Submitted:
24 July 2024
Posted:
25 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
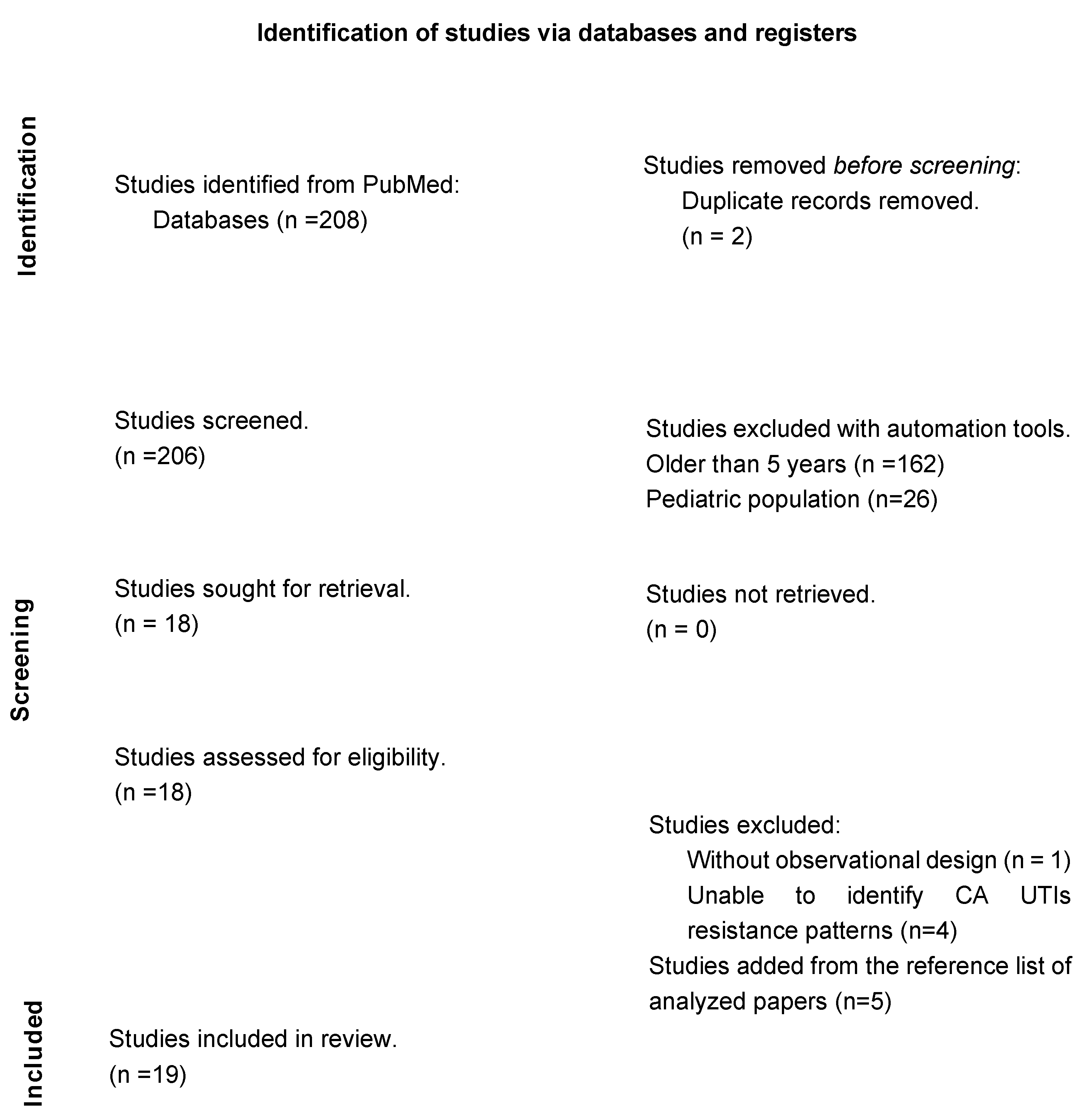
2. Materials and Methods
3. Results
4. Discussion
4.1. Reported Resistance Rates and in Community Acquired Uncomplicated Urinary Tract Infections
4.2. Plasmidic Fluoroquinolone Resistance Genes and Phylotypes Prevalence
4.3. Risk Factors Identified for Resistance
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Takhar, S.S.; Moran, G.J. Diagnosis and management of urinary tract infection in the emergency department and outpatient settings. Infect Dis Clin North Am 2014, 28, 33–48. [Google Scholar] [CrossRef]
- Akgoz, M.; Akman, I.; Ates, A.B.; Celik, C.; Keskin, B.; Ozmen Capin, B.B.; Karahan, Z.C. Plasmidic Fluoroquinolone Resistance Genes in Fluoroquinolone-Resistant and/or Extended Spectrum Beta-Lactamase-Producing Escherichia coli Strains Isolated from Pediatric and Adult Patients Diagnosed with Urinary Tract Infection. Microb Drug Resist 2020, 26, 1334–1341. [Google Scholar] [CrossRef]
- Bazaid, A.S.; Saeed, A.; Alrashidi, A.; Alrashidi, A.; Alshaghdali, K.; S, A.H.; Alreshidi, T.; Alshammary, M.; Alarfaj, A.; Thallab, R.; et al. Antimicrobial Surveillance for Bacterial Uropathogens in Ha’il, Saudi Arabia: A Five-Year Multicenter Retrospective Study. Infect Drug Resist 2021, 14, 1455–1465. [Google Scholar] [CrossRef]
- Ozturk, R.; Murt, A. Epidemiology of urological infections: a global burden. World J Urol 2020, 38, 2669–2679. [Google Scholar] [CrossRef]
- Chardavoyne, P.C.; Kasmire, K.E. Appropriateness of Antibiotic Prescriptions for Urinary Tract Infections. West J Emerg Med 2020, 21, 633–639. [Google Scholar] [CrossRef]
- Fasugba, O.; Gardner, A.; Mitchell, B.G.; Mnatzaganian, G. Ciprofloxacin resistance in community- and hospital-acquired Escherichia coli urinary tract infections: a systematic review and meta-analysis of observational studies. BMC Infect Dis 2015, 15, 545. [Google Scholar] [CrossRef]
- Gupta, K.; Hooton, T.M.; Naber, K.G.; Wullt, B.; Colgan, R.; Miller, L.G.; Moran, G.J.; Nicolle, L.E.; Raz, R.; Schaeffer, A.J.; et al. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: A 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis 2011, 52, e103–120. [Google Scholar] [CrossRef]
- Han, J.H.; Nachamkin, I.; Tolomeo, P.; Mao, X.; Bilker, W.B.; Lautenbach, E. Temporal changes in resistance mechanisms in colonizing Escherichia coli isolates with reduced susceptibility to fluoroquinolones. Diagn Microbiol Infect Dis 2013, 76, 491–496. [Google Scholar] [CrossRef]
- Jacoby, G.A.; Strahilevitz, J.; Hooper, D.C. Plasmid-mediated quinolone resistance. Microbiol Spectr 2014, 2. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin Microbiol Rev 2018, 31. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Touchon, M.; Perrin, A.; de Sousa, J.A.M.; Vangchhia, B.; Burn, S.; O’Brien, C.L.; Denamur, E.; Gordon, D.; Rocha, E.P. Phylogenetic background and habitat drive the genetic diversification of Escherichia coli. PLoS Genet 2020, 16, e1008866. [Google Scholar] [CrossRef]
- Afsharikhah, S.; Ghanbarpour, R.; Mohseni, P.; Adib, N.; Bagheri, M.; Jajarmi, M. High prevalence of β-lactam and fluoroquinolone resistance in various phylotypes of Escherichia coli isolates from urinary tract infections in Jiroft city, Iran. BMC Microbiol 2023, 23, 114. [Google Scholar] [CrossRef]
- Sohrabi, C.; Franchi, T.; Mathew, G.; Kerwan, A.; Nicola, M.; Griffin, M.; Agha, M.; Agha, R. PRISMA 2020 statement: What’s new and the importance of reporting guidelines. Int J Surg 2021, 88, 105918. [Google Scholar] [CrossRef]
- de Souza da-Silva, A.P.; de Sousa, V.S.; de Araújo Longo, L.G.; Caldera, S.; Baltazar, I.C.L.; Bonelli, R.R.; Santoro-Lopes, G.; Riley, L.W.; Moreira, B.M. Prevalence of fluoroquinolone-resistant and broad-spectrum cephalosporin-resistant community-acquired urinary tract infections in Rio de Janeiro: Impact of Escherichia coli genotypes ST69 and ST131. Infect Genet Evol 2020, 85, 104452. [Google Scholar] [CrossRef]
- Cristea, V.C.; Gheorghe, I.; Czobor Barbu, I.; Popa, L.I.; Ispas, B.; Grigore, G.A.; Bucatariu, I.; Popa, G.L.; Angelescu, M.C.; Velican, A.; et al. Snapshot of Phylogenetic Groups, Virulence, and Resistance Markers in Escherichia coli Uropathogenic Strains Isolated from Outpatients with Urinary Tract Infections in Bucharest, Romania. Biomed Res Int 2019, 2019, 5712371. [Google Scholar] [CrossRef]
- Dobbyn, D.; Zeggil, T.; Kudrowich, B.; Beahm, N.P. Ciprofloxacin resistances rates in Escherichia coli across Canada (CREAC): a longitudinal analysis 2015-2019. Int J Antimicrob Agents 2022, 59, 106532. [Google Scholar] [CrossRef]
- Jacoby, G.A. Mechanisms of resistance to quinolones. Clin Infect Dis 2005, 41 Suppl 2, S120–126. [Google Scholar] [CrossRef]
- KOT, B. Antibiotic Resistance Among Uropathogenic. Polish Journal of Microbiology 2019, 68, 403–415. [Google Scholar] [CrossRef]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect Drug Resist 2019, 12, 3903–3910. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Vila, J.; Moreno-Martinez, A.; Ruiz, J.; Martinez, J.A.; Sanchez, M.; Soriano, E.; Mensa, J. Molecular epidemiology and evolution of resistance to quinolones in Escherichia coli after prolonged administration of ciprofloxacin in patients with prostatitis. J Antimicrob Chemother 2002, 49, 55–59. [Google Scholar] [CrossRef]
- Soucy, J.R.; Schmidt, A.M.; Quach, C.; Buckeridge, D.L. Fluoroquinolone Use and Seasonal Patterns of Ciprofloxacin Resistance in Community-Acquired Urinary Escherichia coli Infection in a Large Urban Center. Am J Epidemiol 2020, 189, 215–223. [Google Scholar] [CrossRef]
- Uppala, A.; King, E.A.; Patel, D. Cefazolin versus fluoroquinolones for the treatment of community-acquired urinary tract infections in hospitalized patients. Eur J Clin Microbiol Infect Dis 2019, 38, 1533–1538. [Google Scholar] [CrossRef]
- Fromer, D.L.; Cheng, W.Y.; Gao, C.; Mahendran, M.; Hilts, A.; Duh, M.S.; Joshi, A.V.; Mulgirigama, A.; Mitrani-Gold, F.S. Likelihood of Antimicrobial Resistance in Urinary E coli Isolates among US Female Patients with Recurrent vs Non-Recurrent Uncomplicated Urinary Tract Infection. Urology 2024. [Google Scholar] [CrossRef]
- Zavala-Cerna, M.G.; Segura-Cobos, M.; Gonzalez, R.; Zavala-Trujillo, I.G.; Navarro-Perez, S.F.; Rueda-Cruz, J.A.; Satoscoy-Tovar, F.A. The Clinical Significance of High Antimicrobial Resistance in Community-Acquired Urinary Tract Infections. Can J Infect Dis Med Microbiol 2020, 2020, 2967260. [Google Scholar] [CrossRef]
- Guzmán, M.; Salazar, E.; Cordero, V.; Castro, A.; Villanueva, A.; Rodulfo, H.; De Donato, M. Multidrug resistance and risk factors associated with community-acquired urinary tract infections caused by Escherichia coli in Venezuela. Biomedica 2019, 39, 96–107. [Google Scholar] [CrossRef]
- Martos, I.; Colucci Camusso, G.; Albornoz, M.; Barros Nores, J.; Juaneda, R.; Belisle, D.F.; Furiasse, D. [Etiological profile and antimicrobial sensitivity in 1740 urinary infections of the community in the city of Córdoba, Argentina.]. Arch Esp Urol 2021, 74, 645–651. [Google Scholar]
- Findlay, J.; Gould, V.C.; North, P.; Bowker, K.E.; Williams, M.O.; MacGowan, A.P.; Avison, M.B. Characterization of cefotaxime-resistant urinary Escherichia coli from primary care in South-West England 2017-18. J Antimicrob Chemother 2020, 75, 65–71. [Google Scholar] [CrossRef]
- Leforestier, A.; Vibet, M.A.; Gentet, N.; Javaudin, F.; Le Bastard, Q.; Montassier, E.; Batard, E. Modeling the risk of fluoroquinolone resistance in non-severe community-onset pyelonephritis. Eur J Clin Microbiol Infect Dis 2020, 39, 1123–1127. [Google Scholar] [CrossRef]
- Naber, K.G.; Wagenlehner, F.; Kresken, M.; Cheng, W.Y.; Catillon, M.; Duh, M.S.; Yu, L.; Khanal, A.; Mulgirigama, A.; Joshi, A.V.; et al. Escherichia coli resistance, treatment patterns and clinical outcomes among females with uUTI in Germany: a retrospective physician-based chart review study. Sci Rep 2023, 13, 12077. [Google Scholar] [CrossRef]
- Jurałowicz, E.; Bartoszko-Tyczkowska, A.; Tyczkowska-Sieroń, E.; Kurnatowska, I. Etiology and bacterial susceptibility to antibiotics in patients with recurrent lower urinary tract infections. Pol Arch Intern Med 2020, 130, 373–381. [Google Scholar] [CrossRef]
- Demirci, M.; Ünlü, Ö.; İstanbullu Tosun, A. Detection of O25b-ST131 clone, CTX-M-1 and CTX-M-15 genes via real-time PCR in Escherichia coli strains in patients with UTIs obtained from a university hospital in Istanbul. J Infect Public Health 2019, 12, 640–644. [Google Scholar] [CrossRef]
- Islam, M.A.; Islam, M.R.; Khan, R.; Amin, M.B.; Rahman, M.; Hossain, M.I.; Ahmed, D.; Asaduzzaman, M.; Riley, L.W. Prevalence, etiology and antibiotic resistance patterns of community-acquired urinary tract infections in Dhaka, Bangladesh. PLoS One 2022, 17, e0274423. [Google Scholar] [CrossRef]
- Kotb, D.N.; Mahdy, W.K.; Mahmoud, M.S.; Khairy, R.M.M. Impact of co-existence of PMQR genes and QRDR mutations on fluoroquinolones resistance in Enterobacteriaceae strains isolated from community and hospital acquired UTIs. BMC Infect Dis 2019, 19, 979. [Google Scholar] [CrossRef]
- Sun, J.; Du, L.; Yan, L.; Dai, W.; Wang, Z.; Xu, X. Eight-Year Surveillance of Uropathogenic Escherichia coli in Southwest China. Infect Drug Resist 2020, 13, 1197–1202. [Google Scholar] [CrossRef]
- Stapleton, A.E.; Wagenlehner, F.M.E.; Mulgirigama, A.; Twynholm, M. Escherichia coli Resistance to Fluoroquinolones in Community-Acquired Uncomplicated Urinary Tract Infection in Women: a Systematic Review. Antimicrob Agents Chemother 2020, 64. [Google Scholar] [CrossRef]
- Seija, V.; Fratchez, V.; Ventura, V.; Pintos, M.; González, M. [Risk factors for community-acquired urinary tract infection caused by fluoroquinolone resistant E. coli]. Rev Chilena Infectol 2014, 31, 400–405. [Google Scholar] [CrossRef]
- Betitra, Y.; Teresa, V.; Miguel, V.; Abdelaziz, T. Determinants of quinolone resistance in Escherichia coli causing community-acquired urinary tract infection in Bejaia, Algeria. Asian Pac J Trop Med 2014, 7, 462–467. [Google Scholar] [CrossRef]
- Halaji, M.; Fayyazi, A.; Rajabnia, M.; Zare, D.; Pournajaf, A.; Ranjbar, R. Phylogenetic Group Distribution of Uropathogenic Escherichia coli and Related Antimicrobial Resistance Pattern: A Meta-Analysis and Systematic Review. Front Cell Infect Microbiol 2022, 12, 790184. [Google Scholar] [CrossRef]
- Thompson, D.; Xu, J.; Ischia, J.; Bolton, D. Fluoroquinolone resistance in urinary tract infections: Epidemiology, mechanisms of action and management strategies. BJUI Compass 2024, 5, 5–11. [Google Scholar] [CrossRef]
- van Driel, A.A.; Notermans, D.W.; Meima, A.; Mulder, M.; Donker, G.A.; Stobberingh, E.E.; Verbon, A. Antibiotic resistance of Escherichia coli isolated from uncomplicated UTI in general practice patients over a 10-year period. Eur J Clin Microbiol Infect Dis 2019, 38, 2151–2158. [Google Scholar] [CrossRef]
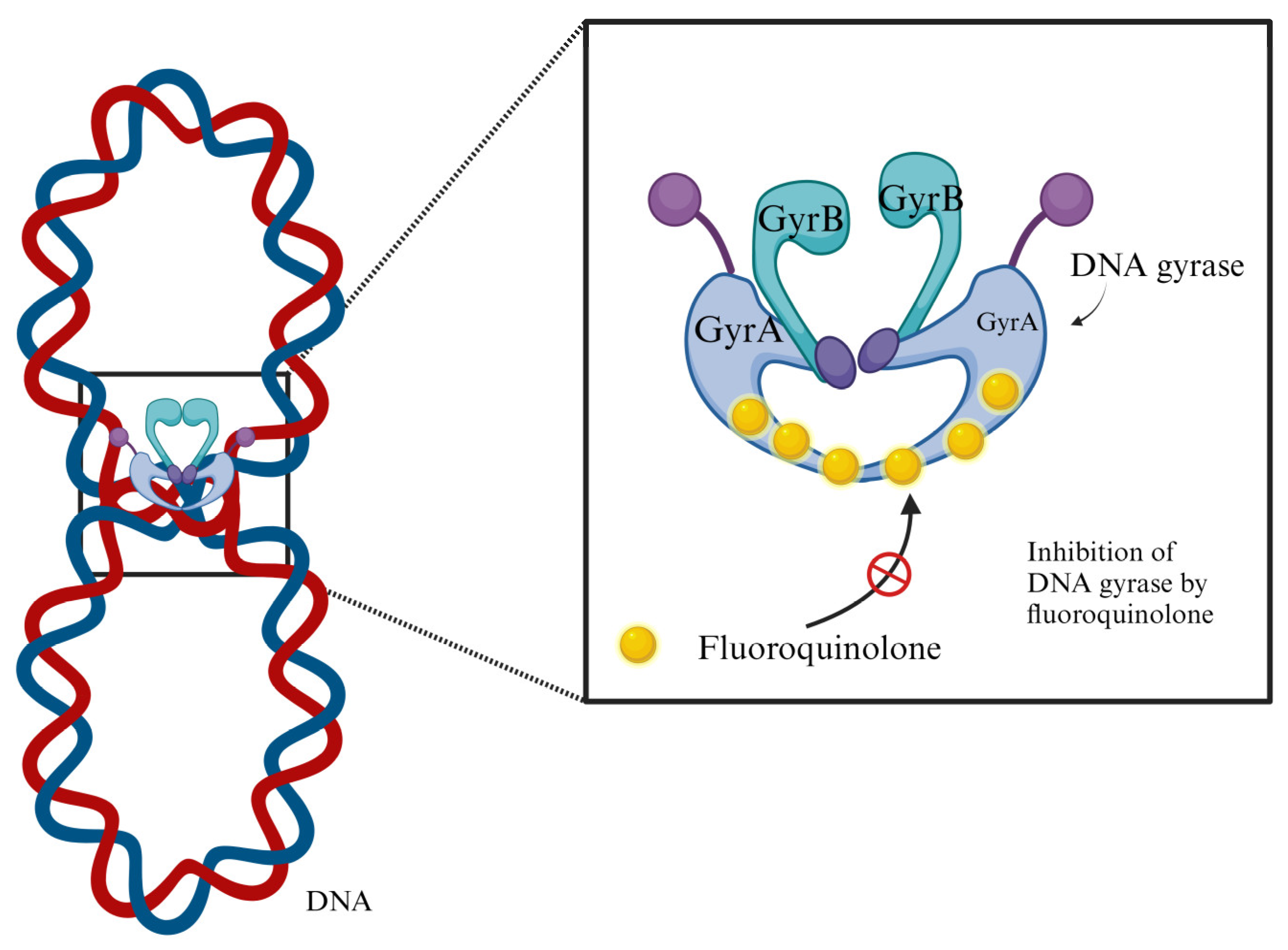
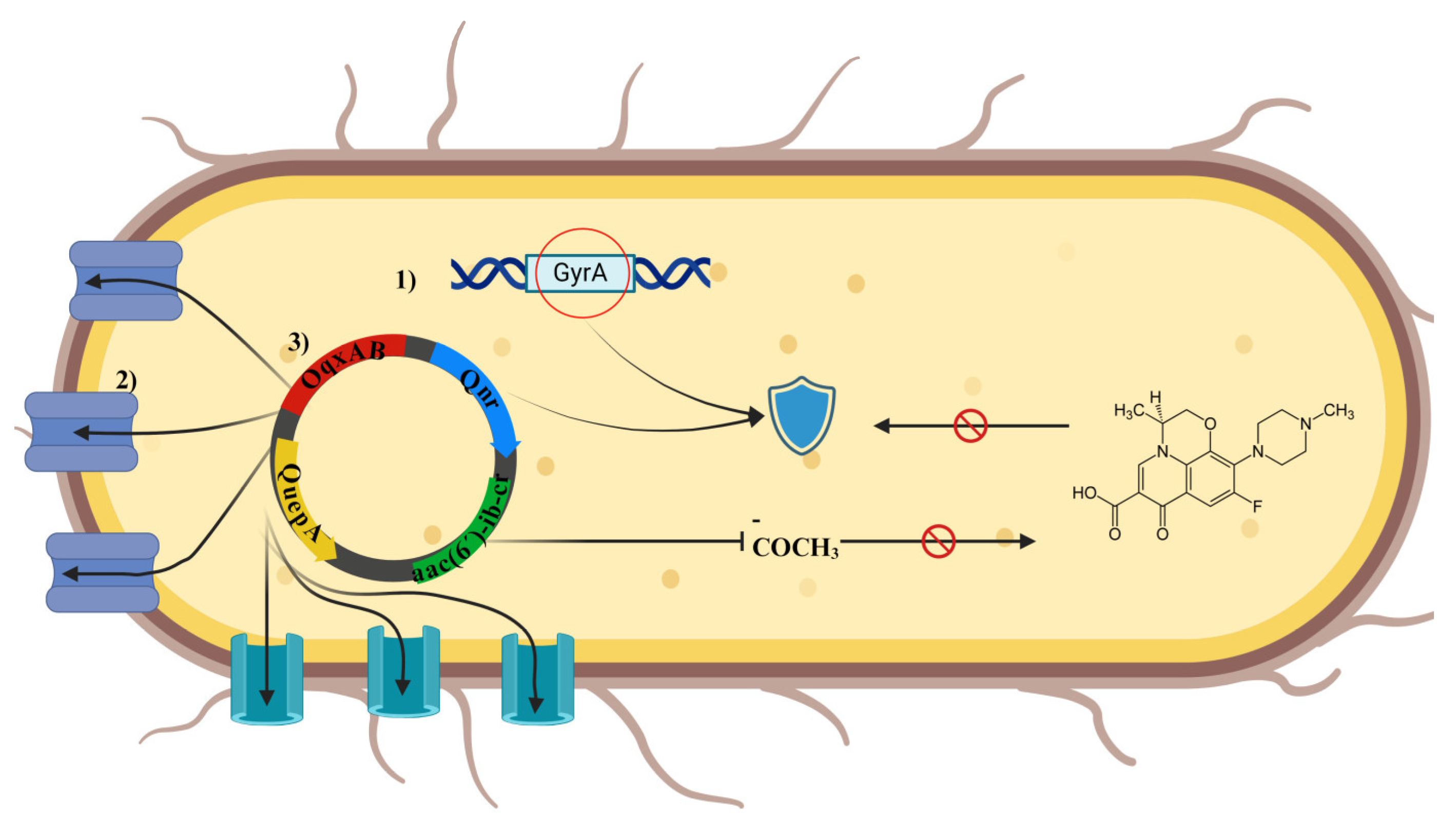
| Country / Year | Population | Period | Isolates | Resistance to FQ | Phylotype | AR gene | ESBL |
|---|---|---|---|---|---|---|---|
| Canada 2020 [22] | 11,333 isolates from community acquired UTIs (507 nosocomial) | April 2010 - Dec 2014 | E. Coli | CIP: 18.4% | NA | NA | NA |
| Canada 2022 [17] | 591 antibiograms from patients with CA UTIs | 2015 – 2019 | E. Coli | FQ: 11-63% | NA | NA | NA |
| USA 2019 [23] | 73 cases of CA-UTIs | April 2015 to May 2016 | E. Coli (59%) | CIP: 18% | NA | NA | NA |
| USA 2024 [24] |
68,033 cases of non-recurrent uUTI and 12,234 cases of recurrent UTIs |
October 2015 - February 2020 | E. Coli | Recurrent UTIs FQ: 14.2% Non-recurrent uUTIs FQ: 8.6% |
NA | NA | Recurrent UTIs 5.9% and non-recurrent UTIs 4.1% |
| Mexico 2020 [25] | 296 in patients with community acquired UTI. | 2018-2019 | E. Coli | CIP: 30% | NA | NA | 25.6% of which 89% were resistant to CIP |
| Venezuela 2019 [26] | 43 isolates from patients with uUTIs and 60 complicated UTIs | January - June 2014 | E. Coli | CIP: 29.7% | NA |
blaTEM ( 65.4%) blaCTX-M (34.6%) blaSHV (23.1%) |
20.4% |
| Brazil 2020 [15] | 499 isolates from patients with CA-UTIs | November 2015 | E. Coli | CIP: 20 % | B2 (30%) D (23%) A (13%) F (12%) B1 (9%) C (8%) E (3%) |
blaTEM (10%) blaSHV (3%) blaCTX-M (93%) |
8% |
| Argentina 2021 [27] | 1740 cases of CA-UTI’s | January 2016 to December 2017 | E. Coli (80%) | CIP: 15.2 % | B2 D |
blaCTX-M | 0.2% of which 56.9% were resistant to CIP |
| United Kingdom 2020 [28] | 836 E. coli isolates for resistance |
September 2017 – August 2018 | E. Coli | CIP: 50.7% | B2 D |
blaCTX-M pAmpC blaCMY blaDHA |
NA |
| France 2020 [29] | 190 Women with non-severe community-onset pyelonephritis. | Mar - Aug 2018, and Apr – Aug 2019 | E. Coli (84%) | FQ: 3-17% | NA | NA | NA |
| Germany 2023 [30] | 386 isolates from female patients with uUTIs | Jan 2017 -Dec 2019 | E. Coli | FQ: 5.2% CIP: 8.2% |
NA | NA | NA |
| Poland 2020 [31] | 796 Isolates from 332 patients with recurrent lower UTIs | 2016 - 2018 | E. coli (40%) | CIP: 39.9% | NA | NA | 9% |
| Romania 2019 [16] | 787 patients with CA-UTIs | June 2018 | E. coli (91%) | LEV: 14.86% CIP: 14.99% |
B2 (35%) B1 (27%) D (16%) A (22%). |
blaCTX-M (42.3%) blaTEM (38.0%) blaSHV (19.7%) fimH (93.9%) hlyD (44.3%) afaBC (38.2%) hlyA (12.4%) cnf-1 (7.7%). |
9% |
| Turkey 2019 [32] |
101 from CA UTIs and nosocomial UTIs | April - August 2018 |
E. Coli | CIP: 50.98% | NA |
blaCTX-M-1 (73%) blaCTX-M-15 (37%) O25b-ST131 (22%) |
50.49% |
| Turkey 2020 [2] | 141 adult outpatients with UTI | 01-06-2015 to 31-03-2016 | E. Coli | FQ: 35.92% | NA |
qnrS (67.4%) aac(6’)-1b-cr (42.4%) qnrB (7.6%) |
51.78%, of which 29.12% were resistant to FQ. |
| Bangladesh 2022 [33] | 4,500 patients with community-acquired UTIs | September 2016 – November 2018 | E. Coli (51.6%) | CIP: 69% |
NA | NA | NA |
| Egypt 2019 [34] | 440 Isolates from patients with UTIs | July 2016 -March 2017 | E. Coli (64%) | CIP: 19.2% NOR: 19.2% OFX: 19.2% |
NA |
qnrB (62.9%) qnrS (46.9%) qepA (6.3%) |
NA |
| Iran 2023 [13] | 168 CA uncomplicated UTI | Spring season of 2021 | E. Coli | CIP: 55.6% | B2 (29%) D 17.9% E 14.1% F 9.4% C 6.6 % |
blaTEM (89.6%) blaCTX-M (44.3%) blaSHV (6.6%) blaCMY (0.9%) |
52.8% |
| Saudi Arabia 2021 [3] | 428 patients with a positive urine culture (≥10^5 CFU/mL). |
Jan 2015 - Dec 2019 |
E. Coli (45%) |
CIP: 53.1% | NA | bla_CTX-M | 23.7%, of which 47.8% were resistant to CIP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





