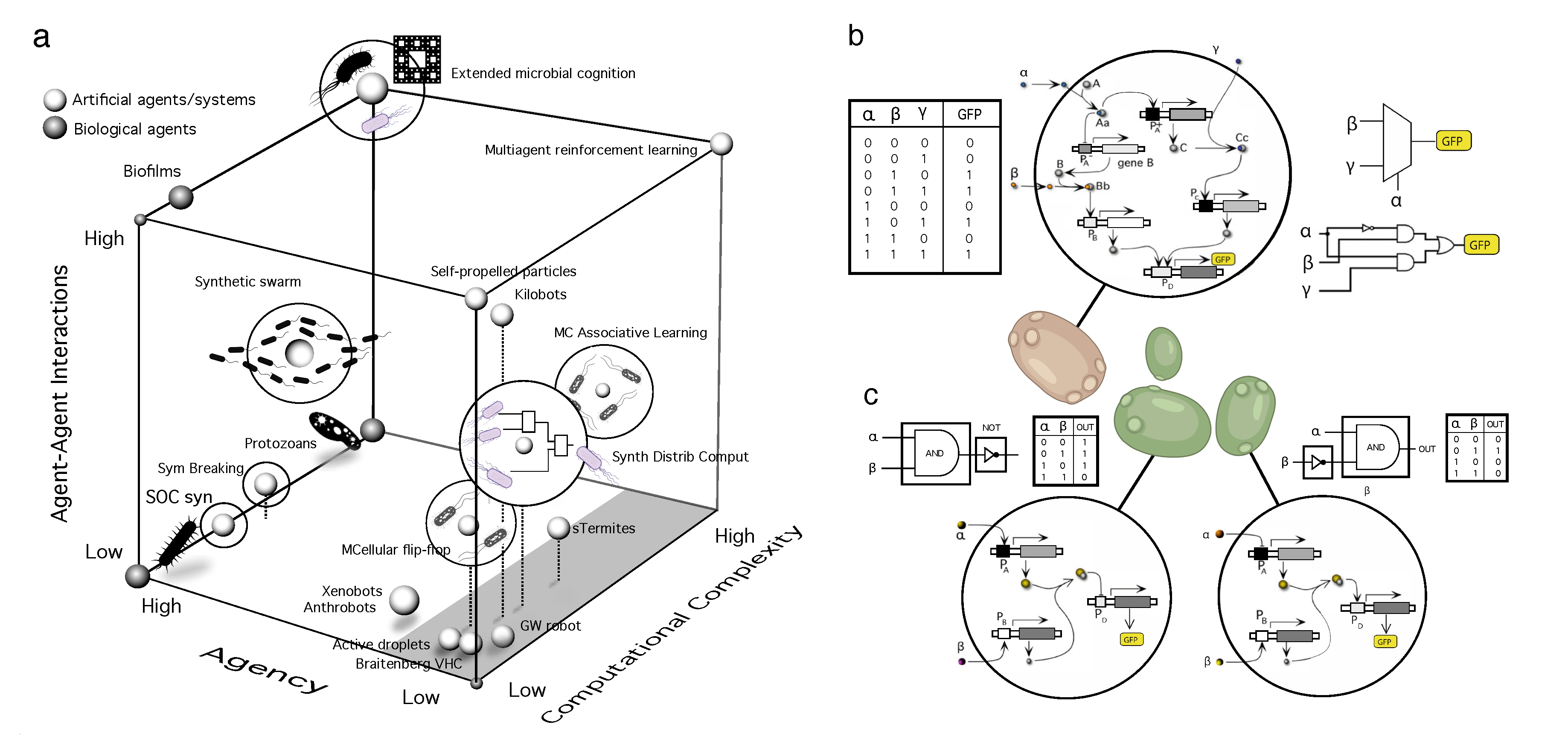
- Synthetic multicellular circuits. This class involves cellular circuits that have been modified or introduced through genetic engineering within living cells, typically used as a chassis [35,36,37,38,39]. Many designs within this domain rely on a modular approach to circuit complexity based on standard combinatorial circuit design [40,41,42]. Cellular consortia have been used as MC implementations of all kinds of simple responses, from combining Boolean gates [43,44,45,46] to pattern formation [4,47]. These designs involve strains interacting through chemical signals propagating in a liquid medium or diffusing over short distances on an agar plate.
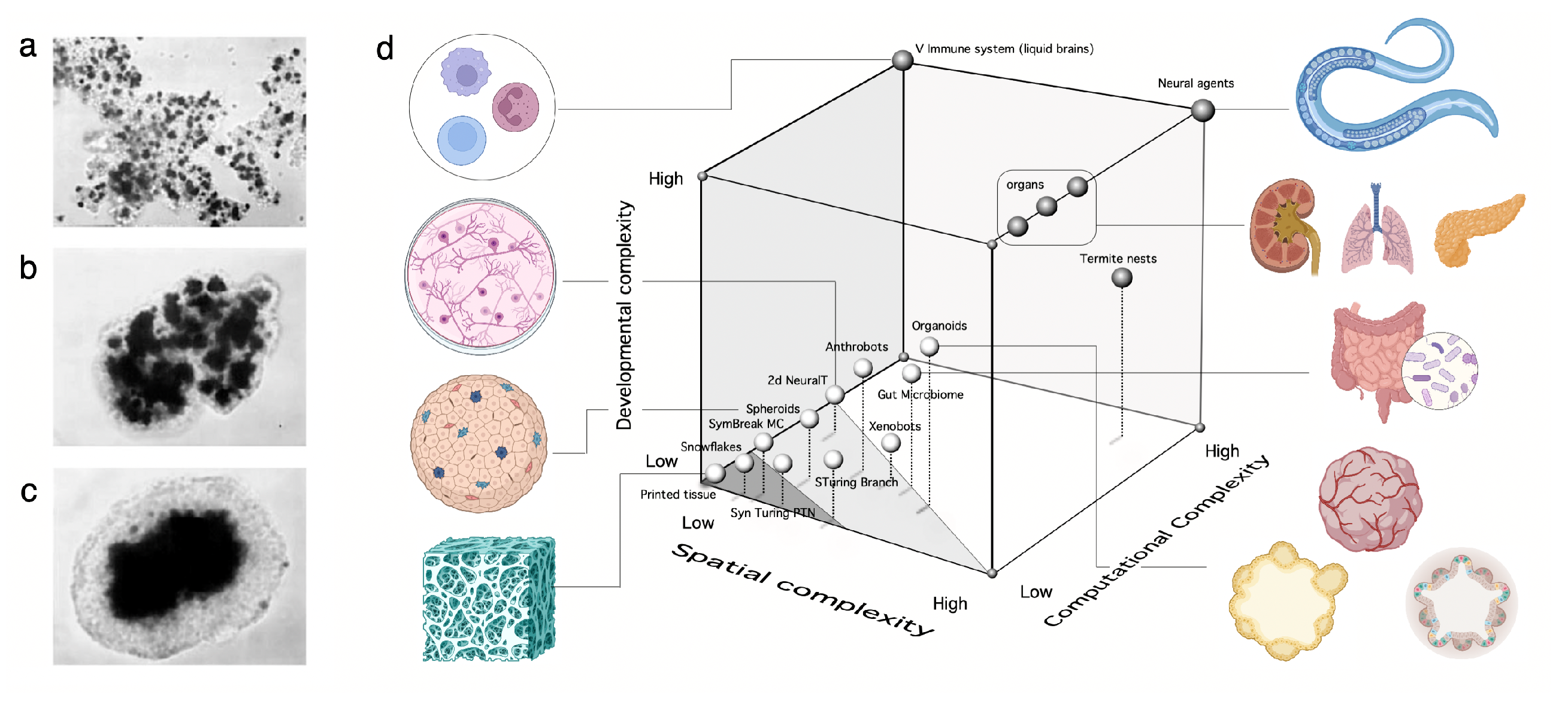
- Programmable synthetic assemblies. The next step towards engineering MC systems exploits the predictable properties displayed by adhesion-driven spatial morphodynamics. Again, this bottom-up engineering allows predicting (i. e. programming) the outcome of the final spatial structure. It was early understood that cell sorting due to different adhesion energies could easily explain the self-organized aggregation of a set of randomly mixed cells [48,49]. Despite the self-organized nature of the process, it is possible to make some predictions concerning the spatial arrangements at steady state.
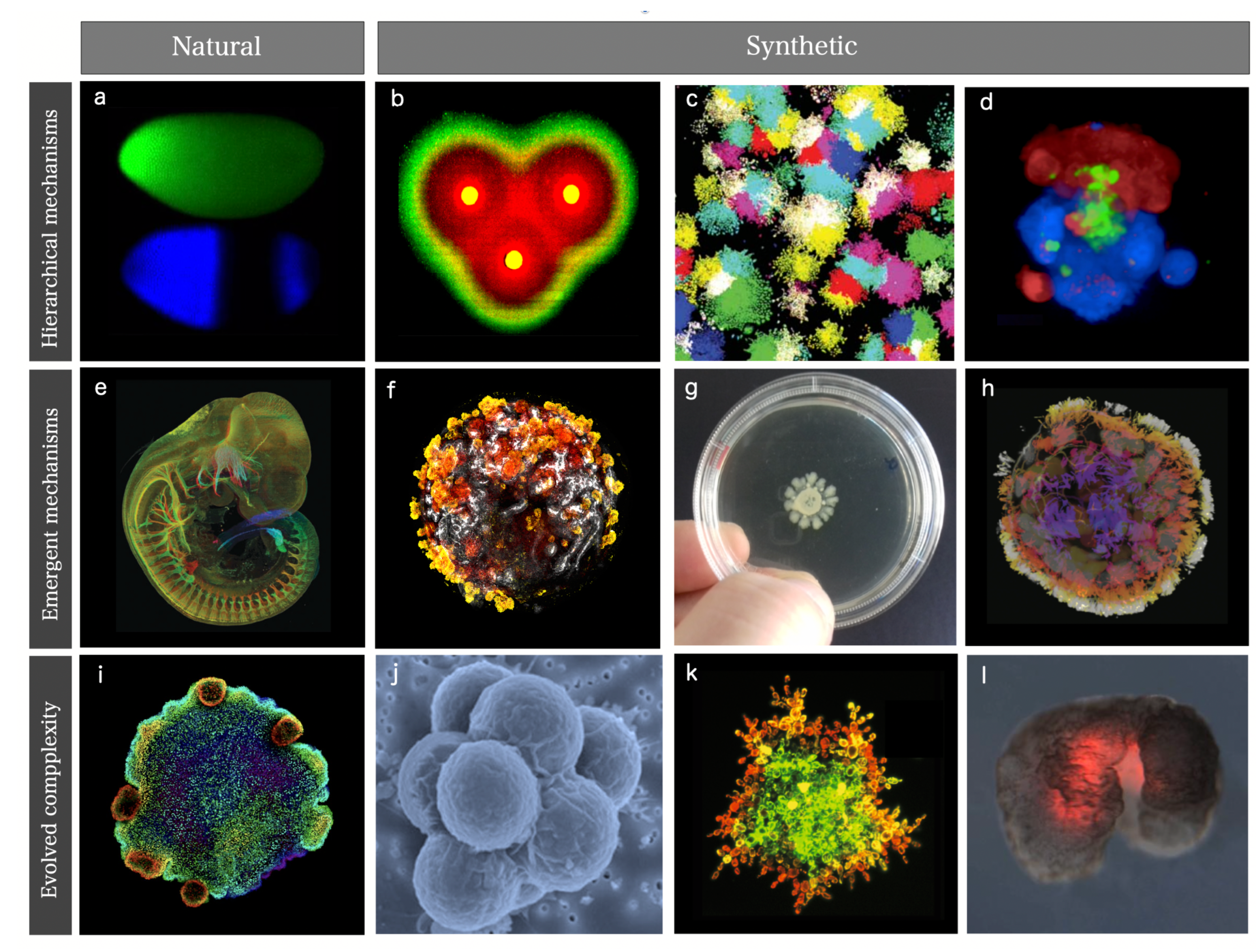
- Synthetic morphology and agential materials. One way of moving beyond cell-level engineering involves considering cell collectives as agential materials. These systems exhibit emergent properties at the system level that cannot be understood in terms of the properties of the constituents (genes and cells). This approach takes advantage of higher-order properties of embodied living matter (such as memory, context-sensitive navigation of problem spaces and homeostasis) to perform computations and design morphologies beyond the bottom-up principles of synthetic biology [29,50]. This class includes organoids and biobots and other MC assemblies capable of collective responses in space and time and novel forms of behaviour.
II. Synthetic Multicellular Classes
A. Synthetic Multicellular Circuits
B. Programmable Synthetic Assemblies

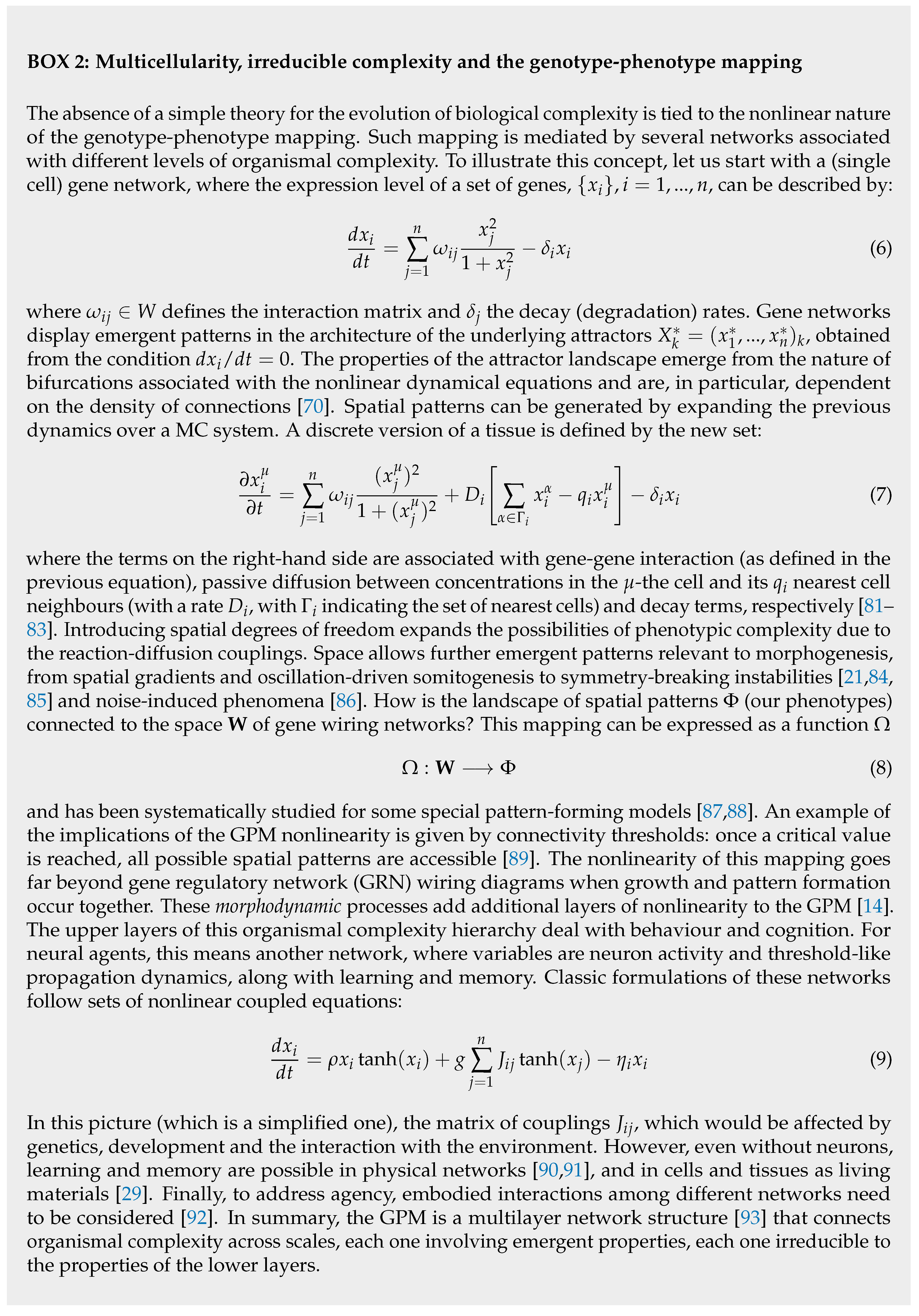
C. Synthetic Morphology and Agential Materials
III. Open Problems
IV. Discussion
Acknowledgments
References
- Knoll, A.H. The multiple origins of complex multicellularity. Annual Review of Earth and Planetary Sciences 2011, 39, 217–239. [Google Scholar] [CrossRef]
- Grosberg, R.K.; Strathmann, R.R. The evolution of multicellularity: a minor major transition? Annu. Rev. Ecol. Evol. Syst. 2007, 38, 621–654. [Google Scholar] [CrossRef]
- Levin, M. The Computational Boundary of a “Self”: Developmental Bioelectricity Drives Multicellularity and Scale-Free Cognition. Frontiers in Psychology 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Gerchman, Y.; Collins, C.H.; Arnold, F.H.; Weiss, R. A synthetic multicellular system for programmed pattern formation. Nature 2005, 434, 1130–1134. [Google Scholar] [CrossRef]
- Zhu, R.; del Rio-Salgado, J.M.; Garcia-Ojalvo, J.; Elowitz, M.B. Synthetic multistability in mammalian cells. Science 2022, 375. [Google Scholar] [CrossRef]
- Wauford, N.; Patel, A.; Tordoff, J.; Enghuus, C.; Jin, A.; Toppen, J.; Kemp, M.L.; Weiss, R. Synthetic symmetry breaking and programmable multicellular structure formation. Cell Systems 2023, 14, 806–818. [Google Scholar] [CrossRef] [PubMed]
- Herron, M.D.; Borin, J.M.; Boswell, J.C.; Walker, J.; Chen, I.C.K.; Knox, C.A.; Boyd, M.; Rosenzweig, F.; Ratcliff, W.C. De novo origins of multicellularity in response to predation. Scientific reports 2019, 9, 2328. [Google Scholar] [CrossRef] [PubMed]
- Bozdag, G.O.; Libby, E.; Pineau, R.; Reinhard, C.T.; Ratcliff, W.C. Oxygen suppression of macroscopic multicellularity. Nature communications 2021, 12, 2838. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Trillo, I.; Burger, G.; Holland, P.W.; King, N.; Lang, B.F.; Roger, A.J.; Gray, M.W. The origins of multicellularity: a multi-taxon genome initiative. TRENDS in Genetics 2007, 23, 113–118. [Google Scholar] [CrossRef]
- Ruiz-Trillo, I.; Nedelcu, A.M. Evolutionary transitions to multicellular life: principles and mechanisms; Vol. 2, Springer, 2015; pp. 53–0763. [CrossRef]
- Sebé-Pedrós, A.; Degnan, B.M.; Ruiz-Trillo, I. The origin of Metazoa: a unicellular perspective. Nature Reviews Genetics 2017, 18, 498–512. [Google Scholar] [CrossRef]
- Newman, S.A.; Bhat, R. Dynamical patterning modules: a “pattern language” for development and evolution of multicellular form. International Journal of Developmental Biology 2009, 53, 693. [Google Scholar] [CrossRef] [PubMed]
- Newman, S.A. Physico-genetic determinants in the evolution of development. Science 2012, 338, 217–219. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Ciudad, I.; Newman, S.; Solé, R. Phenotypic and dynamical transitions in model genetic networks I. Emergence of patterns and genotype-phenotype relationships. Evolution & development 2001, 3, 84–94. [Google Scholar] [CrossRef]
- Ratcliff, W.C.; Denison, R.F.; Borrello, M.; Travisano, M. Experimental evolution of multicellularity. Proceedings of the National Academy of Sciences 2012, 109, 1595–1600. [Google Scholar] [CrossRef] [PubMed]
- Kriegman, S.; Blackiston, D.; Levin, M.; Bongard, J. A scalable pipeline for designing reconfigurable organisms. Proceedings of the National Academy of Sciences 2020, 117, 1853–1859. [Google Scholar] [CrossRef]
- Blackiston, D.; Kriegman, S.; Bongard, J.; Levin, M. Biological Robots: Perspectives on an Emerging Interdisciplinary Field. Soft Robotics 2023, 10, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, B. How the leopard changed its spots: The evolution of complexity; Vol. 24, Princeton University Press, 2001.
- Alberch, P. The logic of monsters: Evidence for internal constraint in development and evolution. Geobios 1989, 22, 21–57. [Google Scholar] [CrossRef]
- Alberch, P. From genes to phenotype: dynamical systems and evolvability. Genetica 1991, 84, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Turing, A.M. The chemical basis of morphogenesis. Bulletin of mathematical biology 1990, 52, 153–197. [Google Scholar] [CrossRef]
- Huang, S. The molecular and mathematical basis of Waddington’s epigenetic landscape: A framework for post-Darwinian biology? Bioessays 2012, 34, 149–157. [Google Scholar] [CrossRef]
- Furusawa, C.; Kaneko, K. Origin of complexity in multicellular organisms. Physical review letters 2000, 84, 6130. [Google Scholar] [CrossRef] [PubMed]
- Nadell, C.D.; Bucci, V.; Drescher, K.; Levin, S.A.; Bassler, B.L.; Xavier, J.B. Cutting through the complexity of cell collectives. Proceedings of the Royal Society B: biological sciences 2013, 280, 20122770. [Google Scholar] [CrossRef] [PubMed]
- McMillen, P.; Levin, M. Collective intelligence: A unifying concept for integrating biology across scales and substrates. Communications Biology 2024, 7, 378. [Google Scholar] [CrossRef] [PubMed]
- Solé, R. Synthetic transitions: towards a new synthesis. Philosophical Transactions of the Royal Society B: Biological Sciences 2016, 371, 20150438. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimkhani, M.R.; Ebisuya, M. Synthetic developmental biology: build and control multicellular systems. Current opinion in chemical biology 2019, 52, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Ara, G.; Stapornwongkul, K.S.; Ebisuya, M. Scaling up complexity in synthetic developmental biology. Science 2022, 378, 864–868. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Levin, M. Synthetic morphology with agential materials. Nature Reviews Bioengineering 2023, 1, 46–59. [Google Scholar] [CrossRef]
- Levin, M. Darwin’s agential materials: evolutionary implications of multiscale competency in developmental biology. Cellular and Molecular Life Sciences 2023, 80. [Google Scholar] [CrossRef] [PubMed]
- Kamm, R.D.; Bashir, R. Creating Living Cellular Machines. Annals of Biomedical Engineering 2013, 42, 445–459. [Google Scholar] [CrossRef]
- Kamm, R.D.; Bashir, R.; Arora, N.; Dar, R.D.; Gillette, M.U.; others. Perspective: The promise of multi-cellular engineered living systems. APL Bioengineering 2018, 2. [Google Scholar] [CrossRef]
- Bongard, J.; Levin, M. Living Things Are Not (20th Century) Machines: Updating Mechanism Metaphors in Light of the Modern Science of Machine Behavior. Frontiers in Ecology and Evolution 2021, 9. [Google Scholar] [CrossRef]
- Bongard, J.; Levin, M. There’s Plenty of Room Right Here: Biological Systems as Evolved, Overloaded, Multi-Scale Machines. Biomimetics 2023, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Bashor, C.J.; Horwitz, A.A.; Peisajovich, S.G.; Lim, W.A. Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems. Annual review of biophysics 2010, 39, 515–537. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.A.; Der, B.S.; Shin, J.; Vaidyanathan, P.; Paralanov, V.; Strychalski, E.A.; Ross, D.; Densmore, D.; Voigt, C.A. Genetic circuit design automation. Science 2016, 352, aac7341. [Google Scholar] [CrossRef] [PubMed]
- Cameron, D.E.; Bashor, C.J.; Collins, J.J. A brief history of synthetic biology. Nature Reviews Microbiology 2014, 12, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Budde, M.W.; Mayalu, M.N.; Zhu, J.; Lu, A.C.; Murray, R.M.; Elowitz, M.B. Synthetic mammalian signaling circuits for robust cell population control. Cell 2022, 185, 967–979. [Google Scholar] [CrossRef] [PubMed]
- Youk, H.; Lim, W.A. Secreting and Sensing the Same Molecule Allows Cells to Achieve Versatile Social Behaviors. Science 2014, 343. [Google Scholar] [CrossRef] [PubMed]
- Amos, M. Cellular computing; Systems Biology, 2004. [CrossRef]
- Grozinger, L.; Amos, M.; Gorochowski, T.E.; Carbonell, P.; Oyarzún, D.A.; Stoof, R.; Fellermann, H.; Zuliani, P.; Tas, H.; Goñi-Moreno, A. Pathways to cellular supremacy in biocomputing. Nature communications 2019, 10, 5250. [Google Scholar] [CrossRef] [PubMed]
- Goñi-Moreno, Á. Biocomputation: Moving Beyond Turing with Living Cellular Computers. Communications of the ACM 2024, 67, 70–77. [Google Scholar] [CrossRef]
- Tamsir, A.; Tabor, J.J.; Voigt, C.A. Robust multicellular computing using genetically encoded NOR gates and chemical `wires’. Nature 2011, 469, 212–215. [Google Scholar] [CrossRef]
- Regot, S.; Macia, J.; Conde, N.; Furukawa, K.; Kjellén, J.; Peeters, T.; Hohmann, S.; de Nadal, E.; Posas, F.; Solé, R. Distributed biological computation with multicellular engineered networks. Nature 2011, 469, 207–211. [Google Scholar] [CrossRef]
- Moon, T.S.; Lou, C.; Tamsir, A.; Stanton, B.C.; Voigt, C.A. Genetic programs constructed from layered logic gates in single cells. Nature 2012, 491, 249–253. [Google Scholar] [CrossRef]
- Macia, J.; Manzoni, R.; Conde, N.; Urrios, A.; de Nadal, E.; Solé, R.; Posas, F. Implementation of Complex Biological Logic Circuits Using Spatially Distributed Multicellular Consortia. PLOS Computational Biology 2016, 12, e1004685. [Google Scholar] [CrossRef]
- Song, H.; Ding, M.Z.; Jia, X.Q.; Ma, Q.; Yuan, Y.J. Synthetic microbial consortia: from systematic analysis to construction and applications. Chemical Society Reviews 2014, 43, 6954–6981. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, M.S. Differential adhesion in morphogenesis: a modern view. Current opinion in genetics & development 2007, 17, 281–286. [Google Scholar] [CrossRef]
- Krens, S.G.; Heisenberg, C.P. Cell sorting in development. Current topics in developmental biology 2011, 95, 189–213. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.A. Synthetic morphology: prospects for engineered, self-constructing anatomies. Journal of anatomy 2008, 212, 707–719. [Google Scholar] [CrossRef]
- Urrios, A.; Macia, J.; Manzoni, R.; Conde, N.; Bonforti, A.; de Nadal, E.; Posas, F.; Solé, R. A Synthetic Multicellular Memory Device. ACS Synthetic Biology 2016, 5, 862–873. [Google Scholar] [CrossRef] [PubMed]
- Macia, J.; Vidiella, B.; Solé, R.V. Synthetic associative learning in engineered multicellular consortia. Journal of The Royal Society Interface 2017, 14, 20170158. [Google Scholar] [CrossRef]
- Macia, J.; Sole, R. How to Make a Synthetic Multicellular Computer. PLoS ONE 2014, 9, e81248. [Google Scholar] [CrossRef]
- Hopfield, J. Physics, computation, and why biology looks so different. Journal of Theoretical Biology 1994, 171, 53–60. [Google Scholar] [CrossRef]
- Voigt, C.A. Genetic parts to program bacteria. Current opinion in biotechnology 2006, 17, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Ruder, W.C.; Lu, T.; Collins, J.J. Synthetic biology moving into the clinic. Science 2011, 333, 1248–1252. [Google Scholar] [CrossRef]
- Benenson, Y. Biomolecular computing systems: principles, progress and potential. Nature Reviews Genetics 2012, 13, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Brophy, J.A.; Voigt, C.A. Principles of genetic circuit design. Nature methods 2014, 11, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Dannenfelser, R.; Allen, G.M.; VanderSluis, B.; Koegel, A.K.; Levinson, S.; Stark, S.R.; Yao, V.; Tadych, A.; Troyanskaya, O.G.; Lim, W.A. Discriminatory Power of Combinatorial Antigen Recognition in Cancer T Cell Therapies. Cell Systems 2020, 11, 215–228. [Google Scholar] [CrossRef]
- Vidiella, B.; Guillamon, A.; Sardanyés, J.; Maull, V.; Pla, J.; Conde, N.; Solé, R. Engineering self-organized criticality in living cells. Nature communications 2021, 12, 4415. [Google Scholar] [CrossRef]
- Solé, R.; Amor, D.R.; Duran-Nebreda, S.; Conde-Pueyo, N.; Carbonell-Ballestero, M.; Montañez, R. Synthetic collective intelligence. Biosystems 2016, 148, 47–61. [Google Scholar] [CrossRef]
- Solé, R.V.; Macia, J. Expanding the landscape of biological computation with synthetic multicellular consortia. Natural Computing 2013, 12, 485–497. [Google Scholar] [CrossRef]
- Solé, R.; Valverde, S. Evolving complexity: how tinkering shapes cells, software and ecological networks. Philosophical Transactions of the Royal Society B: Biological Sciences 2020, 375, 20190325. [Google Scholar] [CrossRef]
- Koza, J.R.; Keane, M.A.; Streeter, M.J. Evolving inventions. Scientific American 2003, 288, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Kitney, R.I.; Joly, N.; Buck, M. Engineering modular and orthogonal genetic logic gates for robust digital-like synthetic biology. Nature communications 2011, 2, 508. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Etxeberria, A. Agency in natural and artificial systems. Artificial Life 2005, 11, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A. On minimal autonomous agency: natural and artificial. Complex Systems 2018, 27, 289–313. [Google Scholar] [CrossRef]
- Bonner, J.T. The evolution of complexity by means of natural selection; Princeton University Press, 1988. [CrossRef]
- Valentine, J.W.; Collins, A.G.; Meyer, C.P. Morphological complexity increase in metazoans. Paleobiology 1994, 20, 131–142. [Google Scholar] [CrossRef]
- Kauffman, S.A. Metabolic stability and epigenesis in randomly constructed genetic nets. Journal of theoretical biology 1969, 22, 437–467. [Google Scholar] [CrossRef] [PubMed]
- Huang, S. Multistability and multicellularity: cell fates as high-dimensional attractors of gene regulatory networks. In Computational Systems Biology; Elsevier, 2006; pp. 293–326. [CrossRef]
- Koseska, A.; Zaikin, A.; Kurths, J.; García-Ojalvo, J. Timing Cellular Decision Making Under Noise via Cell–Cell Communication. PLoS ONE 2009, 4, e4872. [Google Scholar] [CrossRef] [PubMed]
- Gardner, T.S.; Cantor, C.R.; Collins, J.J. Construction of a genetic toggle switch in Escherichia coli. Nature 2000, 403, 339–342. [Google Scholar] [CrossRef]
- Hopfield, J.J. Neural networks and physical systems with emergent collective computational abilities. Proceedings of the National Academy of Sciences 1982, 79, 2554–2558. [Google Scholar] [CrossRef]
- Zhou, J.X.; Huang, S. Understanding gene circuits at cell-fate branch points for rational cell reprogramming. Trends in Genetics 2011, 27, 55–62. [Google Scholar] [CrossRef]
- Deutsch, A.; Dormann, S. Mathematical modeling of biological pattern formation; Springer, 2005; pp. 49–61. [CrossRef]
- Ollé-Vila, A.; Duran-Nebreda, S.; Conde-Pueyo, N.; Montañez, R.; Solé, R. A morphospace for synthetic organs and organoids: the possible and the actual. Integrative Biology 2016, 8, 485–503. [Google Scholar] [CrossRef]
- Mombach, J.C.; Glazier, J.A.; Raphael, R.C.; Zajac, M. Quantitative comparison between differential adhesion models and cell sorting in the presence and absence of fluctuations. Physical Review Letters 1995, 75, 2244. [Google Scholar] [CrossRef]
- Glass, D.S.; Riedel-Kruse, I.H. A synthetic bacterial cell-cell adhesion toolbox for programming multicellular morphologies and patterns. Cell 2018, 174, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Stevens, A.J.; Harris, A.R.; Gerdts, J.; Kim, K.H.; Trentesaux, C.; Ramirez, J.T.; McKeithan, W.L.; Fattahi, F.; Klein, O.D.; Fletcher, D.A.; others. Programming multicellular assembly with synthetic cell adhesion molecules. Nature 2023, 614, 144–152. [Google Scholar] [CrossRef]
- Mjolsness, E.; Sharp, D.H.; Reinitz, J. A connectionist model of development. Journal of theoretical Biology 1991, 152, 429–453. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Ciudad, I.; Garcia-Fernández, J.; Solé, R.V. Gene networks capable of pattern formation: from induction to reaction–diffusion. Journal of theoretical biology 2000, 205, 587–603. [Google Scholar] [CrossRef]
- Solé, R.V.; Salazar-Ciudad, I.; Garcia-Fernández, J. Common pattern formation, modularity and phase transitions in a gene network model of morphogenesis. Physica A: Statistical Mechanics and its Applications 2002, 305, 640–654. [Google Scholar] [CrossRef]
- Koch, A.J.; Meinhardt, H. Biological pattern formation: from basic mechanisms to complex structures. Reviews of modern physics 1994, 66, 1481. [Google Scholar] [CrossRef]
- Murray, J.D. Mathematical biology II, 3 ed.; Interdisciplinary applied mathematics, Springer: New York, NY, 2002. [Google Scholar]
- Sagués, F.; Sancho, J.M.; García-Ojalvo, J. Spatiotemporal order out of noise. Reviews of Modern Physics 2007, 79, 829–882. [Google Scholar] [CrossRef]
- Munteanu, A.; Sole, R.V. Neutrality and robustness in evo-devo: emergence of lateral inhibition. PLoS computational biology 2008, 4, e1000226. [Google Scholar] [CrossRef]
- Cotterell, J.; Sharpe, J. An atlas of gene regulatory networks reveals multiple three-gene mechanisms for interpreting morphogen gradients. Molecular systems biology 2010, 6, 425. [Google Scholar] [CrossRef] [PubMed]
- Solé, R.V.; Fernández, P.; Kauffman, S.A. Adaptive walks in a gene network model of morphogenesis: insights into the Cambrian explosion. arXiv preprint q-bio/0311013, -8. [CrossRef]
- Stern, M.; Murugan, A. Learning without neurons in physical systems. Annual Review of Condensed Matter Physics 2023, 14, 417–441. [Google Scholar] [CrossRef]
- Stern, M.; Liu, A.J.; Balasubramanian, V. Physical effects of learning. Physical Review E 2024, 109, 024311. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, R.; Scheier, C. Understanding intelligence; MIT press, 2001. [CrossRef]
- De Domenico, M.; Solé-Ribalta, A.; Cozzo, E.; Kivelä, M.; Moreno, Y.; Porter, M.A.; Gómez, S.; Arenas, A. Mathematical formulation of multilayer networks. Physical Review X 2013, 3, 041022. [Google Scholar] [CrossRef]
- Ball, P. Organisms as agents of evolution. John Templeton Foundation: West Conshohocken, PA, USA.
- Corrò, C.; Novellasdemunt, L.; Li, V.S. A brief history of organoids. American Journal of Physiology-Cell Physiology 2020, 319, C151–C165. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef] [PubMed]
- Sasai, Y. Cytosystems dynamics in self-organization of tissue architecture. Nature 2013, 493, 318–326. [Google Scholar] [CrossRef]
- Moris, N.; Anlas, K.; van den Brink, S.C.; Alemany, A.; Schröder, J.; Ghimire, S.; Balayo, T.; van Oudenaarden, A.; Martinez Arias, A. An in vitro model of early anteroposterior organization during human development. Nature 2020, 582, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Beccari, L.; Moris, N.; Girgin, M.; Turner, D.A.; Baillie-Johnson, P.; Cossy, A.C.; Lutolf, M.P.; Duboule, D.; Arias, A.M. Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 2018, 562, 272–276. [Google Scholar] [CrossRef]
- Lane, N. Life ascending: the ten great inventions of evolution; WW Norton: New York, NY, 2010. [Google Scholar]
- Llinás, R.R. I of the vortex: From neurons to self; A Bradford Book, Bradford Books: Cambridge, MA, 2002. [Google Scholar]
- Jablonka, E.; Lamb, M.J. The evolution of information in the major transitions. Journal of theoretical biology 2006, 239, 236–246. [Google Scholar] [CrossRef]
- Gunawardena, J. Learning Outside the Brain: Integrating Cognitive Science and Systems Biology. Proceedings of the IEEE 2022, 110, 590–612. [Google Scholar] [CrossRef]
- Gumuskaya, G.; Srivastava, P.; Cooper, B.G.; Lesser, H.; Semegran, B.; Garnier, S.; Levin, M. Motile Living Biobots Self-Construct from Adult Human Somatic Progenitor Seed Cells. Advanced Science 2024, 11, 2303575. [Google Scholar] [CrossRef] [PubMed]
- Fields, C.; Levin, M. Competency in Navigating Arbitrary Spaces as an Invariant for Analyzing Cognition in Diverse Embodiments. Entropy 2022, 24, 819. [Google Scholar] [CrossRef] [PubMed]
- Chernet, B.T.; Levin, M. Transmembrane voltage potential is an essential cellular parameter for the detection and control of tumor development in aXenopusmodel. Disease Models & Mechanisms 2013, 6, 595–607. [Google Scholar] [CrossRef]
- Levin, M. Bioelectrical approaches to cancer as a problem of the scaling of the cellular self. Progress in Biophysics and Molecular Biology 2021, 165, 102–113. [Google Scholar] [CrossRef]
- Smith, J.M.; Burian, R.; Kauffman, S.; Alberch, P.; Campbell, J.; Goodwin, B.; Lande, R.; Raup, D.; Wolpert, L. Developmental constraints and evolution: a perspective from the Mountain Lake conference on development and evolution. The Quarterly Review of Biology 1985, 60, 265–287. [Google Scholar] [CrossRef]
- Ferrell, J.E. Bistability, Bifurcations, and Waddington’s Epigenetic Landscape. Current Biology 2012, 22, R458–R466. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Manicka, S.; Hoel, E.; Levin, M. Gene regulatory networks exhibit several kinds of memory: Quantification of memory in biological and random transcriptional networks. iScience 2021, 24, 102131. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Clawson, W.; Levin, M. Learning in Transcriptional Network Models: Computational Discovery of Pathway-Level Memory and Effective Interventions. International Journal of Molecular Sciences 2022, 24, 285. [Google Scholar] [CrossRef]
- Yang, C.Y.; Bialecka-Fornal, M.; Weatherwax, C.; Larkin, J.W.; Prindle, A.; Liu, J.; Garcia-Ojalvo, J.; Süel, G.M. Encoding Membrane-Potential-Based Memory within a Microbial Community. Cell Systems 2020, 10, 417–423. [Google Scholar] [CrossRef]
- Gabalda-Sagarra, M.; Carey, L.B.; Garcia-Ojalvo, J. Recurrence-based information processing in gene regulatory networks. Chaos: An Interdisciplinary Journal of Nonlinear Science 2018, 28, 106313. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Saez, M.S.; Vilarroya, O.; Garcia-Ojalvo, J. Biological computation through recurrence. Biochemical and Biophysical Research Communications 2024, 728, 150301. [Google Scholar] [CrossRef]
- Wilson, E.O. The insect societies; Harvard University Press: London, England, 1971. [Google Scholar]
- Bonabeau, E.; Theraulaz, G.; Deneubourg, J.L.; Aron, S.; Camazine, S. Self-organization in social insects. Trends in ecology & evolution 1997, 12, 188–193. [Google Scholar] [CrossRef]
- Gordon, D.M. Ants at work: how an insect society is organized; Simon and Schuster, 1999.
- Bonabeau, E.; Dorigo, M.; Theraulaz, G. Swarm intelligence: from natural to artificial systems; Oxford university press, 1999; pp. I–XII, 1–307. [CrossRef]
- Prindle, A.; Liu, J.; Asally, M.; Ly, S.; Garcia-Ojalvo, J.; Süel, G.M. Ion channels enable electrical communication in bacterial communities. Nature 2015, 527, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.W.; Zhai, X.; Kikuchi, K.; Redford, S.E.; Prindle, A.; Liu, J.; Greenfield, S.; Walczak, A.M.; Garcia-Ojalvo, J.; Mugler, A.; Süel, G.M. Signal Percolation within a Bacterial Community. Cell Systems 2018, 7, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Corral, R.; Liu, J.; Prindle, A.; Süel, G.M.; Garcia-Ojalvo, J. Metabolic basis of brain-like electrical signalling in bacterial communities. Philosophical Transactions of the Royal Society B 2019, 374, 20180382. [Google Scholar] [CrossRef]
- Libby, E.; Rainey, P.B. A conceptual framework for the evolutionary origins of multicellularity. Physical biology 2013, 10, 035001. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Moriya, S.; Ide, K.; Hayakawa, T.; Akima, H.; Sato, S.; Kubota, S.; Tanii, T.; Niwano, M.; Teller, S.; Soriano, J.; Hirano-Iwata, A. Impact of modular organization on dynamical richness in cortical networks. Science Advances 2018, 4. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Spitzner, F.P.; Takemuro, T.; Buendía, V.; Murota, H.; Morante, C.; Konno, T.; Sato, S.; Hirano-Iwata, A.; Levina, A.; Priesemann, V.; Muñoz, M.A.; Zierenberg, J.; Soriano, J. Modular architecture facilitates noise-driven control of synchrony in neuronal networks. Science Advances 2023, 9. [Google Scholar] [CrossRef]
- Kriegman, S.; Blackiston, D.; Levin, M.; Bongard, J. Kinematic self-replication in reconfigurable organisms. Proceedings of the National Academy of Sciences 2021, 118, e2112672118. [Google Scholar] [CrossRef]
- Gregory, T.R. The evolution of complex organs. Evolution: Education and Outreach 2008, 1, 358–389. [Google Scholar] [CrossRef]
- Theraulaz, G.; Bonabeau, E.; Deneubourg, J.L. The origin of nest complexity in social insects. Complexity 1998, 3, 15–25. [Google Scholar] [CrossRef]
- Bonabeau, E.; Theraulaz, G.; Deneubourg, J.L.; Franks, N.R.; Rafelsberger, O.; Joly, J.L.; Blanco, S. A model for the emergence of pillars, walls and royal chambers in termite nests. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 1998, 353, 1561–1576. [Google Scholar] [CrossRef]
- Turner, J.S. Termites as models of swarm cognition. Swarm intelligence 2011, 5, 19–43. [Google Scholar] [CrossRef]
- Foo, J.L.; Ling, H.; Lee, Y.S.; Chang, M.W. Microbiome engineering: Current applications and its future. Biotechnology journal 2017, 12, 1600099. [Google Scholar] [CrossRef] [PubMed]
- Dou, J.; Bennett, M.R. Synthetic biology and the gut microbiome. Biotechnology journal 2018, 13, 1700159. [Google Scholar] [CrossRef]
- Solé, R.V.; Montañez, R.; Duran-Nebreda, S. Synthetic circuit designs for earth terraformation. Biology Direct 2015, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Conde-Pueyo, N.; Vidiella, B.; Sardanyés, J.; Berdugo, M.; Maestre, F.T.; De Lorenzo, V.; Solé, R. Synthetic biology for terraformation lessons from mars, earth, and the microbiome. life 2020, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Foster, K.R.; Schluter, J.; Coyte, K.Z.; Rakoff-Nahoum, S. The evolution of the host microbiome as an ecosystem on a leash. Nature 2017, 548, 43–51. [Google Scholar] [CrossRef]
- Zhang, F.; Guo, Z.; Li, Z.; Luan, H.; Yu, Y.; Zhu, A.T.; Ding, S.; Gao, W.; Fang, R.H.; Zhang, L.; others. Biohybrid microrobots locally and actively deliver drug-loaded nanoparticles to inhibit the progression of lung metastasis. Science Advances 2024, 10, eadn6157. [Google Scholar] [CrossRef]
- Kunitomo, H.; Sato, H.; Iwata, R.; Satoh, Y.; Ohno, H.; Yamada, K.; Iino, Y. Concentration memory-dependent synaptic plasticity of a taste circuit regulates salt concentration chemotaxis in Caenorhabditis elegans. Nature Communications 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Hiroki, S.; Yoshitane, H.; Mitsui, H.; Sato, H.; Umatani, C.; Kanda, S.; Fukada, Y.; Iino, Y. Molecular encoding and synaptic decoding of context during salt chemotaxis in C. elegans. Nature Communications 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Saez, M.S.; Vilarroya, O.; Garcia-Ojalvo, J. A multiscale sensorimotor model of experience-dependent behavior in a minimal organism. Biophysical Journal 2024, 123, 1654–1667. [Google Scholar] [CrossRef] [PubMed]
- Lyon, P.; Keijzer, F.; Arendt, D.; Levin, M. Reframing cognition: getting down to biological basics. Philosophical Transactions of the Royal Society B: Biological Sciences 2021, 376, 20190750. [Google Scholar] [CrossRef] [PubMed]
- Bongard, J.C. Evolutionary robotics: Taking a biologically inspired approach to the design of autonomous, adaptive machines. Communications of the ACM 2013, 56, 74–83. [Google Scholar] [CrossRef]
- Nolfi, S.; Bongard, J.; Husbands, P.; Floreano, D. , Evolutionary Robotics. In Springer Handbooks; Springer International Publishing, 2016; p. 2035–2068. [CrossRef]
- Seilacher, A. Fossil behavior. Scientific American 1967, 217, 72–83. [Google Scholar] [CrossRef]
- Plotnick, R.E. Behavioral biology of trace fossils. Paleobiology 2012, 38, 459–473. [Google Scholar] [CrossRef]
- Andrianantoandro, E.; Basu, S.; Karig, D.K.; Weiss, R. Synthetic biology: new engineering rules for an emerging discipline. Molecular systems biology 2006, 2, 2006–0028. [Google Scholar] [CrossRef] [PubMed]
- Lopatkin, A.J.; Collins, J.J. Predictive biology: modelling, understanding and harnessing microbial complexity. Nature Reviews Microbiology 2020, 18, 507–520. [Google Scholar] [CrossRef]
- Lazebnik, Y. Can a biologist fix a radio?—Or, what I learned while studying apoptosis. Cancer cell 2002, 2, 179–182. [Google Scholar] [CrossRef]
- Lobo, D.; Solano, M.; Bubenik, G.A.; Levin, M. A linear-encoding model explains the variability of the target morphology in regeneration. Journal of The Royal Society Interface 2014, 11, 20130918. [Google Scholar] [CrossRef] [PubMed]
- Corominas-Murtra, B.; Petridou, N. Viscoelastic Networks: Forming Cells and Tissues. Frontiers in Physics 2021, 9. [Google Scholar] [CrossRef]
- Petridou, N.I.; Corominas-Murtra, B.; Heisenberg, C.P.; Hannezo, E. Rigidity percolation uncovers a structural basis for embryonic tissue phase transitions. Cell 2021, 184, 1914–1928. [Google Scholar] [CrossRef] [PubMed]
- Lenne, P.F.; Trivedi, V. Sculpting tissues by phase transitions. Nature Communications 2022, 13, 664. [Google Scholar] [CrossRef] [PubMed]
| 1 | Plants follow a very different organization plant and developmental trajectories, with no fixed numbers of organs, such as leaves, that are highly redundant parts. |
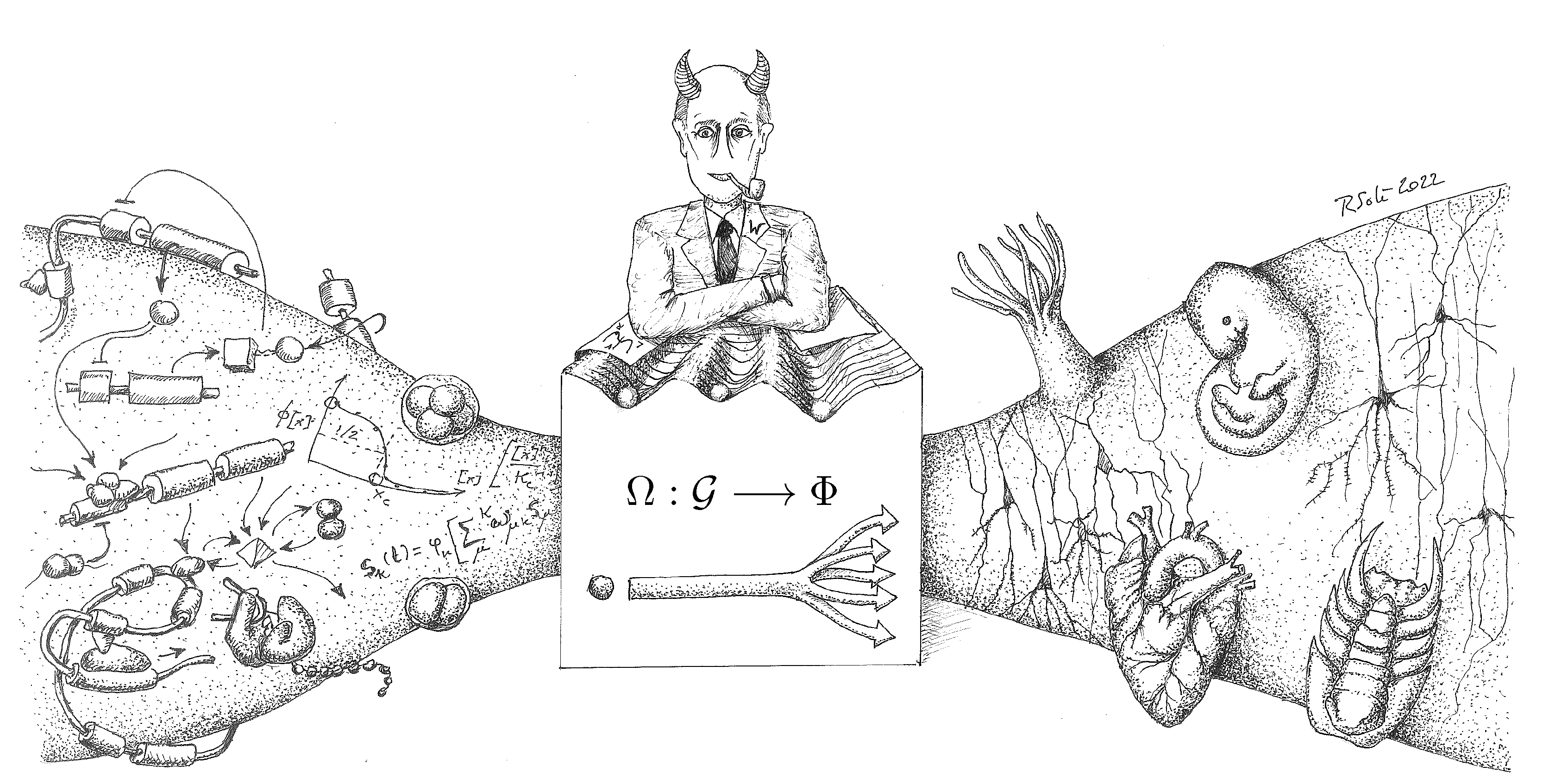
| 2 | This hypothetical creature is inspired by Laplace’s Demon, proposed by Pierre-Simon Laplace, capable of knowing the precise location and momentum of every atom in the universe. With this information, it could predict the past and future of every particle, demonstrating a deterministic universe where the future is entirely predictable given complete knowledge of the present. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




