Submitted:
24 July 2024
Posted:
25 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
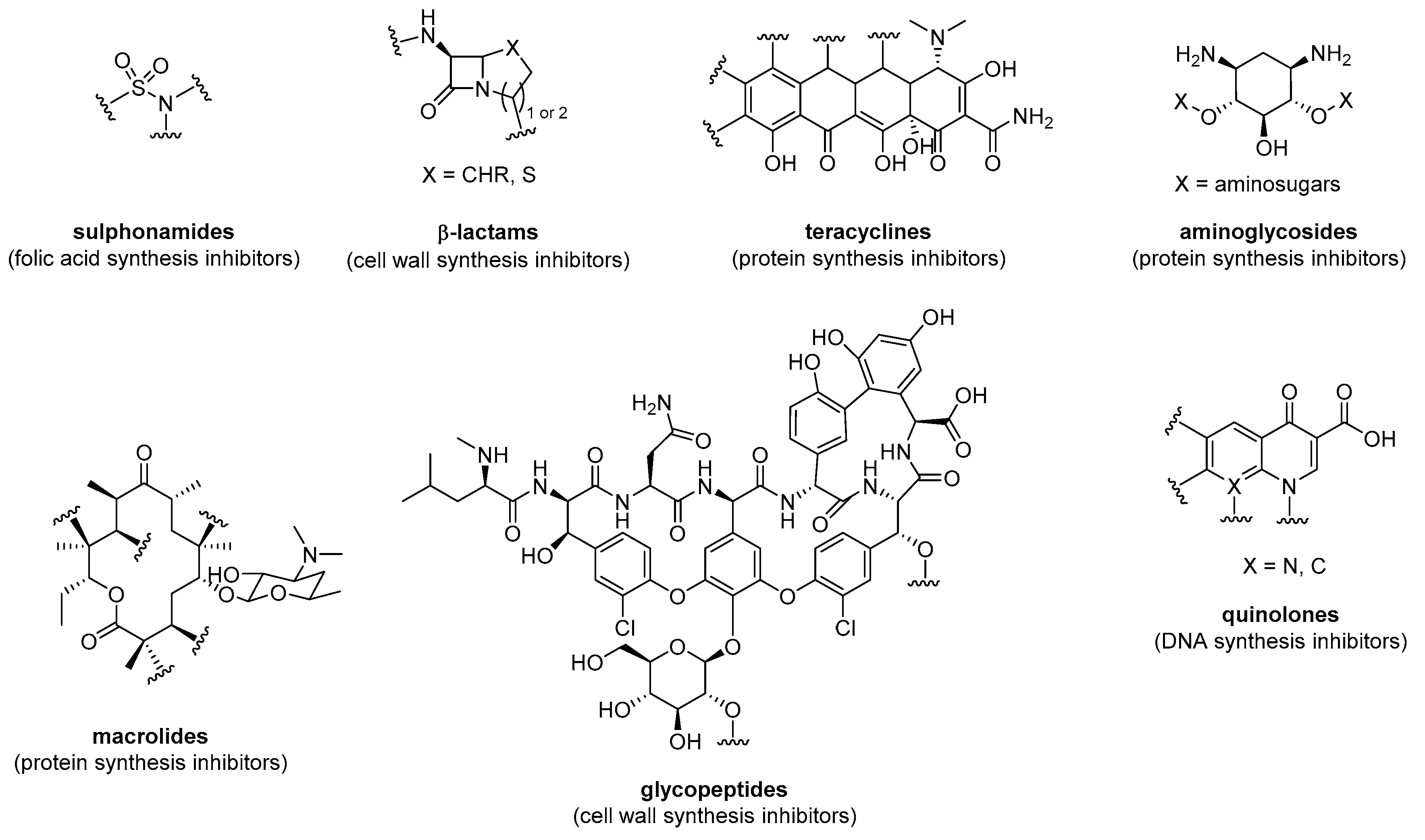
2. Small Molecule Antibiotics
3. Antimicrobial Peptides (AMPs)
4. Antibiotic-AMP Conjugates
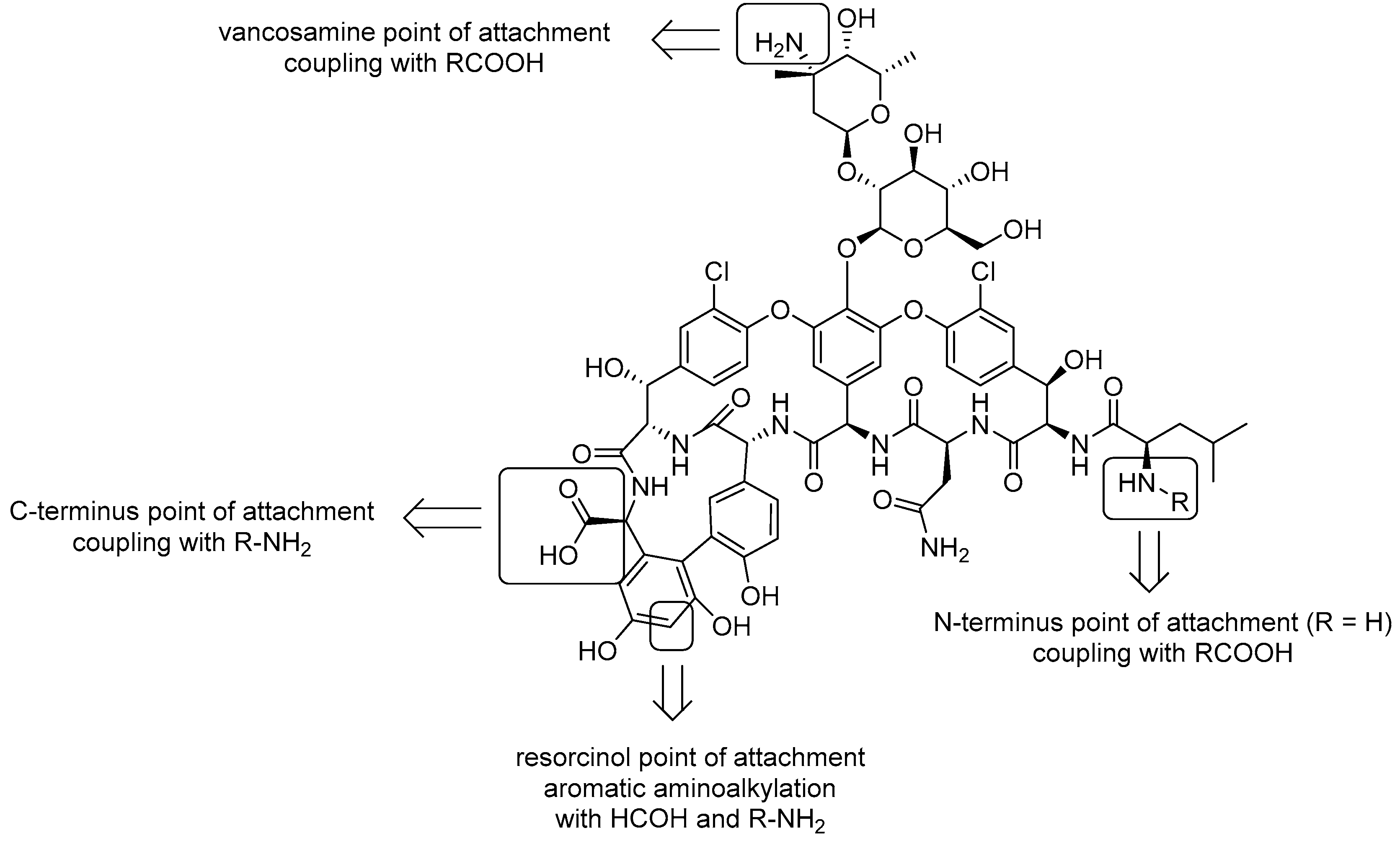
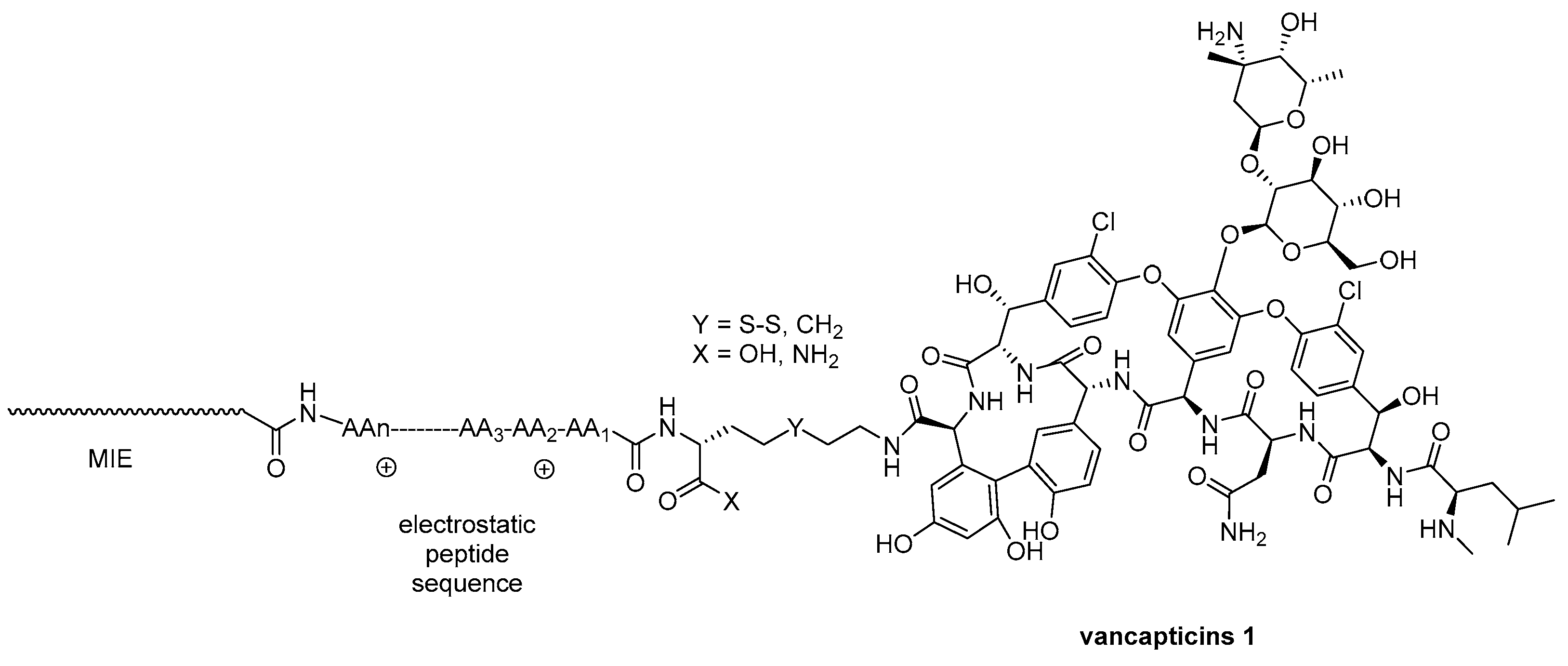
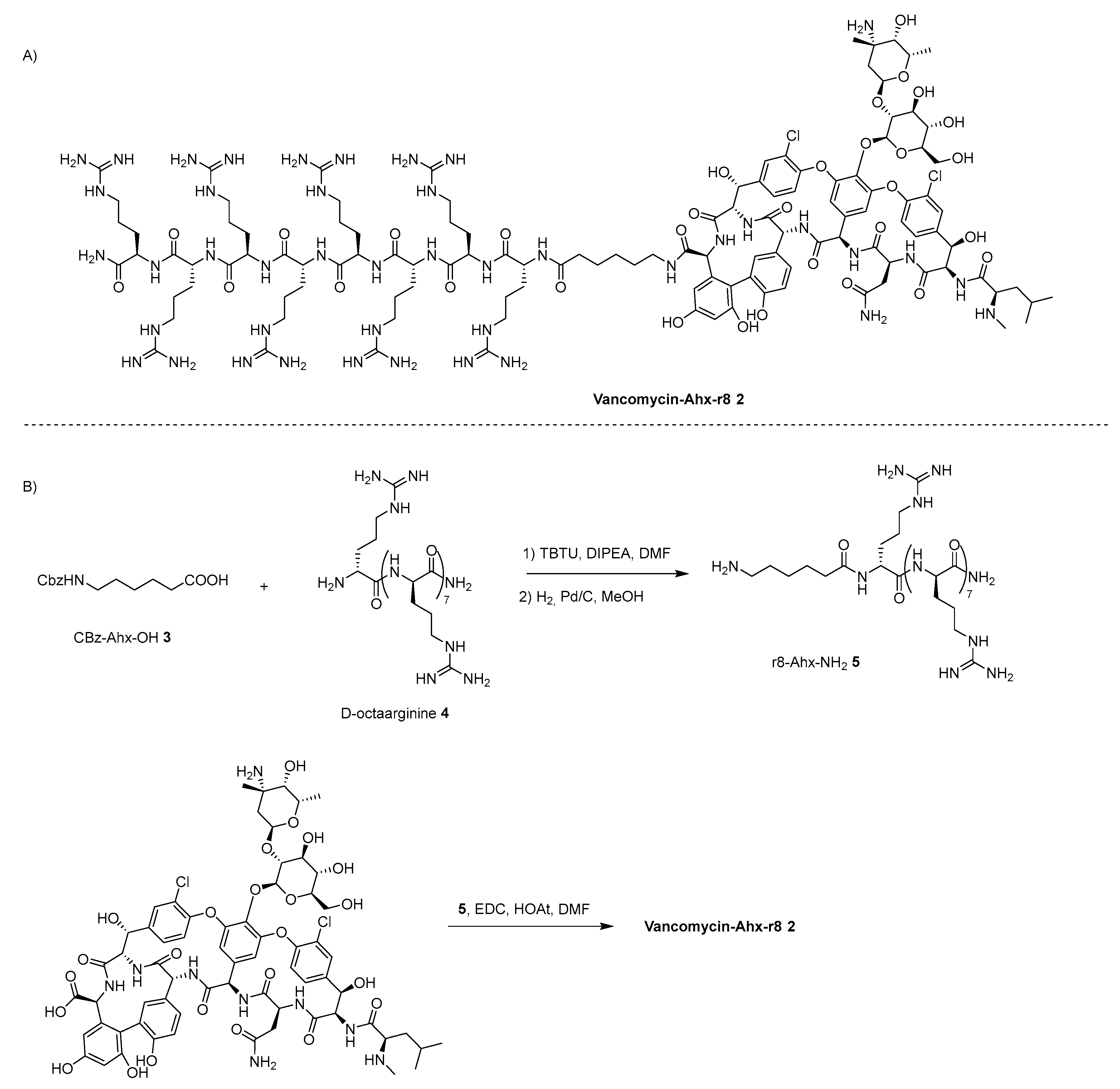
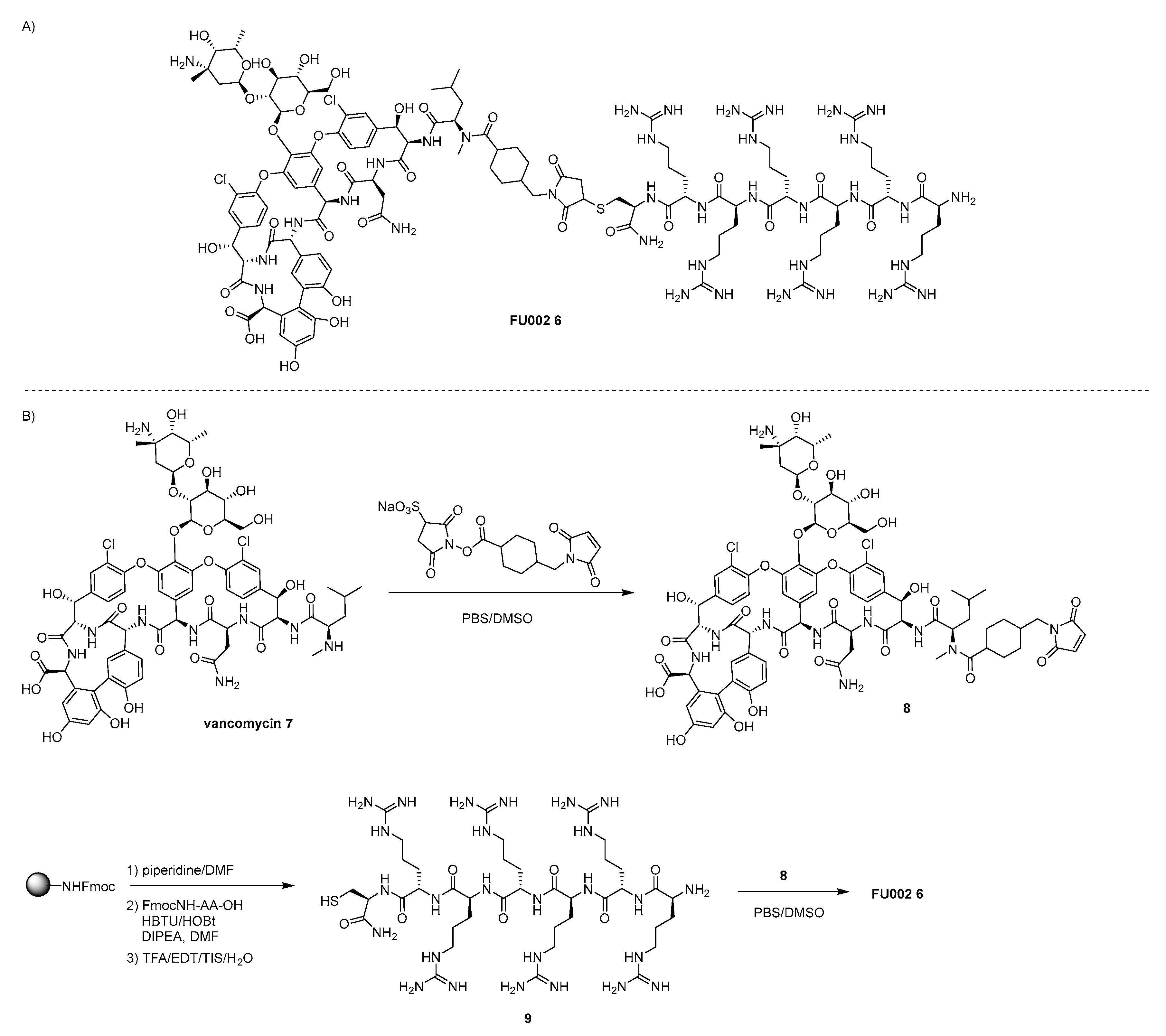
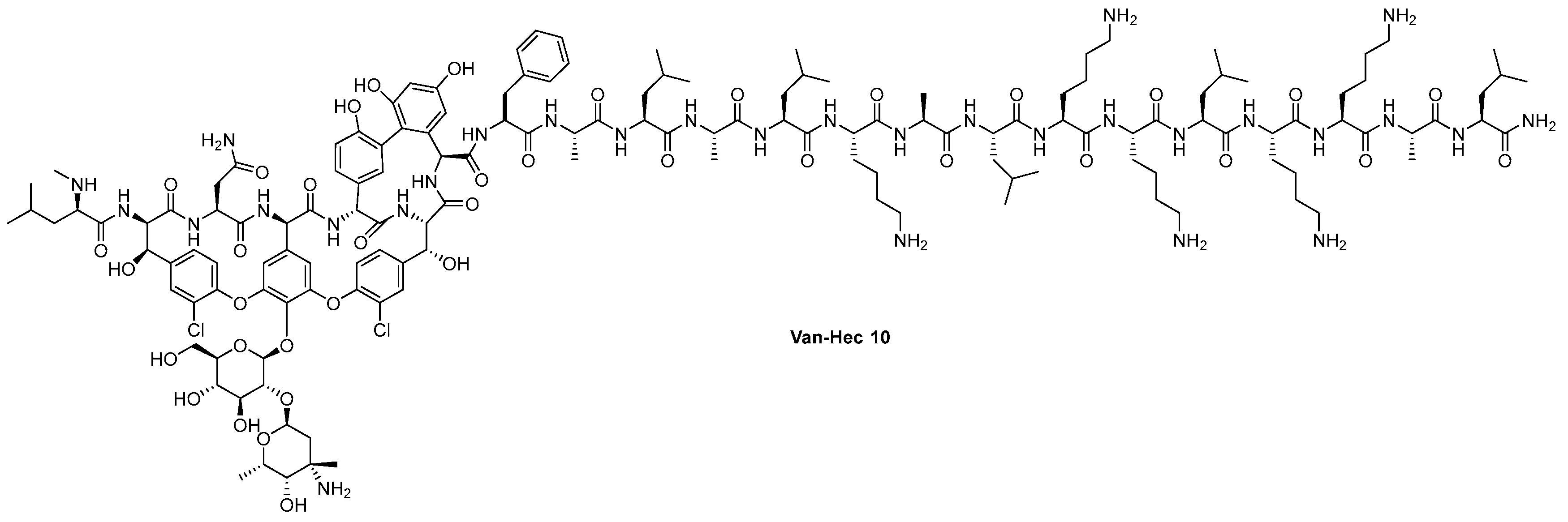
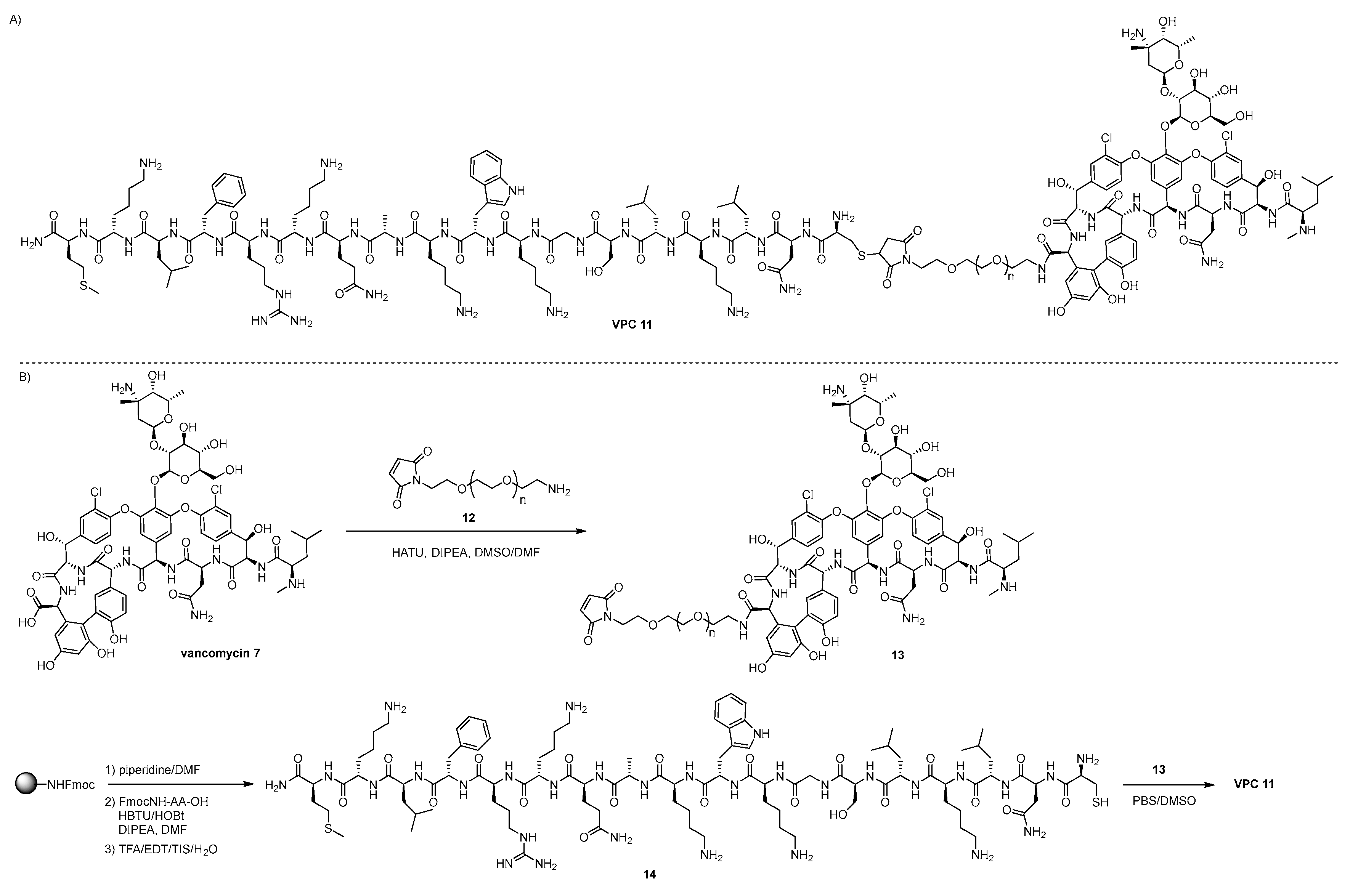
4.1. Vanconomycin-AMP Conjugates
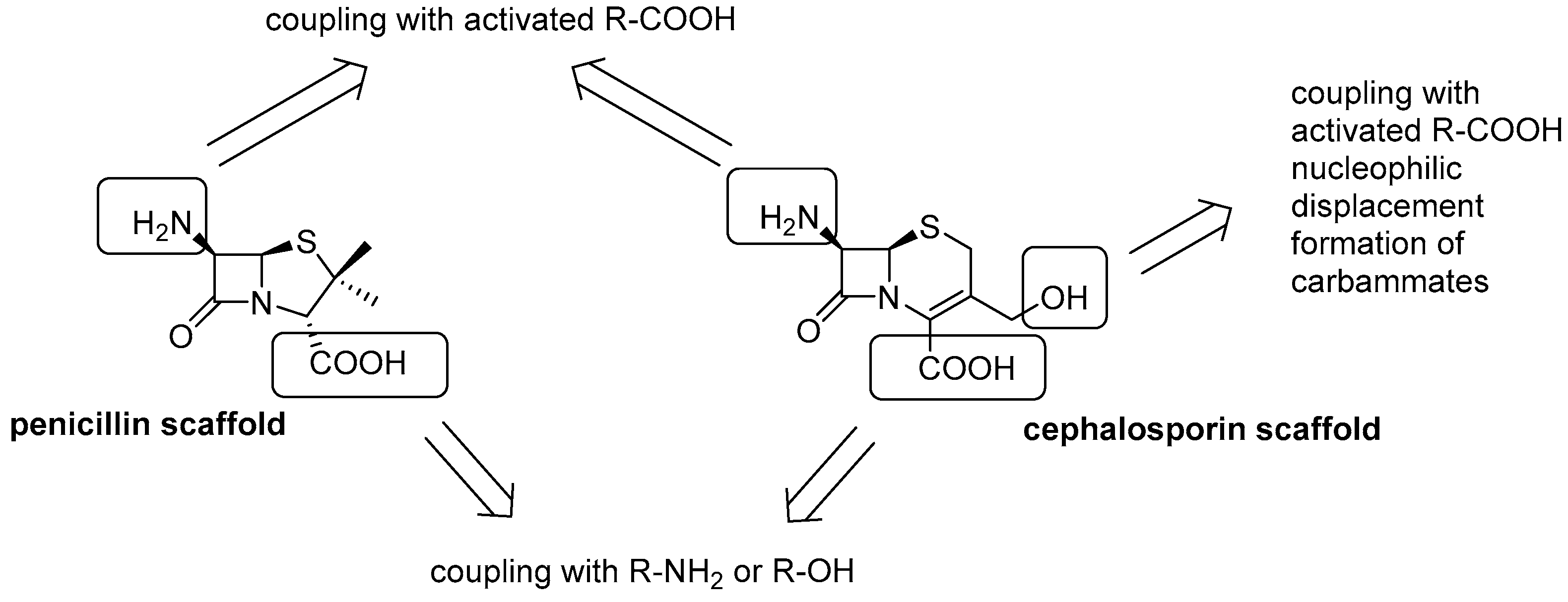
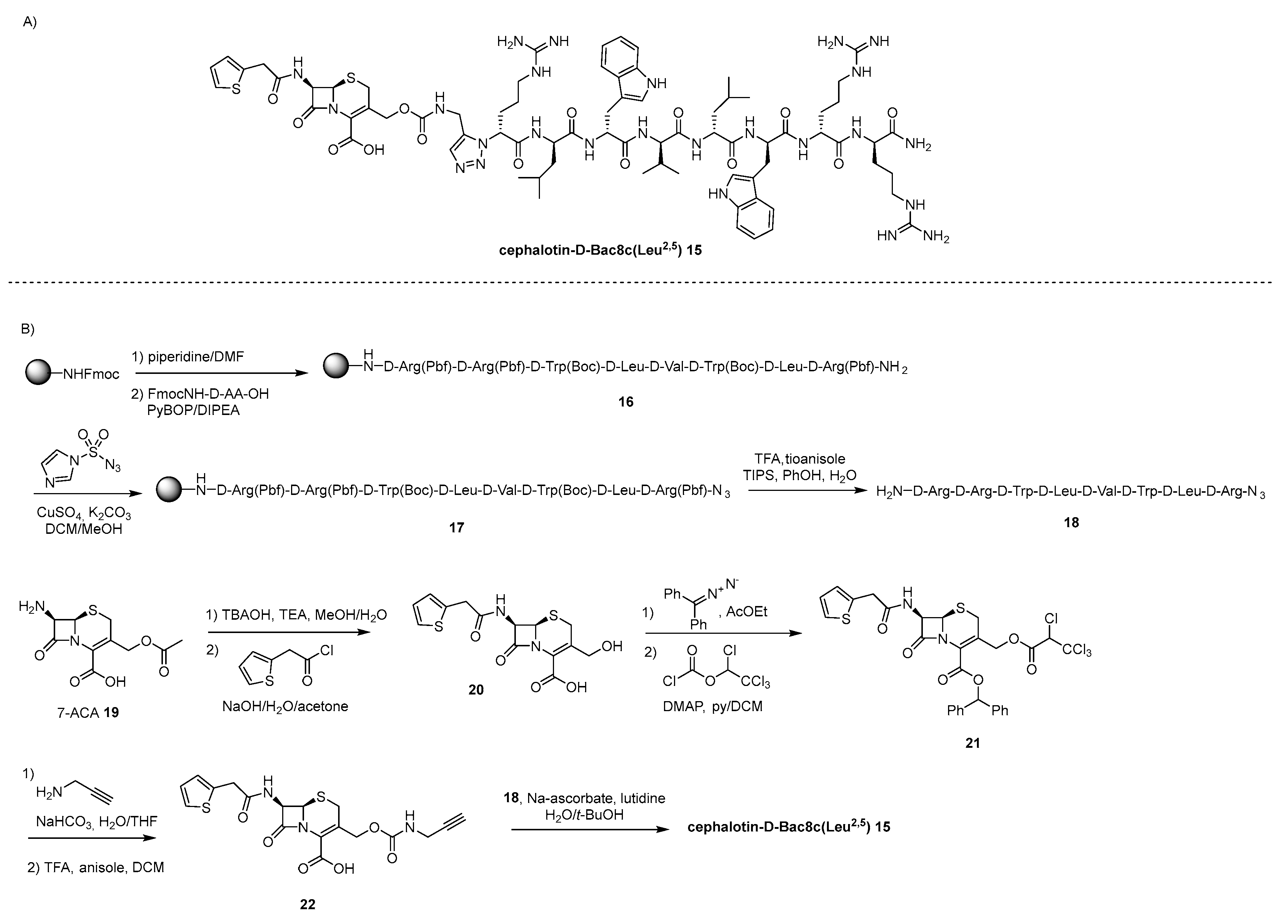
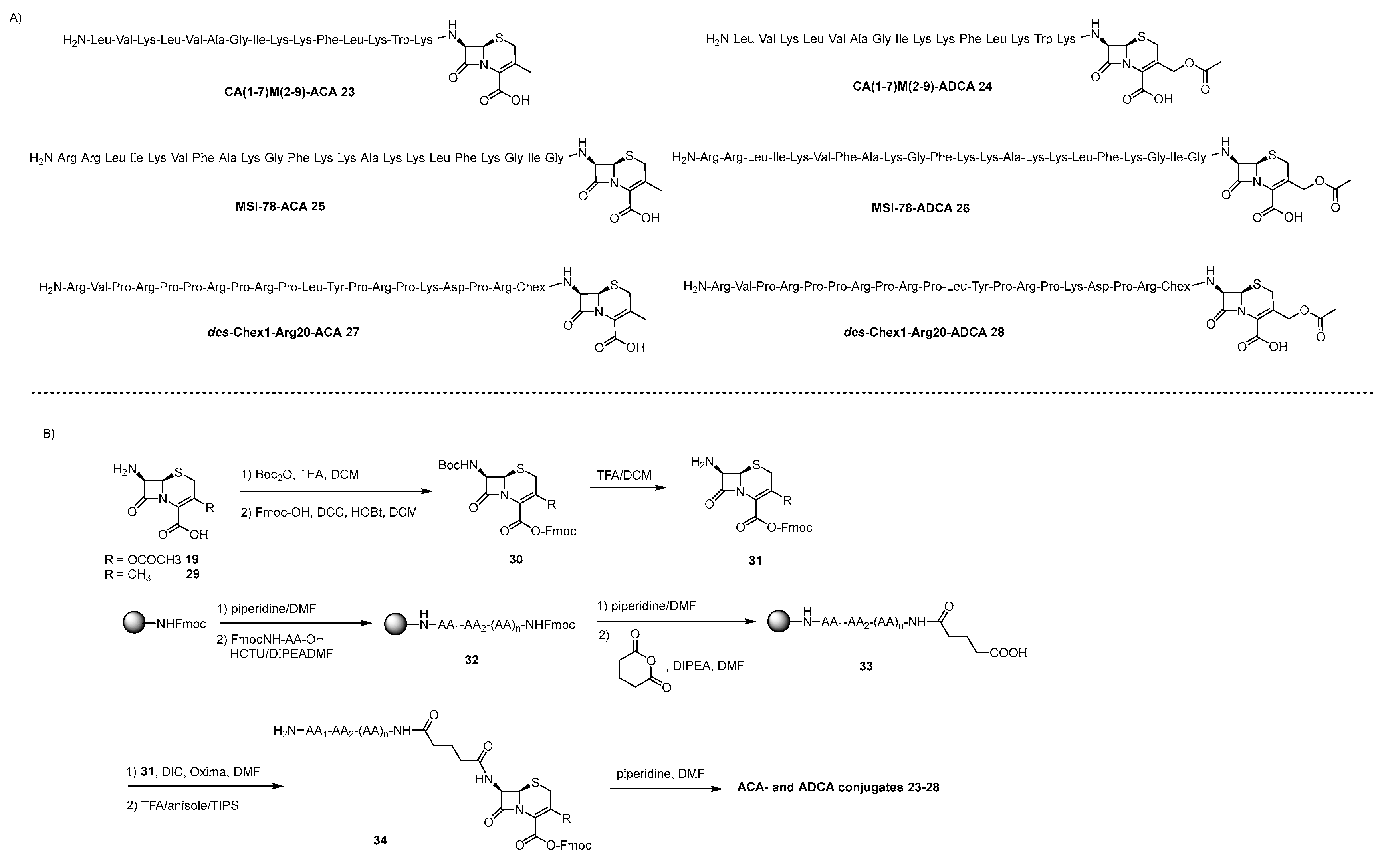
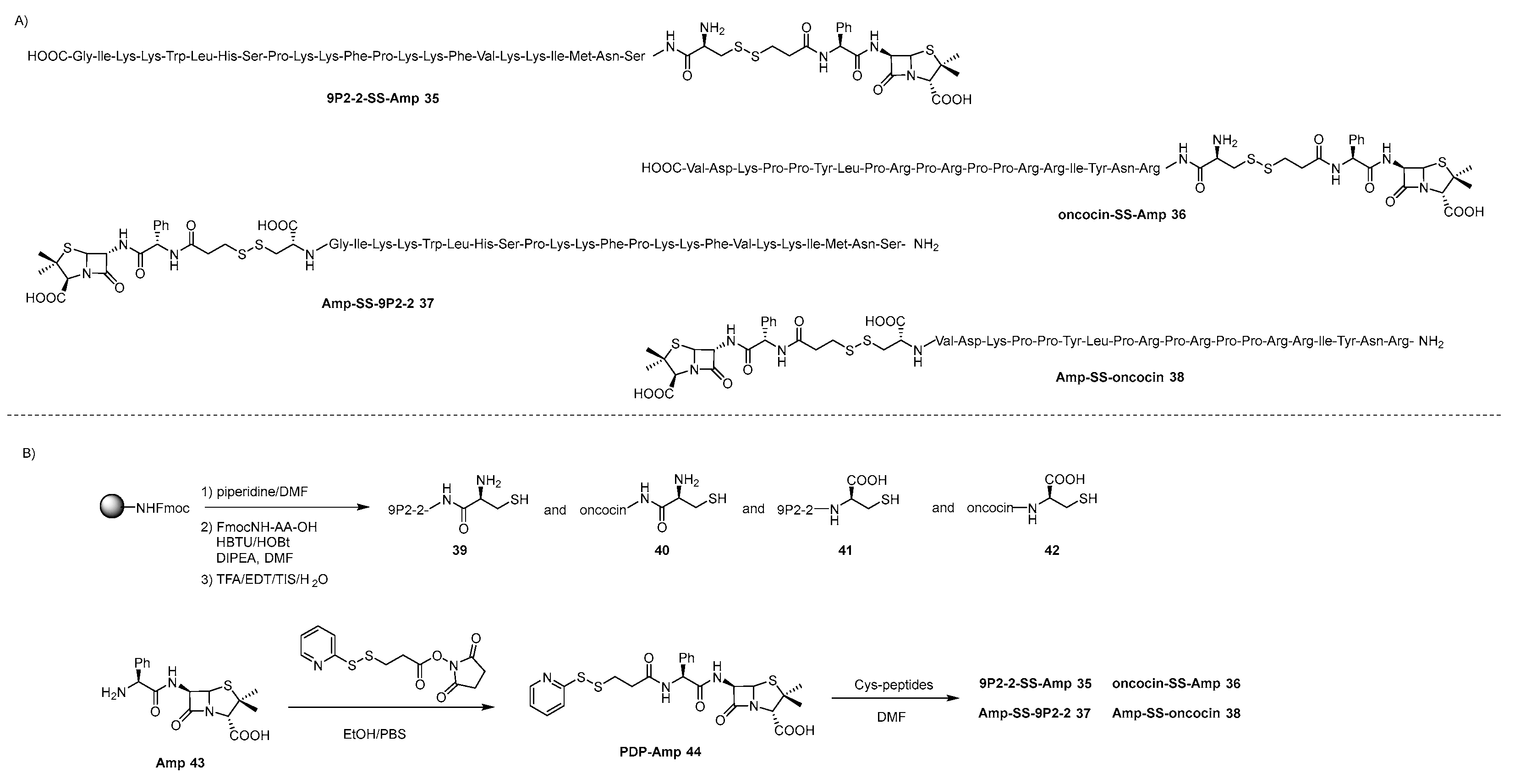
4.2. lactams-AMP Conjugates
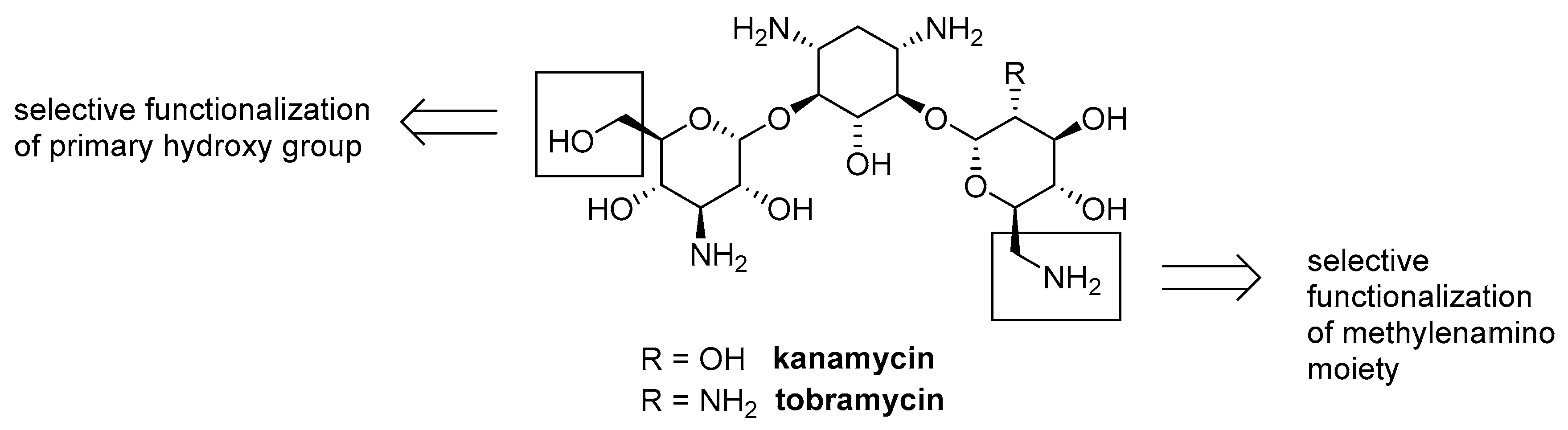
4.3. Aminoglycoside-AMP Conjugates
4.4. Miscellanous
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Theuretzbacher, U.; Gottwalt, S.; Beyer, P.; Butler, M.; Czaplewski, L.; Lienhardt, C.; Moja, L.; Paul, M.; Paulin, S.; Rex, J.H.; Silver, L.L.; Spigelman, M.; Thwaites, G.E.; Paccaud, J.P.; Harbarth, S. Analysis of the clinical antibacterial and antituberculosis pipeline. Lancet Infect Dis. 2019, 19, e40–e50. [Google Scholar] [CrossRef] [PubMed]
- Gould, K. Antibiotics: from prehistory to the present day. J. Antimocrob. Chemother. 2016, 71, 572–575. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.B. Drugs that changed society: history and current status of the early antibiotics: salvarsan, sulfonamides, and b-lactams. Molecules 2021, 26, 6057. [Google Scholar] [CrossRef] [PubMed]
- Baran, A.; Kwiatkowska, A.; Potocki, L. Antibiotics and bacterial resistance – A short story of an endless arms race. Int. J. Mol. Sci. 2023, 24, 5777. [Google Scholar] [CrossRef]
- Halawa, E.M.; Fadel, M.; Al-Rabia, M.W.; Behairy, A.; Nouh, N.A.; Abdo, M.; Olga, R.; Fericean, L.; Atwa, A.M.; El-Nablaway, M.; Abdeen, A. Antibiotic action and resistance: updated review of mechanisms, spread, influencing factors, and alternative approaches for combating resistance. Front. Pharmacol. 2024, 14, 1305294. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Redwan Matin Zidan, B.M.; Mitra, S.; Emra, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; Hossain, M.J.; Koirala, N. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 12, 1750–1766. [Google Scholar] [CrossRef] [PubMed]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular mechanisms of antibiotic resistance revisited. Nat Rev Microbiol 2023, 21, 280–295. [Google Scholar] [CrossRef] [PubMed]
- Kaye, K.S.; Pogue, J.M. Infections caused by resistant gram-negative bacteria: epidemiology and management. Pharmacotherapy 2015, 35, 949–962. [Google Scholar] [CrossRef]
- Cascioferro, S.; Parrino, B.; Carbone, D.; Pecoraro, C.; Diana, P. Novel strategies in the war against antibiotic resistance. Future Med. Chem. 2021, 13, 529–531. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; Ouellette, M.; Outterson, K.; Patel, J.; Cavaleri, M.; Cox, E.M.; Houchens, C.R.; Grayson, M.L.; Hansen, P.; Singh, N.; Theuretzbacher, U.; Magrini, N.; Aboderin, A.O.; Al-Abri, S.S.; Awang Jalil, N.; Benzonana, N.; Bhattacharya, S.; Brink, A.J.; Burkert, F.R.; Cars, O.; Cornaglia, G.; Dyar, O.J.; Friedrich, A.W.; Gales, A.C.; Gandra, S.; Giske, C.G.; Goff, D.A.; Goossens, H.; Gottlieb, T.; Guzman Blanco, M.; Hryniewicz, W.; Kattula, D.; Jinks, T.; Kanj, S.S.; Kerr, L.; Kieny, M.P.; Kim, Y.S.; Kozlov, R.S.; Labarca, J.; Laxminarayan, R.; Leder, K.; Leibovici, L.; Levy-Hara, G.; Littman, J.; Malhotra-Kumar, S.; Manchanda, V.; Moja, L.; Ndoye, B.; Pan, A.; Paterson, D.L.; Paul, M.; Qiu, H.; Ramon-Pardo, P.; Rodríguez-Baño, J.; Sanguinetti, M.; Sengupta, S.; Sharland, M.; Si-Mehand, M.; Silver, L.L.; Song, W.; Steinbakk, M.; Thomsen, J.; Thwaites, G.E.; van der Meer, J.W.; Van Kinh, N.; Vega, S.; Villegas, M.V.; Wechsler-Fördös, A.; Wertheim, H.F.L.; Wesangula, E.; Woodford, N.; Yilmaz, F.O.; Zorzet, A. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.J.; Gwynn, M.N.; Holmes, D. J.; Pompliano, D.L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discovery 2007, 6, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Tommasi, R.; Brown, D.G.; Walkup, G.K.; Manchester, J.I.; Miller, A.A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discovery 2015, 14, 529–42. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, R.; Moser, H.E. Physicochemical Properties of Antibacterial Compounds: Implications for Drug Discovery. J. Med. Chem. 2008, 51, 2871–2878. [Google Scholar] [CrossRef] [PubMed]
- Olsufyeva, E.N.; Yankovskaya, V.S. Main trends in the design of semi-synthetic antibiotics of a new generation. Russ. Chem. Rev. 2020, 89, 339–387. [Google Scholar] [CrossRef]
- Wright, G.D. Opportunities for natural products in 21st-century antibiotic discovery. Nat. Prod. Rep. 2017, 34, 694–701. [Google Scholar] [CrossRef]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007, 6, 29–40. [Google Scholar] [CrossRef]
- Renwick, M.J.; Brogan, D.M.; Mossialos, E. A systematic review and critical assessment of incentive strategies for discovery and development of novel antibiotics. J. Antibiot. 2016, 69, 73–88. [Google Scholar] [CrossRef]
- Silver, L.L. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 2011, 24, 71–109. [Google Scholar] [CrossRef]
- Outterson, K.; Powers, J.H.; Daniel, G.W.; McClellan, M.B. Repairing the broken market for antibiotic innovation. Health Aff. 2015, 34, 277–285. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: part 1: causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Shi, Z.; Zhang, J.; Tia, L.; Xin, L.; Liang, C.; Ren, X.; Li, M. A comprehensive overview of the antibiotics approved in the last two decades: retrospects and prospects. Molecules 2023, 28, 1762. [Google Scholar] [CrossRef] [PubMed]
- Coates, A.R.M.; Hu, Y.; Holt, J.; Yeh, P. Antibiotic combination therapy against resistant bacterial infections: synergy, rejuvenation, and resistance reduction. Expert Rev. Anti-Infect. Ther. 2020, 18, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, G.J.; Delgado, N.N.; Maharjan, R.; Cain, A.K. How antibiotics work together: molecular mechanisms behind combination therapy. Curr. Opin. Microbiol. 2020, 57, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.; Greenstein, T.; Colindres, R.V.; Aldridge, B.B. Leveraging laboratory and clinical studies to design effective antibiotic combination therapy. Curr. Opin. Microbiol. 2021, 64, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Alkhzem, A.H.; Woodman, T.J.; Blagbrought, S.I. Design and synthesis of hybrid compounds as novel drugs and medicines. RSC Adv. 2022, 12, 19470–19484. [Google Scholar] [CrossRef] [PubMed]
- Cavaco, M.; Castanho, M.A.R.B.; Neves, V. The Use of Antibody-Antibiotic Conjugates to Fight Bacterial Infections. Front. Microbiol. 2022, 13, 835677. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.D.; Wright, G.D. Antibacterial drug discovery in the resistance era. Nature 2016, 17, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Raileanu, M.; Borlan, R.; Campu, A.; Janosi, L.; Turcu, I.; Focsan, M.; Bacalum, M. No country for old antibiotics! Antimicrobial peptides (AMP) as next-generation treatment for skin and soft tissue infection. Int. J. Pharm. 2023, 642, 123169. [Google Scholar] [CrossRef]
- Strempel, N.; Strehmel, J.; Overhage, J. Potential application of antimicrobial peptides in the treatment of bacterial biofilm infections. Curr. Pharm. Des. 2015, 21, 67–84. [Google Scholar] [CrossRef]
- David, A.A.; Park, S.E.; Parag, K.; Tiwari, R.K. Antibiotics-peptide conjugates: against multidrug-resistant bacterial pathogens. Curr. Top. Med. Chem. 2018, 18, 1926–1936. [Google Scholar] [CrossRef] [PubMed]
- Le, C.-F.; Fang, C.-M.; Sekaran, S.D. Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother. 2017, 61, e02340–16. [Google Scholar] [CrossRef]
- Hadjicharalambous, A.; Bournakas, N.; Newman, H.; Skynner, M.J.; Beswick, P. Antimicrobial and cell-penetrating peptides: understanding penetration for the design of novel conjugate antibiotics. Antibiotics 2022, 11, 1636. [Google Scholar] [CrossRef] [PubMed]
- Gould, K. Antibiotics: from prehistory to the present day. J. Antimicrob. Chemother. 2016, 71, 572–575. [Google Scholar] [CrossRef] [PubMed]
- Gelpi, A.; Glberston, A.; Tucker, J.D. Magic bullet: Paul Ehrlich, Salvarsan and the birth of venereology. Sex. Transm. Infect. 2015, 91, 68–69. [Google Scholar] [CrossRef] [PubMed]
- Hare, R. New light on the history of penicillin. Med. Hist. 1982, 26, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T.; Wencewicz, T.A. Prospects for new antibiotics: a molecule-centered perspective. J. Antibiot. 2014, 67, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Zessel, K.; Mohring, S.; Hamscher, G.; Kietzmann, M.; Stahl, J. Biocompatibility and antibacterial activity of photolytic products of sulfonamides. Chemosphere 2014, 100, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Azevedo-Barbosa, H.; Dias, D.F.; Franco, L.L.; Hawkes, J.A.; Carvalho, D.T. From Antibacterial to Antitumour Agents: A Brief Review on The Chemical and Medicinal Aspects of Sulfonamides. Mini-Rev. Med. Chem. 2020, 20, 2052–2066. [Google Scholar] [CrossRef]
- Lima, L.M.; Silva, B.N.M.; Barbosa, G.; Barreiro, E.J. β-Lactam antibiotics: An overview from a medicinal chemistry perspective. Eur. J. Med. Chem. 2020, 208, 112829. [Google Scholar] [CrossRef]
- Yocum, R.R.; Rasmussen, J.R.; Strominger, J.L. The mechanism of action of penicillin. Penicillin acylates the active site of Bacillus stearothermophilus D-alanine carboxypeptidase. J. Biol. Chem. 1980, 255, 3977e3986. [Google Scholar]
- Waxman, D.J.; Strominger, J.L. Sequence of active site peptides from the penicillin-sensitive D-alanine carboxypeptidase of Bacillus subtilis. Mechanism of penicillin action and sequence homology to beta-lactamases. J. Biol. Chem. 1980, 255, 3964e3976. [Google Scholar]
- Amyes, S.G.B. Resistance to β-Lactams - The Permutations. Journal of Chemotherapy 2003, 15, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M. Can β-lactams be re-engineered to beat MRSA? CMI 2006, 12, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liao, Y.X.; Ding, T.; Ahn, J. Role of β-Lactamase Inhibitors as Potentiators in Antimicrobial Chemotherapy Targeting Gram-Negative Bacteria. Antibiotics 2023, 13, 260. [Google Scholar] [CrossRef] [PubMed]
- Chukwudi, C.U. rRNA binding sites and the molecular mechanism of action of the tetracyclines. Antimicrob. Agents Chemother. 2016, 60, 4433–4441. [Google Scholar] [CrossRef] [PubMed]
- LaPlante, K.L.; Dhand, A.; Wright, K.; Lauterio, M. Re-establishing the utility of tetracycline-class antibiotics for current challenges with antibiotic resistance. Annals of Medicine 2022, 54, 1686–1700. [Google Scholar] [CrossRef]
- Davies, J.; Wright, G.D. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 1997, 5, 234. [Google Scholar] [CrossRef] [PubMed]
- Houghton, J.L.; Green, K.D.; Chen, W.; Garneau-Tsodikova, S. The future of aminoglycosides: the end or renaissance? ChemBioChem 2010, 11, 880–902. [Google Scholar] [CrossRef]
- McCoy, L.S.; Xie, Y.; Tor, Y. Antibiotics that target protein synthesis. Wiley Interdiscip. Rev.: RNA. 2011, 2, 209–232. [Google Scholar] [CrossRef]
- Franҫois, B.; Russell, R.J.; Murray, J.B.; Aboul-ela, F.; Masquida, B.; Vicens, Q.; Westhof, E. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res. 2005, 33, 5677–5690. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.B.; Terry, D.S.; Altman, R.B.; Blanchard, S.C. Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nat. Chem. Biol. 2010, 6, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Garneau-Tsodikova, S.; Labby, K.J. Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. Med. Chem. Comm. 2016, 7, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Bera, S.; Mondal, D.; Palit, S.; Schweizer, F. Structural modifications of the neomycin class of aminoglycosides. Med. Chem. Comm. 2016, 7, 1499–1534. [Google Scholar] [CrossRef]
- Chandrika, N.T.; Garneau-Tsodikova, S. Comprehensive review of chemical strategies for the preparation of new aminoglycosides and their biological activities. Chem. Soc. Rev. 2018, 47, 1189–1249. [Google Scholar] [CrossRef]
- Fosso, M.Y.; Li, Y.; Garneau-Tsodikova, S. New trends in the use of aminoglycosides. Med. Chem. Commun. 2014, 5, 1075–1091. [Google Scholar] [CrossRef] [PubMed]
- Bellucci, M.C.; Volonterio, A. Aminoglycosides: From Antibiotics to Building Blocks for the Synthesis and Development of Gene Delivery Vehicles. Antibiotics 2020, 9, 504. [Google Scholar] [CrossRef]
- Brockmann, H.; Hekel, W. Pikromycin, ein bitter schmeckendes antibioticum aus actinomyeceten. Chem Ber 1951, 84, 284–288. [Google Scholar] [CrossRef]
- Retsema, J.; Fu, W. Macrolides: Structures and microbial targets. Int. J. Antimicrob. Agents 2001, 18, S3–S10. [Google Scholar] [CrossRef]
- Jednačak, T.; Mikulandra, I.; Novak, P. Advanced Methods for Studying Structure and Interactions of Macrolide Antibiotics. Int. J. Mol. Sci. 2020, 21, 7799. [Google Scholar] [CrossRef]
- Dinos, P.G. The macrolide antibiotic renaissance. British Journal of Pharmacology 2017, 174, 2967–2983. [Google Scholar] [CrossRef] [PubMed]
- Undheim, K.; Scaffold Modifications in Erythromycin Macrolide Antibiotics. A Chemical Minireview. Molecules 2020, 25, 3941. [Google Scholar] [CrossRef] [PubMed]
- Myers, A.G.; Clark, R.B. Discovery of Macrolide Antibiotics Effective against Multi-Drug Resistant Gram-Negative Pathogens. Acc. Chem. Res. 2021, 54, 1635–1645. [Google Scholar] [CrossRef] [PubMed]
- Blaskovich, M.A.T.; Hansford, K.A.; Butler, M.S.; Jia, Z.G.; Mark, A.E.; Cooper, M.A. Developments in Glycopeptide Antibiotics. ACS Infect. Dis. 2018, 4, 715–735. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, K.C.; Boddy, C.N.; Brase, S.; Winssinger, N. Chemistry, Biology, and Medicine of the Glycopeptide Antibiotics. Angew. Chem., Int. Ed. 1999, 38, 2096–2152. [Google Scholar] [CrossRef]
- Kahne, D.; Leimkuhler, C.; Lu, W.; Walsh, C. Glycopeptide and lipoglycopeptide antibiotics. Chem. Rev. 2005, 105, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Acharya, Y.; Dhanda, G.; Sarkara, P.; Haldar, J. Pursuit of next-generation glycopeptides: a journey with vancomycin. Chem. Commun. 2022, 58, 1881–1897. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.S.; Hansford, K.A.; Blaskovich, M.A.T.; Halai, R.; Cooper, M.A. Glycopeptide antibiotics: Back to the future. J. Antibiot. 2014, 67, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Lesher, G.Y.; Froelich, E.J.; Gruett, M.D.; Bailey, J.H.; Brundage, R.P. 1,8-Naphthyridine Derivatives: A New Class of Chemotherapeutic Agents. Journal of medicinal and pharmaceutical chemistry 1962, 91, 1063–1065. [Google Scholar] [CrossRef] [PubMed]
- Emmerson, A.M.; Jones, A.M. The quinolones: decades of development and use. J Antimicrob. Chemother. 2003, 51, 13–20. [Google Scholar] [CrossRef]
- Sharma, V.; Das, R.; Metha, D.K.; Sharma, D.; Aman, S.; Khan, M.U. Quinolone scaffolds as potential drug candidates against infectious microbes: a review. Mol. Divers. 2024. [Google Scholar] [CrossRef] [PubMed]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of quinolone action and resistance. Biochem. 2014, 53, 1565–1574. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Anuradha, *!!! REPLACE !!!*; Chandra, A.; Tanwar, T.; Sahu, S.K.; Mittal, A. Emerging quinoline- and quinolone-based antibiotics in the light of epidemics. Chem Biol Drug Des. 2022, 100, 765–785. [Google Scholar] [CrossRef] [PubMed]
- Bulet, P.; Stöcklin, R.; Menin, L. Anti-microbial peptides: from invertebrates to vertebrates. Immunol. Rev 2004, 198, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Felício, M.R.; Silva, O.N.; Gonçalves, S.; Santos, N.C.; Franco, O.L. Peptides with dual antimicrobial and anticancer activities. Front. Chem 2017, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Ghimire, J.; Hart, R.J.; Soldano, A.; Chen, C.H.; Guha, S.; Hoffmann, J.P.; Wimley, W.C. Optimization of host cell-compatible, antimicrobial peptides effective against biofilms and clinical isolates of drug-resistant bacteria. ACS Infect. Dis. 2003, 9, 952–965. [Google Scholar] [CrossRef] [PubMed]
- Erdem Büyükkiraz, M.; Kesmen, Z. Antimicrobial peptides (AMPs): A promising class of antimicrobial compounds. J. App. Microbiol. 2022, 132(3), 1573–1596. [Google Scholar] [CrossRef]
- Chen, C.H.; Lu, T.K. Development and challenges of antimicrobial peptides for therapeutic applications. Antibiotics 2020, 9, 24. [Google Scholar] [CrossRef]
- Gan, B.H.; Gaynord, J.; Rowe, S.M.; Deingruber, T.; Spring, D.R. The multifaceted nature of antimicrobial peptides: Current synthetic chemistry approaches and future directions. Chem. Soc. Rev. 2021, 50, 7820–7880. [Google Scholar] [CrossRef] [PubMed]
- Madani, F.; Lindberg, S.; Langel, U.; Futaki, S.; Graslund, A. Mechanisms of cellular uptake of cell-penetrating peptides. Biophys. J. 2011, 414, 729. [Google Scholar] [CrossRef] [PubMed]
- Jenssen, H.; Hamill, P.; Hancock, R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [PubMed]
- López-Meza, J.E.; Ochoa-Zarzosa, A.; Aguilar, J.A.; Loeza-Lara, P.D. Antimicrobial peptides: diversity and perspectives for their biomedical application. In Biomedical Engineering, Trends, Research and TechnoInc; 2011; pp. 275–304. [Google Scholar]
- Hazam, P.K.; Goyal, R.; Ramakrishnanet, V. Peptide based antimicrobials: design strategies and therapeutic potential. Progress in Biophysics and Molecular Biology 2018, 142, 10–22. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Benfield, A.H.; Henriques, S.T. Mode-of-Action of Antimicrobial Peptides: Membrane Disruption vs. Intracellular Mechanisms. Front. Med. Technol. 2020, 2, 610997. [Google Scholar] [CrossRef]
- Miao, J.; Zhou, X. Interaction between antimicrobial peptides and DNA: Mechanism of action and application in biotechnology. Biotechnol. Adv. 2016, 34, 808–820. [Google Scholar]
- Schenckbecher, K.; Meyer, A.; Ennifar, E. Antibiotics targeting the ribosome. Ribosome Structure and Function 2009, 1–21. [Google Scholar]
- Helmerhorst, E.J.; Troxler, R.F.; Oppenheim, F.G. The human salivary peptide histatin 5 exerts its antifungal activity through the formation of reactive oxygen species. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 14637–14642. [Google Scholar] [CrossRef]
- Regmi, S.; Yoon, S.C.; Yun, H.C.; Young, K.K.; Seung, S.C.; Jin, C.Y. Antimicrobial peptide from Bacillus subtilis CSB138: characterization, killing kinetics, and synergistic potency. Int. Microbiol. 2017, 20, 43–53. [Google Scholar]
- Pletzer, D.; Mansour, S.C.; Hancock, R.E. Synergy between conventional antibiotics and antimicrobial peptides in the treatment of chronic bacterial infections. J. Antibiot. 2018, 71, 87–98. [Google Scholar]
- Zhou, Y.; Peng, Y. Synergistic effect of clinically used antibiotics and peptide antibiotics against Gram-positive and Gram-negative bacteria. Exp. Ther. Med. 2013, 6, 1000–1004. [Google Scholar] [CrossRef] [PubMed]
- Peetla, C.; Stine, A.; Labhasetwar, V. Biophysical interactions with model lipid membranes: applications in drug discovery and drug delivery. Mol. Pharm. 2009, 6, 1264–1276. [Google Scholar] [CrossRef] [PubMed]
- Blaskovich, M.A.T.; Hansford, K.A.; Gong, Y.; Butler, M.S.; Muldoon, C.; Huang, J.X.; Ramu, S.; Silva, A.B.; Cheng, M.; Kavanagh, A.M.; Ziora, Z.; Premraj, R.; Lindahl, F.; Bradford, T.A.; Lee, J.C.; Karoli, T.; Pelingon, R.; Edwards, D.J.; Amado, M.; Elliott, A.G.; Phetsang, W.; Daud, N.H.; Deecke, J.E.; Sidjabat, H.E.; Ramaologa, S.; Zuegg, J.; Betley, J.R.; Beevers, A.P.G.; Smith, R.A.G.; Roberts, J.A.; Paterson, D.L.; Cooper, M.A. Protein-inspired antibiotics active against vancomycin- and daptomycin-resistant bacteria. Nat Commun 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Umstätter, F.; Domhan, C.; Hertlein, T.; Ohlsen, K.; Mühlberg, E.; Kleist, C.; Zimmermann, S.; Beijer, B.; Klika, K.D.; Haberkorn, U.; Mier, W.; Uhl, P. Vancomycin Resistance Is Overcome by Conjugation of Polycationic Peptides. Angew. Chem. Int. Ed. 2020, 59, 8823–8827. [Google Scholar] [CrossRef] [PubMed]
- Rivero-Muller, A.; Vuorenoja, S.; Tuominen, M.; et al. Use of Hecate chorionic gonadotropin beta conjugate in therapy of lutenizing hormone receptor expressing gonadal somatic cell tumors. Mol Cell Endocrinol. 2007, 269, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Jelinkova, P.; Splichal, Z.; Jimenez, A.M.; Haddad, Y.; Mazumdar, A.; Sur, V.P.; Milosavljevic, V.; Kopel, P.; Buchtelova, H.; Guran, R.; Zitka, O.; Richtera, L.; Hegerova, D.; Heger, Z.; Moulick, A.; Adam, V. Novel vancomycin–peptide conjugate as potent antibacterial agent against vancomycin-resistant Staphylococcus aureus. Infection and Drug Resistance 2018, 11, 1807–1817. [Google Scholar] [CrossRef] [PubMed]
- Meredith, T.C.; Aggarwal, P.; Mamat, U.; Lindner, B.; Woodard, R.W. Redefining the requisite lipopolysaccharide structure in Escherichia coli. ACS Chem. Bio. 2006, 1, 33–42. [Google Scholar] [CrossRef]
- Frecer, V.; Ho, B.; Ding, J.L. De. novo design of potent antimicrobial peptides. Antimicrob. Agents Chemother. 2004, 48, 3349–3357. [Google Scholar] [CrossRef]
- Shi, W.; Chen, F.; Zou, X.; Jiao, S.; Wang, S.; Hu, Y.; Lan, L.; Tang, F.; Huang, W. Design, synthesis, and antibacterial evaluation of vancomycin-LPS binding peptide conjugates. Bioorg. Chem. Med. Lett. 2021, 45, 128122. [Google Scholar] [CrossRef]
- Beveridge, E.G.; Martin, A.J. Sodium sulphomethyl derivatives of polymyxins. Br. J. Pharmacol. Chemother. 1967, 29, 125–135. [Google Scholar] [CrossRef]
- Desgranges, S.; Ruddle, C.C.; Burke, L.P.; McFadden, T.M.; O’Brien, J.E.; Fitzgerald-Hughes, D.; Humphreys, H.; Smyth, T.; Devocelle, M. b-Lactam-host defence peptide conjugates as antibiotic prodrug candidates targeting resistant bacteria. RCS Adv. 2012, 2, 2480–2492. [Google Scholar] [CrossRef]
- Li, W.; O’Briean-Simpson, N.M.; Holden, J.A.; Otvos, L.; Reynolds, E.C.; Separovic, F.; Hossian, M.A.; Wade, J.D. Covalent conjugation of cationic antimicrobial peptides with b-lactam antibiotic core. Peptide Science 2018, 110, e24059. [Google Scholar] [CrossRef]
- Yamauchi, R.; Kawano, K.; Yamaoka, Y.; Taniguchi, A.; Yano, Y.; Takasu, K.; Matsusaki, K. Development of antimicrobial peptide-antibiotic conjugates to improve the outer membrane permeability of antibiotics against Gram-negative bacteria. ACS Infect. Dis. 2022, 8, 2339–2347. [Google Scholar] [CrossRef]
- Ritz, D.; Beckwith, J. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 2001, 55, 21–48. [Google Scholar] [CrossRef]
- Azuma, E.; Choda, N.; Odaki, N.; Yano, Y.; Matsuzaki, K. Improvement of therapeutic index by the combination of enhanced peptide cationicity and proline introduction. ACS Infect. Dis. 2020, 6, 2271–2278. [Google Scholar] [CrossRef]
- Knappe, D.; Piantavigna, S.; Hansen, A.; Mechler, A.; Binas, A.; Nolte, O.; Martin, L.L.; Hoffmann, R. Oncocin (VDKPPYLPRPRPPRRIYNR-NH2): a novel antibacterial peptide optimized against gram-negative human pathogens. J. Med. Chem. 2010, 53, 5240–5247. [Google Scholar] [CrossRef]
- Williams, H.C.; Dellavalle, R.P.; Garner, S. Acne vulgaris. Lancet 2012, 379, 361–372. [Google Scholar] [CrossRef]
- Schmidt, N.W.; Deshayes, S.; Hawker, S.; Blacker, A.; Kasko, A.M.; Kim, J.; Wong, G.C.L. Engineering persister-specific antibiotics with synergistic antimicrobial functions. ACS Nano 2014, 8, 8786–8793. [Google Scholar] [CrossRef]
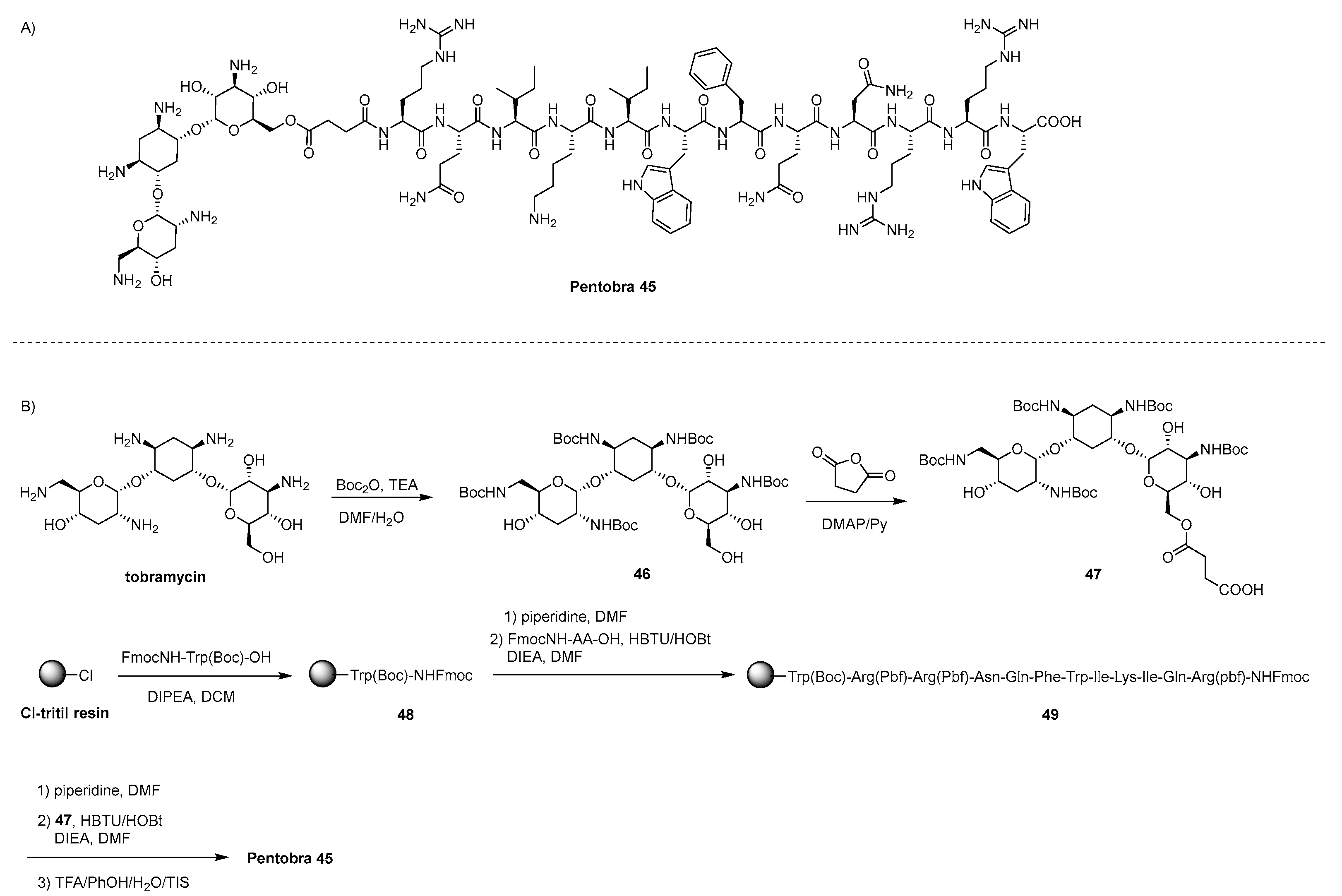
- Schmidt, N.W.; Aga, G.W.; Deshayes, S.; Yu, Y.; Blacker, A.; Champer, J.; Xian, W.; Kasko, A.M.; Kim, J.; Wong, G.C.L. Pentobra: A Potent Antibiotic with Multiple Layers of Selective Antimicrobial Mechanisms against Propionibacterium Acnes. J. Invest. Dermatol. 2015, 135, 1581–1589. [Google Scholar] [CrossRef]
- Schmidt, N.W.; Lis, M.; Zhao, K.; Lai, G.H.; Alexandrova, A.N.; Tew, G.N.; Wong, G.C.L. Molecular basis for nanoscopic membrane curvature generation from quantum mechanical models and synthetic transporter sequences. J. Am. Chem. Soc. 2012, 134, 19207–19216. [Google Scholar] [CrossRef]
- Deshayes, S.; Xian, W.; Schmidt, N.W.; Kordbacheh, S.; Lieng, J.; Wang, J.; Zarmer, S.; St. Germain, S.; Voyen, L.; Thulin, J.; Wong, G.C.L.; Kasko, A.M. Designing hybrid antibiotic peptide conjugates to cross bacterial membranes. Bioconjugate Chem. 2017, 28, 793–804. [Google Scholar] [CrossRef]
- Flannagan, R.S.; Cosío, G.; Grinstein, S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 2009, 7, 355–366. [Google Scholar]
- LaRock, D.L.; Chaudhary, A.; Miller, S.I. Salmonellae interactions with host processes. Nat. Rev. Microbiol. 2015, 13, 191–205. [Google Scholar] [CrossRef]
- Kuriakose, J.; Hernandez-Gordillo, V.; Nepal, M.; Brezden, A.; Pozzi, V.; Seleem, M.N.; Chmielewski, J. Targeting Intracellular Pathogenic Bacteria with Unnatural Proline-Rich Peptides: Coupling Antibacterial Activity with Macrophage Penetration. Angew. Chem., Int. Ed. 2013, 52, 9664–9667. [Google Scholar] [CrossRef]
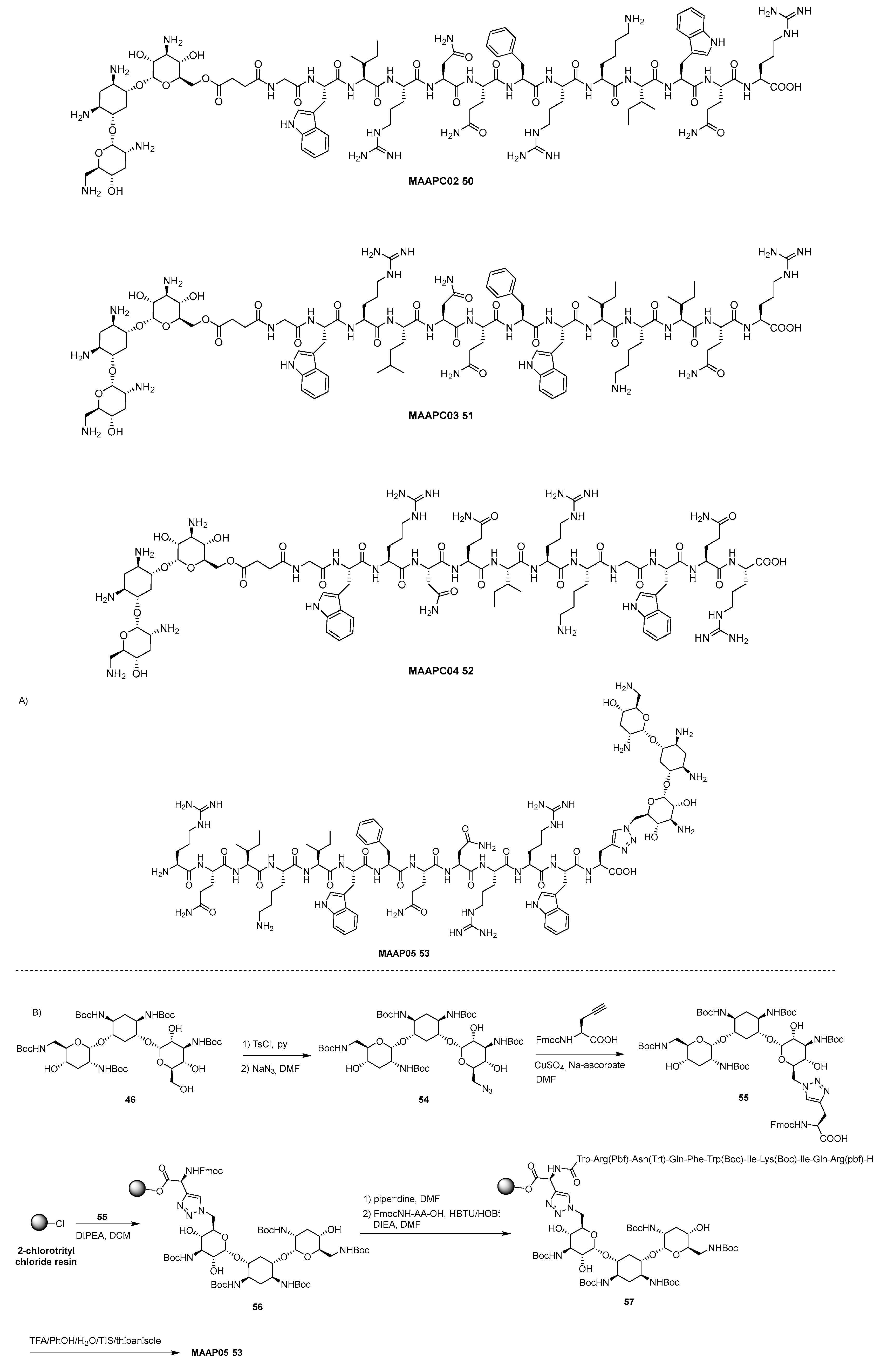
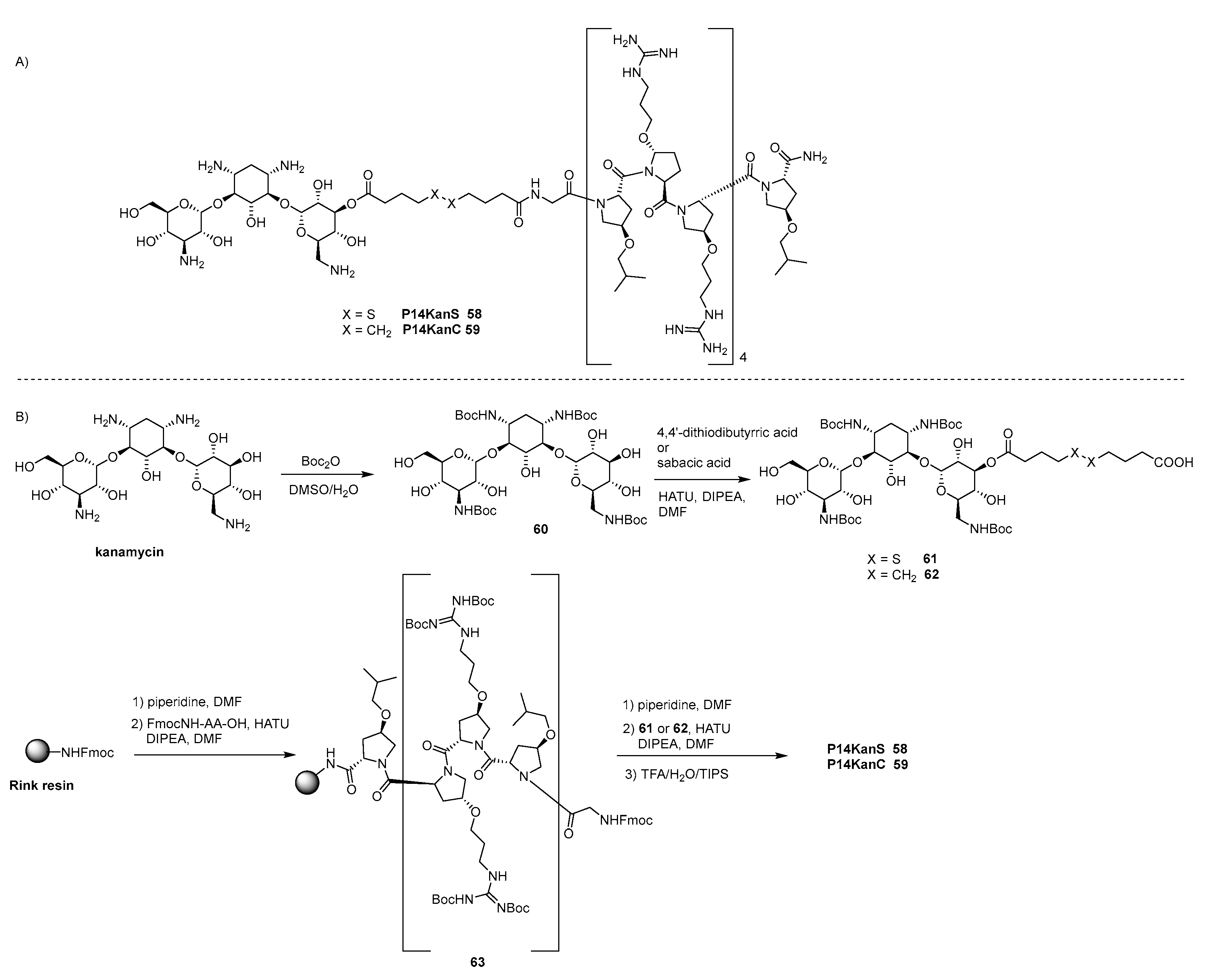
- Brezden, A.; Mohamed, M.F.; Nepal, M.; Harwood, J.S.; Kuriakose, J.; Seleem, M.N.; Chmielewski, J. Dual Targeting of Intracellular Pathogenic Bacteria with a Cleavable Conjugate of Kanamycin and an Antibacterial Cell-Penetrating Peptide. J. Am. Chem. Soc. 2016, 138, 10945–10949. [Google Scholar] [CrossRef]
- Mohamed, M.F.; Brezden, A.; Mohammad, H.; Chmielewski, J.; Seleem, M.N. Targeting biofilms and persisters of ESKAPE pathogens with P14KanS, a kanamycin peptide conjugate. Biochim. Biophys. Acta 2017, 1861, 848–859. [Google Scholar] [CrossRef]
- Scocchi, M.; Tossi, A.; Gennaro, R. Proline-Rich Antimicrobial Peptides: Converging to a Non-Lytic Mechanism of Action. Cell. Mol. Life Sci. 2011, 68, 2317–2330. [Google Scholar] [CrossRef]
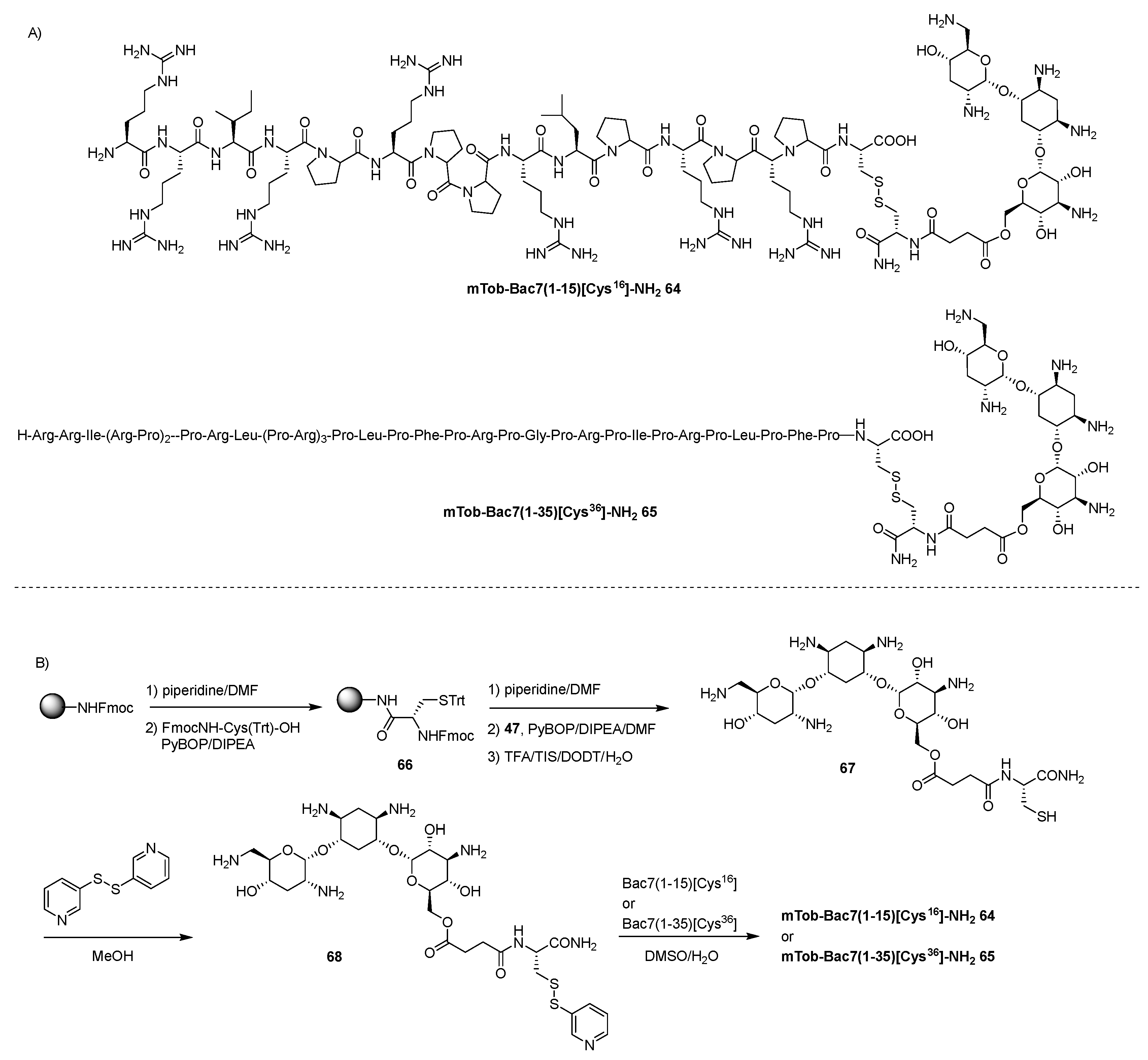
- Guida, F.; Benincasa, M.; Zahariev, S.; Scocchi, M.; Berti, F.; Gennaro, R.; Tossi, A. Effect of Size and N-Terminal Residue Characteristics on Bacterial Cell Penetration and Antibacterial Activity of the Proline-Rich Peptide Bac7. J. Med. Chem. 2015, 58, 1195–1204. [Google Scholar] [CrossRef]
- Gambato, S.; Bellotto, O.; Mardirossian, M.; Di Stasi, A.; Gennaro, R.; Pacor, S.; Caporale, A.; Berti, F.; Scocchi, M.; Tossi, A. Designing New Hybrid Antibiotics: Proline-Rich Antimicrobial Peptides Conjugated to the Aminoglycoside Tobramycin. Bioconjugate Chem. 2023, 34, 1212–1220. [Google Scholar] [CrossRef]
- Lau, Y.H.; De Andrade, P.; Wu, Y.; Spring, D.R. Peptide stapling techniques based on different macrocyclisation chemistries. Chem. Soc. Rev. 2015, 44, 91–102. [Google Scholar] [CrossRef]
- Lourenço, A.L.P.; Rios, T.B.; da Silva, A.P.; Franco, O.L.; Ramada, M.H.S. Peptide Stapling Applied to Antimicrobial Peptides. Antibiotics 2023, 12, 1400. [Google Scholar] [CrossRef]
- Wojciechowska, M.; Macyszyn, J.; Miszkiewicz, J.; Grzela, R.; Trylska, J. Stapled Anoplin as an Antibacterial Agent. Front. Microbiol. 2021, 12, 772038. [Google Scholar] [CrossRef]
- Macyszyn, J.; Burmistrz, M.; Mieczkowski, A.; Wojciechowska, M.; Trylska, J. ACS Omega 2023, 8, 19047–19056. [CrossRef]
- Rüter, C.; Buss, C.; Scharnert, J.; Heusipp, G.; Schmidt, M.A. A newly identified bacterial cell-penetrating peptide that reduces the transcription of pro-inflammatory cytokines. J Cell Sci 2010, 123, 2190–2198. [Google Scholar] [CrossRef]
- Gomarasca, M.; Martins, T.F.; Greune, L.; Hardwidge, P.R.; Schmidt, M.A.; Rüter, C. Bacterium-Derived Cell-Penetrating Peptides Deliver Gentamicin To Kill Intracellular Pathogens. Antimicrob. Agents Chemother. 2017, 61, e02545–16. [Google Scholar] [CrossRef]
- Choi, H.; Lee, D.G. Synergistic effect of antimicrobial peptide arenicin-1 in combination with antibiotics against pathogenic bacteria. Res., Microbiol. 2012, 163, 479–486. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Sun, Y.; Liu, Q.; Wang, X.; Li, Z.; Hao, J. In vitro synergistic activities of antimicrobial peptide brevinin 2CE with five kinds of antibiotics against multidrug-resistant clinical isolates. Curr. Microbiol. 2014, 68, 685–692. [Google Scholar] [CrossRef]
- Selsted, M.E.; Novotny, M.J.; Morris, W.L.; Tang, Y.Q.; Smith, W.; Cullor, J.S. Indolicidin, a novel bactericidal tridecapeptide amide from neutrophils. J. Biol. Chem. 1992, 267, 4292–4295. [Google Scholar] [CrossRef]
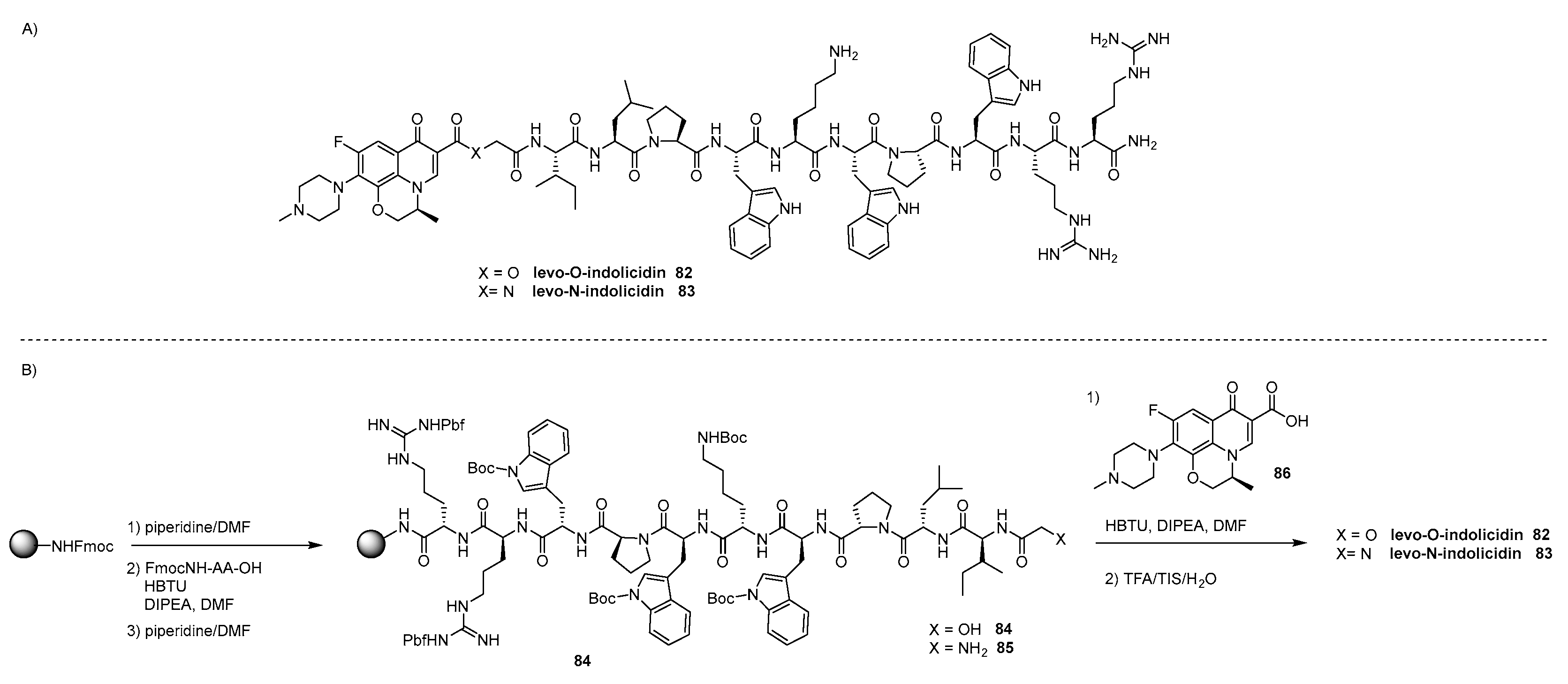
- Ghaffar, K.A.; Hussein, W.M.; Khalil, Z.G.; Capon, R.J.; Skwarczynski, M.; Toth, I. Levofloxacin and indolicidin for combination antimicrobial therapy. Curr. Drug Del. 2015, 12, 108–114. [Google Scholar] [CrossRef]
- Kondori, N.; Baltzer, L.; Dolphin, G.T.; Mattsby-Baltzer, I. Fungicidal Activity of Human Lactoferrin-Derived Peptides Based on the Antimicrobial Aβ Region. Int. J. Antimicrob. Agents 2011, 37, 51–57. [Google Scholar] [CrossRef]
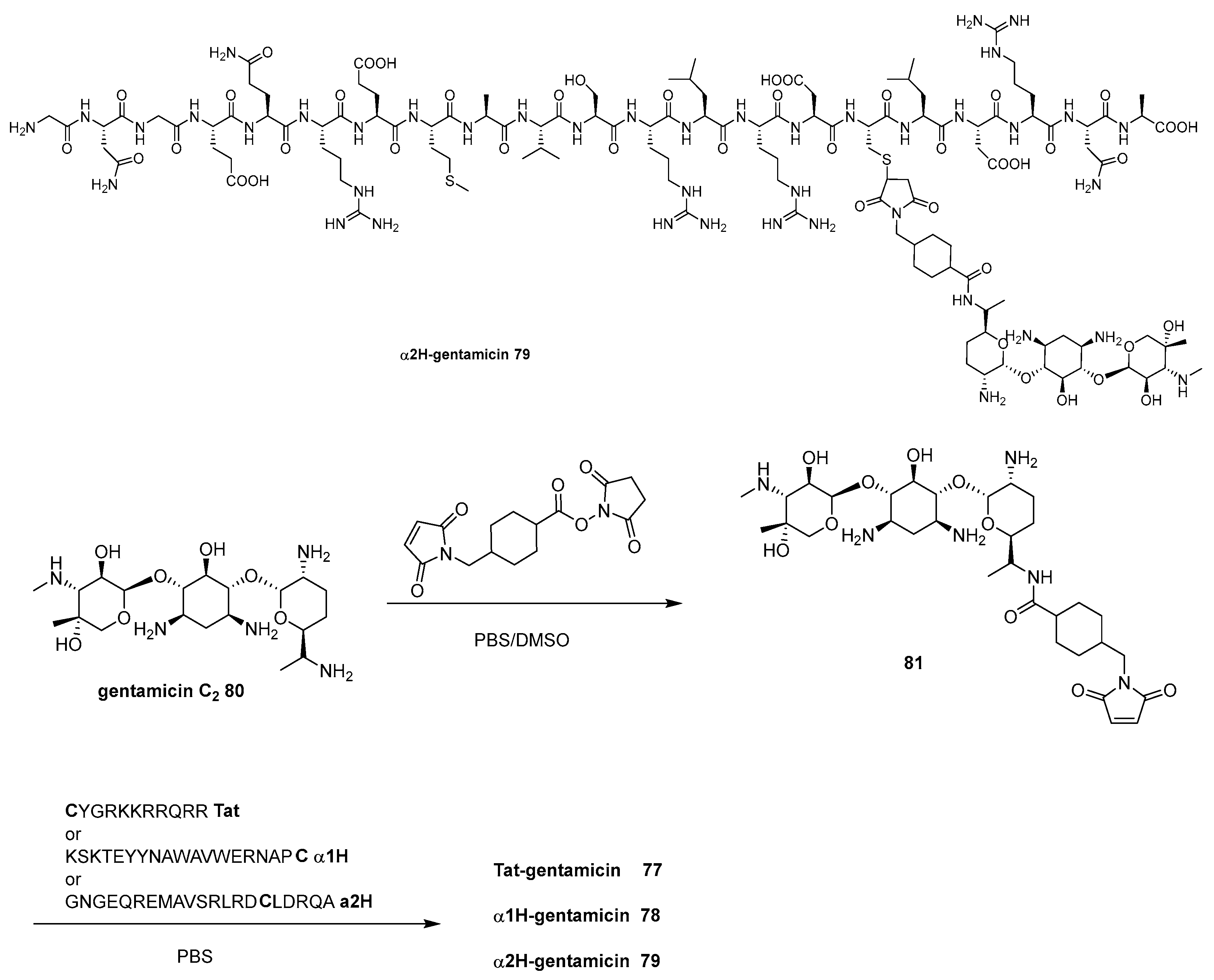
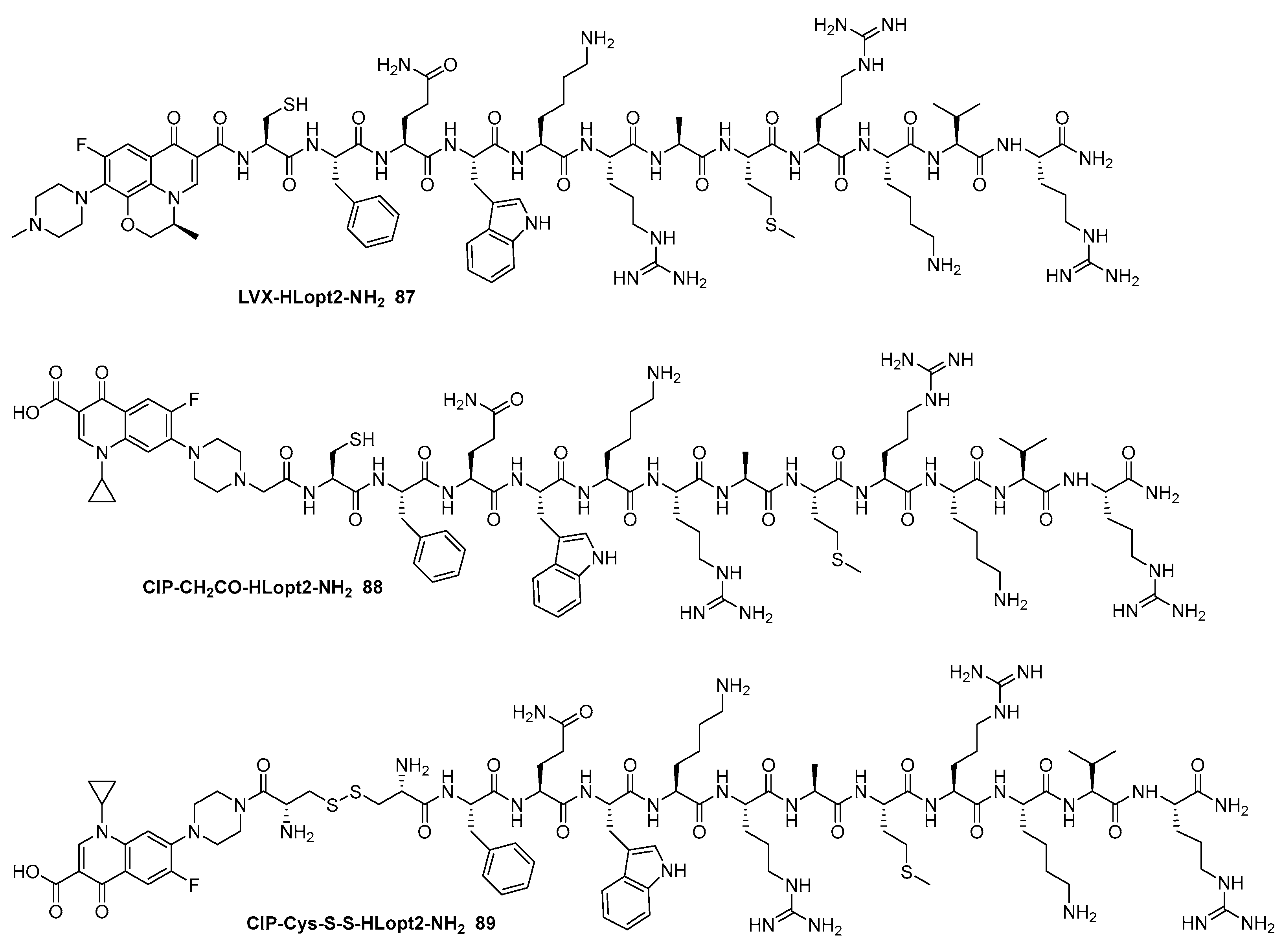
- Ptaszyńska, N.; Gucwa, K.; Olkiewicz, K.; Łȩgowska, A.; Okońska, J.; Ruczyński, J.; Gitlin-Domagalska, A.; Dȩbowski, D.; Milewski, S.; Rolka, K. Antibiotic-Based Conjugates Containing Antimicrobial HLopt2Peptide: Design, Synthesis, Antimicrobial and Cytotoxic Activities. ACS Chem. Biol. 2019, 14, 2233–2242. [Google Scholar]
- Tang, E.N.; Nair, A.; Baker, D.W.; Hum, W.; Zhou, J. In vivo imaging of infection using a bacteria-targeting optical nanoprobe. J. Biomed. Nanotechnol. 2014, 10, 856–863. [Google Scholar] [CrossRef]
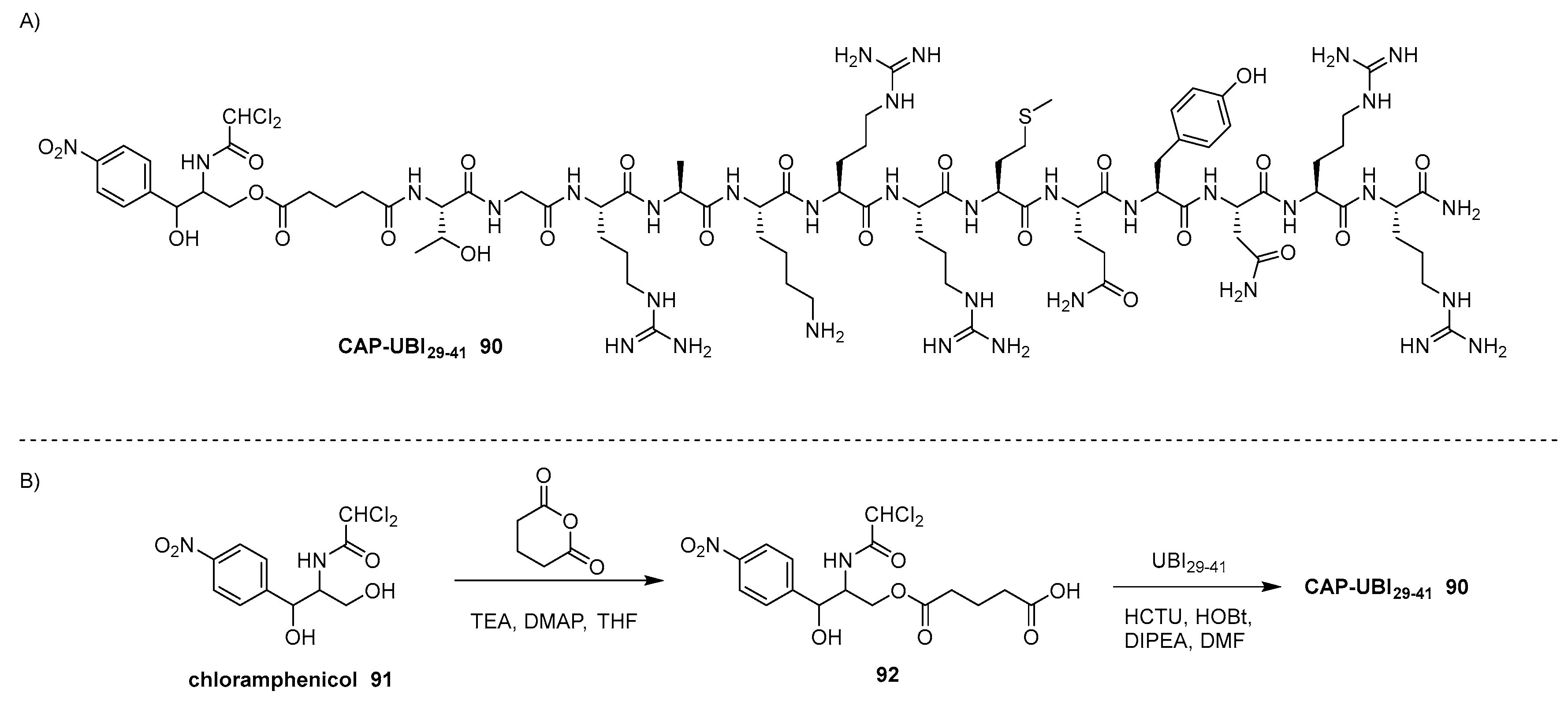
- Dinos, G.P.; Athanassopoulos, C.M.; Missiri, D.A.; Giannopoulou, P.C.; Vlachogiannis, I.A.; Papadopoulos, G.E.; Papaioannou, D.; Kalpaxis, D.L. Chloramphenicol Derivatives as Antibacterial and Anticancer Agents: Historic Problems and Current Solutions. Antibiotics 2016, 5, 20. [Google Scholar] [CrossRef]
- Saeed, S.; Zafar, J.; Khan, B.; Akhtar, A.; Qurieshi, S.; Fatima, S.; Ahmad, N.; Irfanullah, J. Utility of 99mTc-labelled antimicrobial peptide ubiquicidin (29−41) in the diagnosis of diabetic foot infection. Eur. J. Nucl. Med. Mol. Imaging 2013, 40, 737–743. [Google Scholar] [CrossRef]
- Chen, H.; Liu, C.; Chen, D.; Madrid, K.; Peng, S.; Dong, X.; Zhang, M.; Gu, Y. Bacteria-targeting conjugates based on antimicrobial peptide for bacteria diagnosis and therapy. Mol. Pharmaceutics 2015, 12, 2505–2516. [Google Scholar] [CrossRef]
- Phan, L.T.; Jian, T.; Chen, Z.; Qiu, Y.L.; Wang, Z.; Beach, T.; Polemeropoulos, A.; Sun Or, Y. Synthesis and Antibacterial Activity of a Novel Class of 4‘-Substituted 16-Membered Ring Macrolides Derived from Tylosin. J. Med. Chem. 2004, 47, 2965–2968. [Google Scholar] [CrossRef]
- Seefeldt, A.C.; Nguyen, F.; Antunes, S.; Pérébaskine, N.; Graf, M.; et al. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat. Struct. Mol. Biol. 2015, 22, 470–475. [Google Scholar] [CrossRef]
- Khairullina, Z.Z.; Makarov, G.I.; Tereshchenkov, A.G.; Buev, V.S.; Lukianov, D.A.; Polshakov, V.I.; Tashlitsky, V.N.; Osterman, I.A.; Sumbatyan, N.V. Conjugates of Desmycosin with Fragments of Antimicrobial Peptide Oncocin: Synthesis, Antibacterial Activity, Interaction with Ribosome. Biochemistry (Moscow) 2022, 87, 871–889. [Google Scholar] [CrossRef]
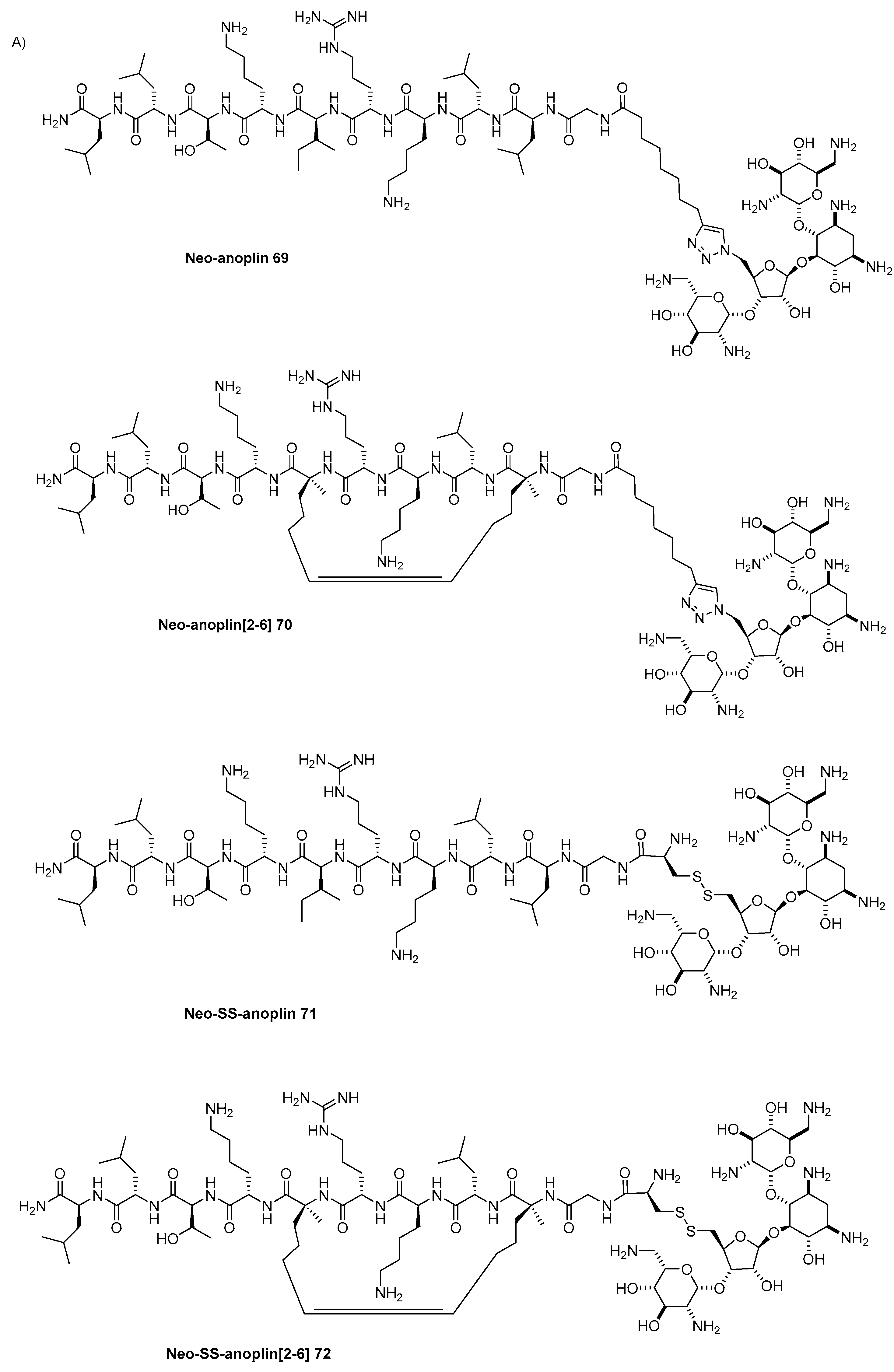
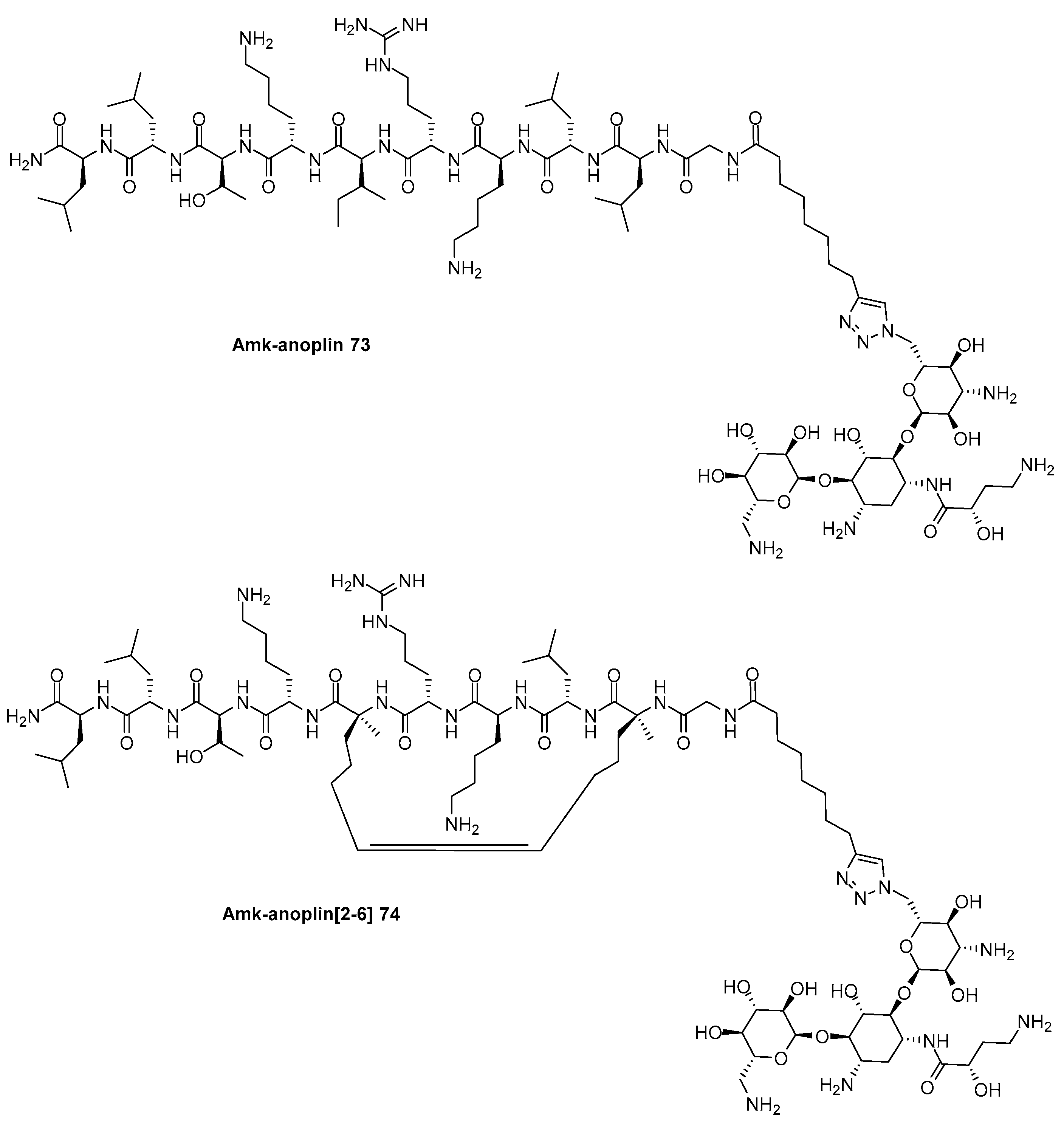
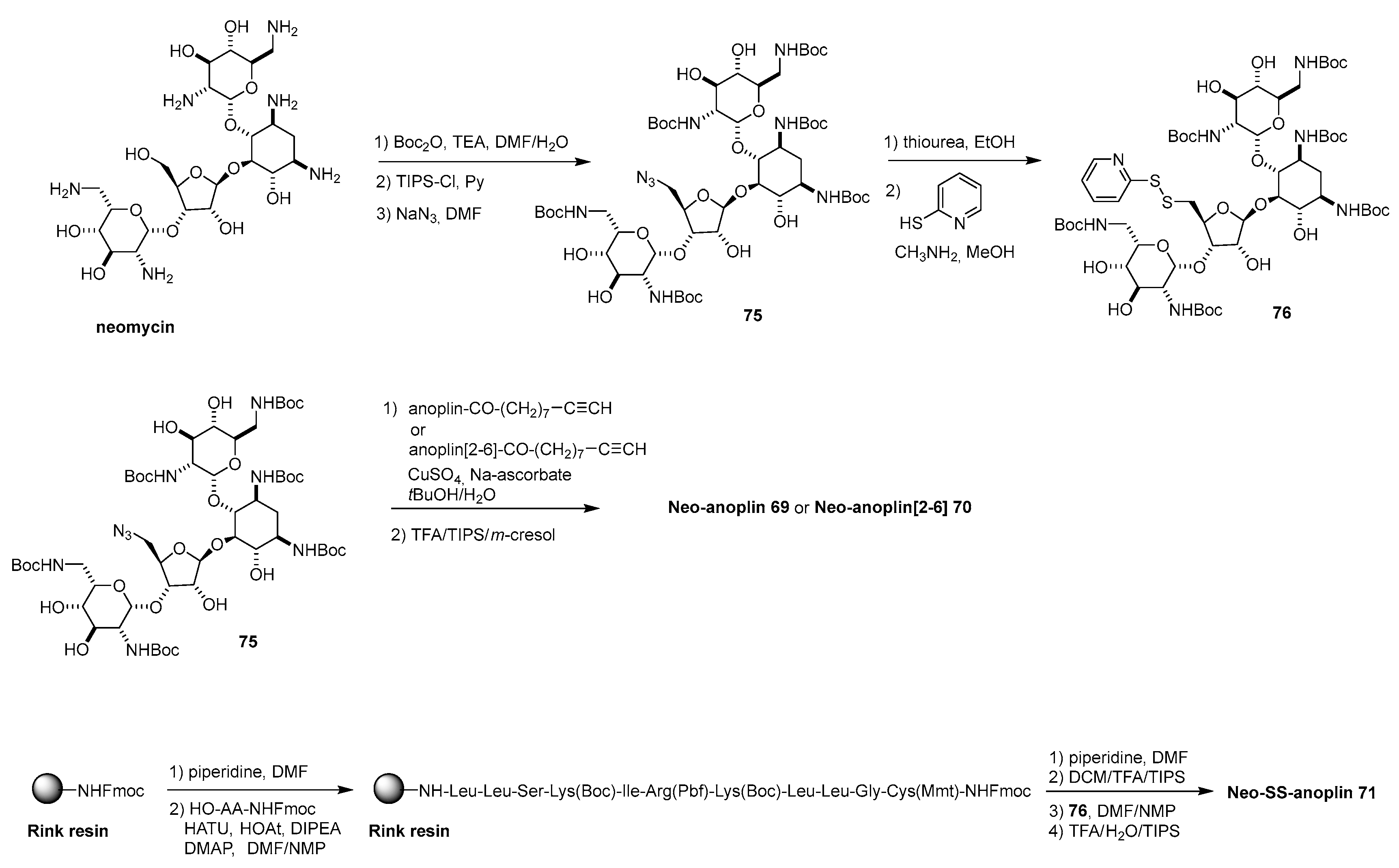
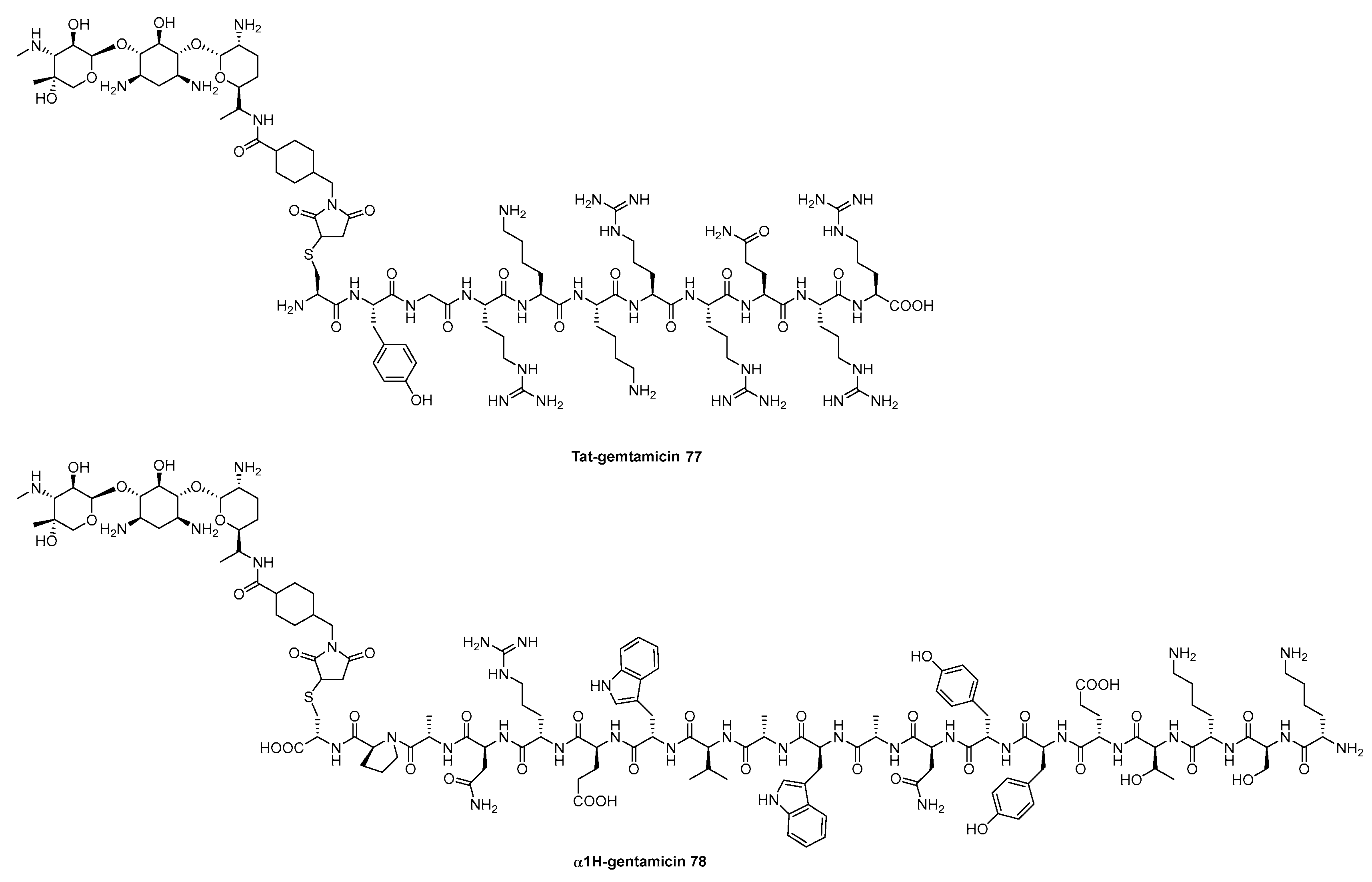
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




