Submitted:
27 July 2024
Posted:
29 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
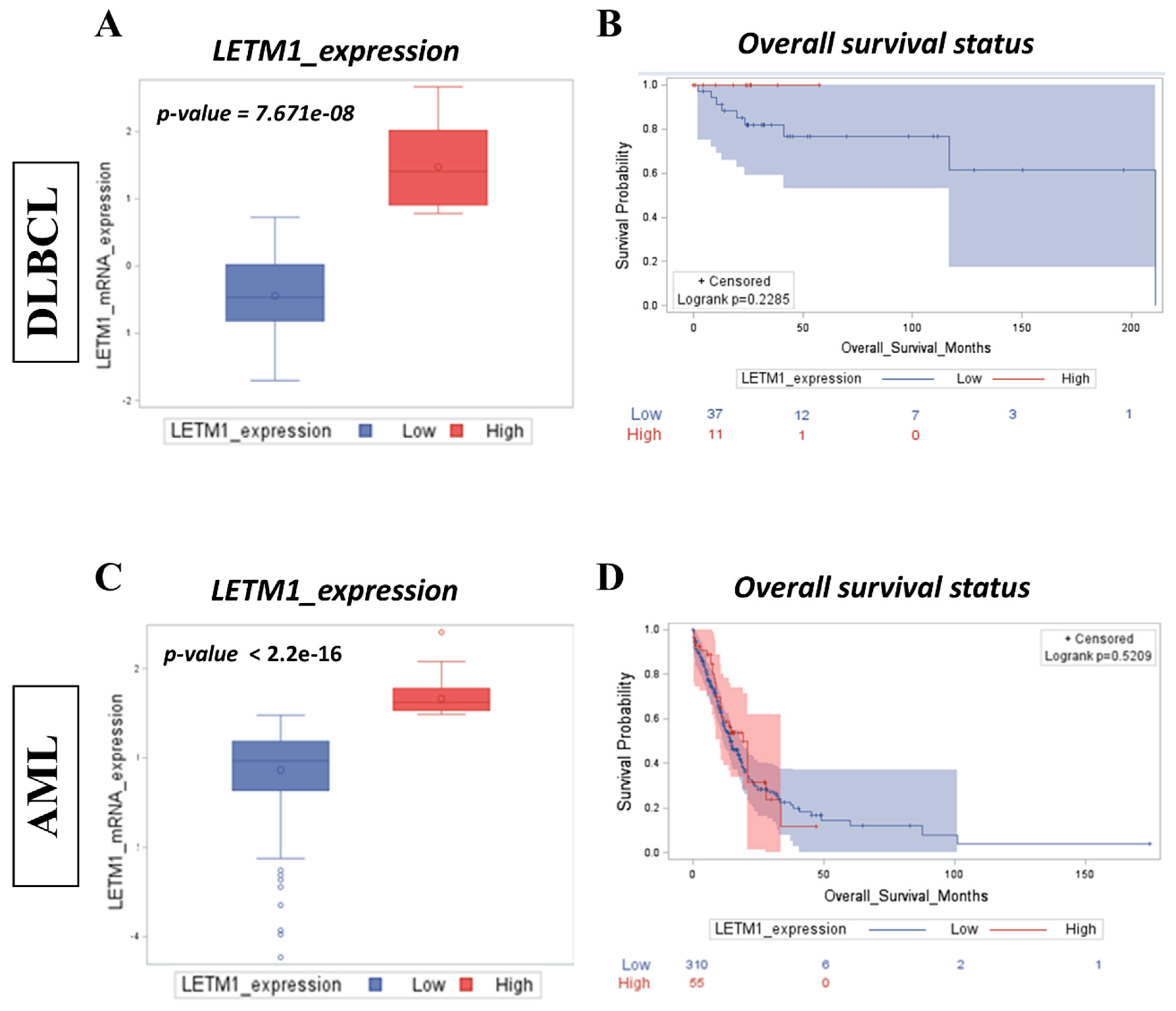
2.1. High LETM1 mRNA Expression Predicts Worse Clinical Outcomes in Both DLBCL and AML Patients
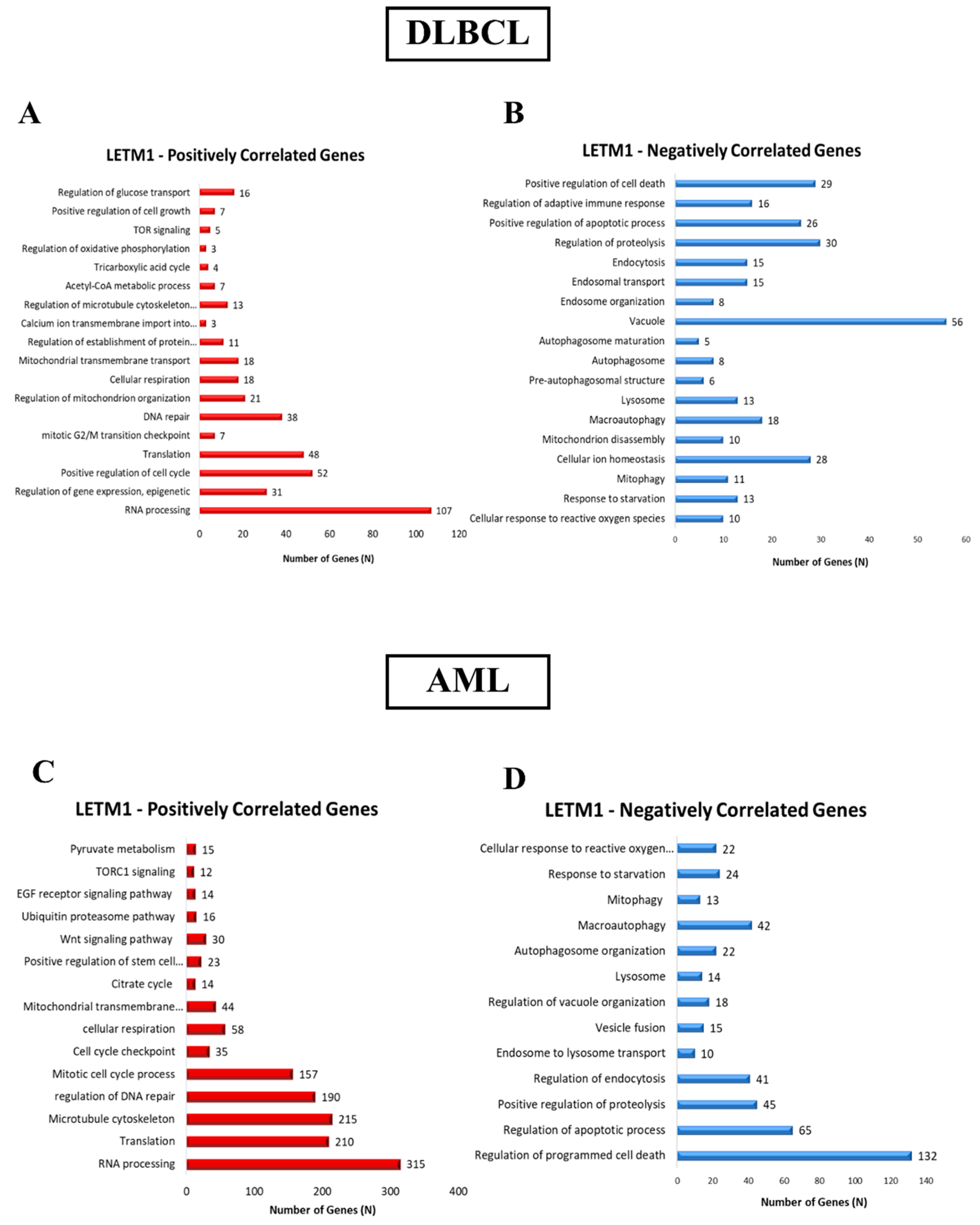
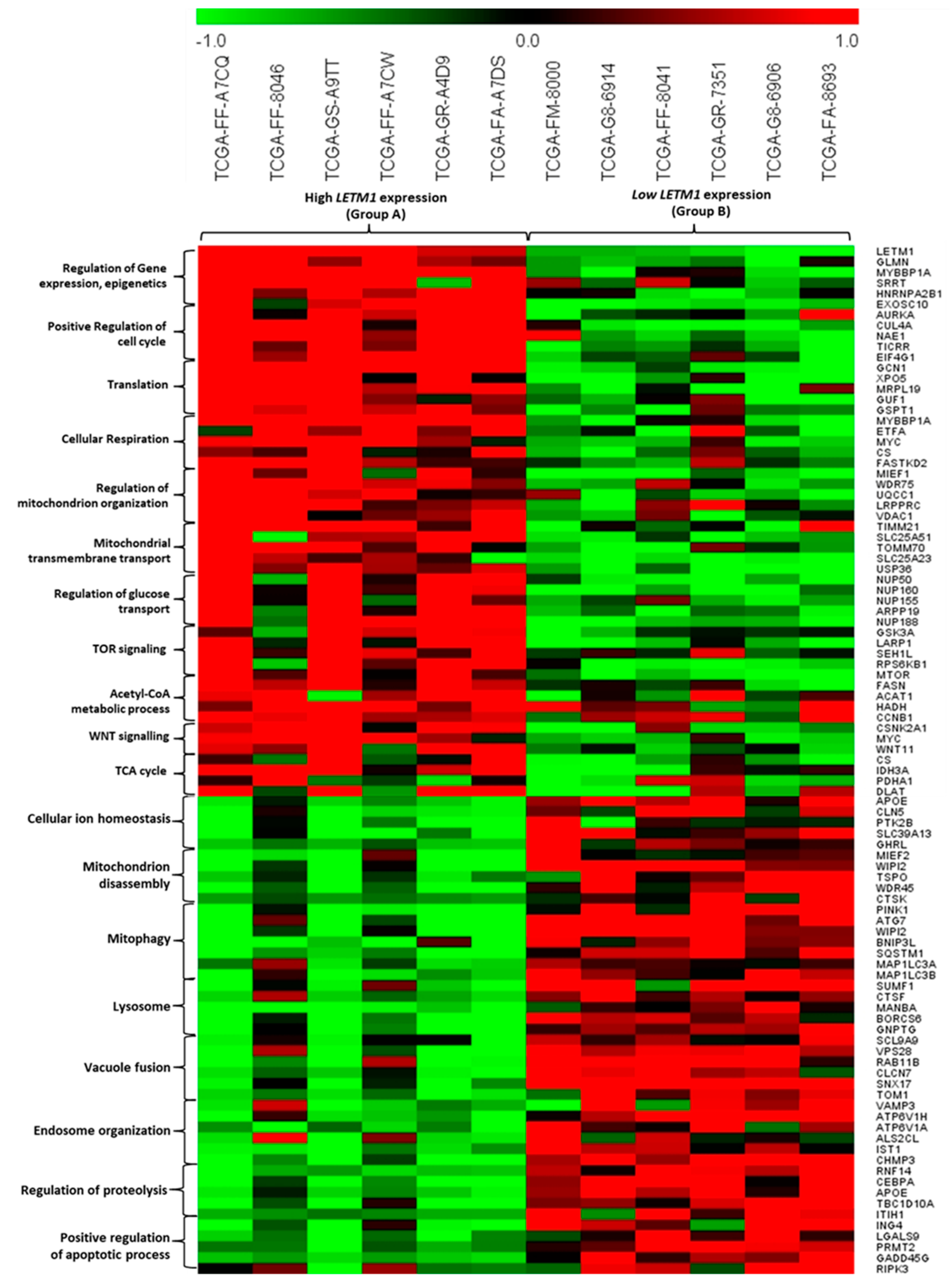
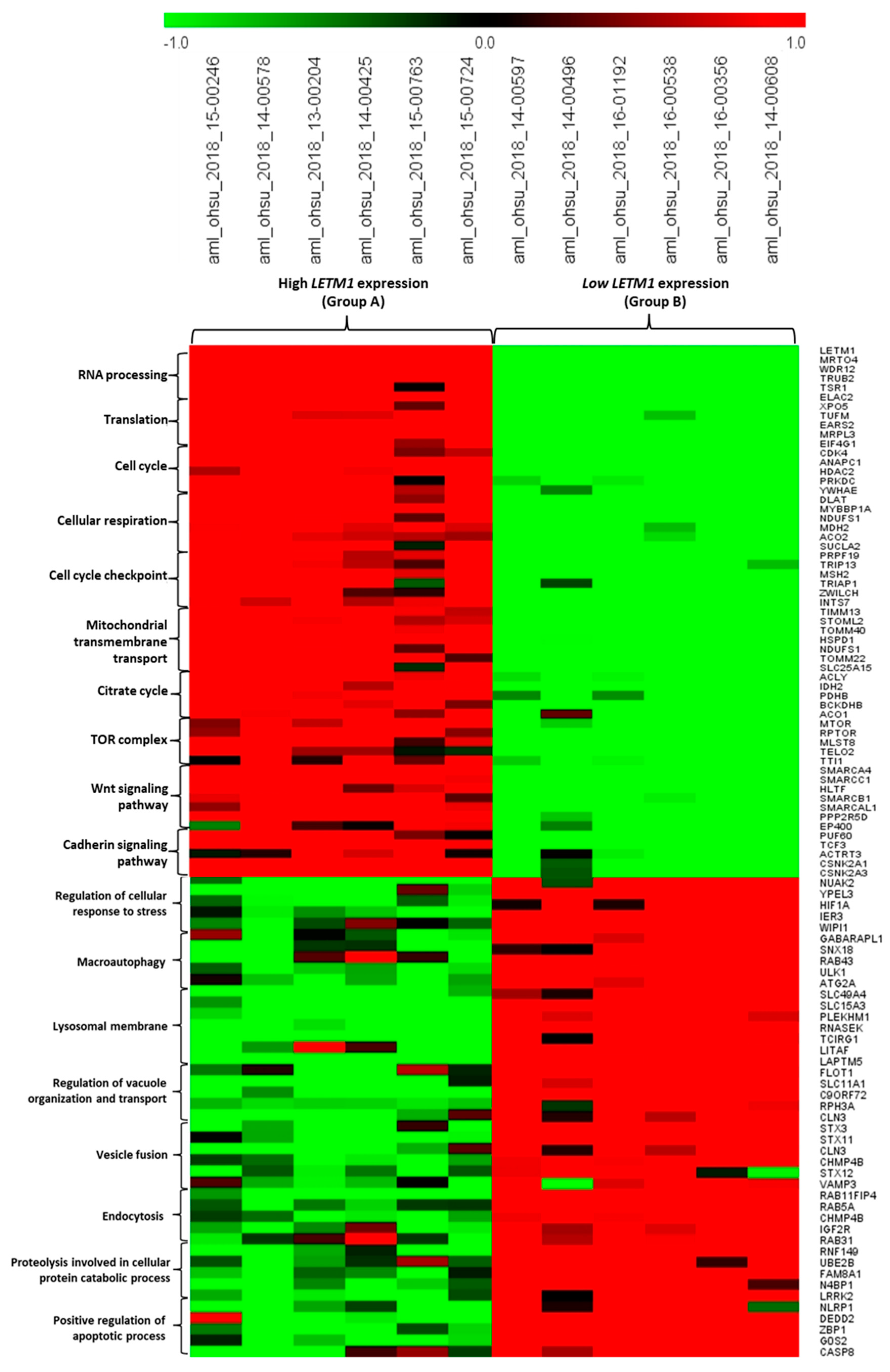
2.2. Identification of DEGs Associated with LETM1 in Both DLBCL and AML Patients
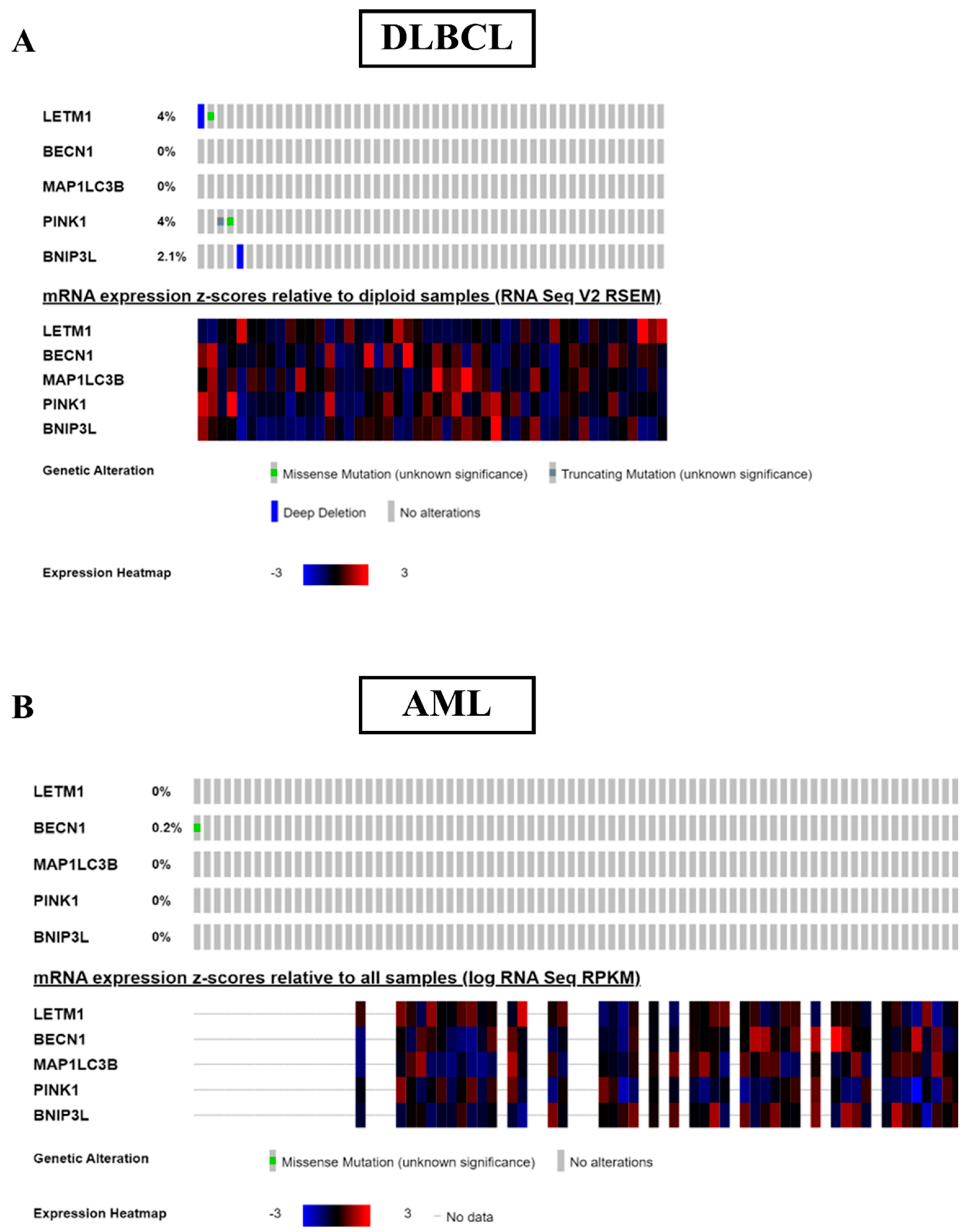
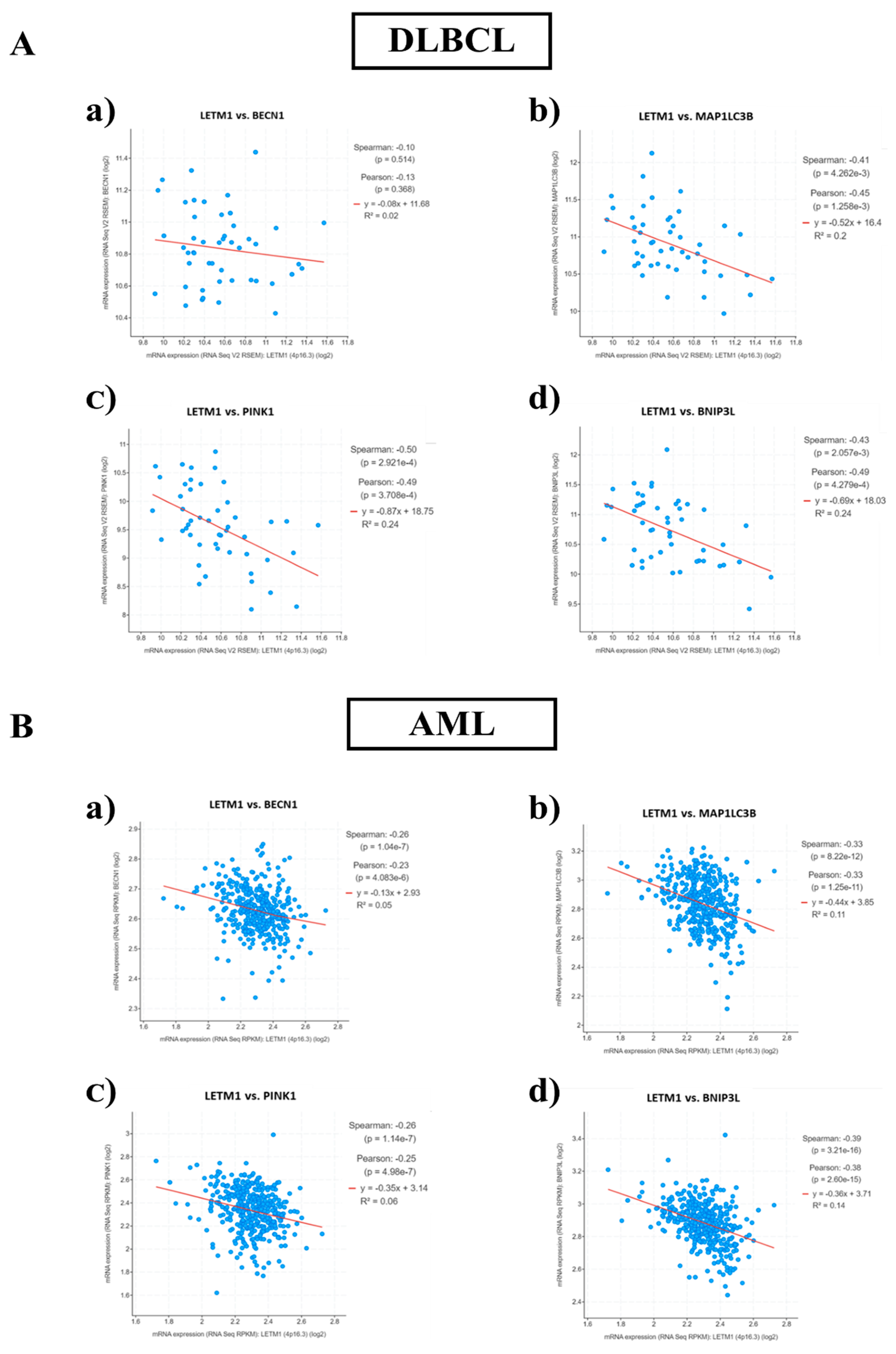
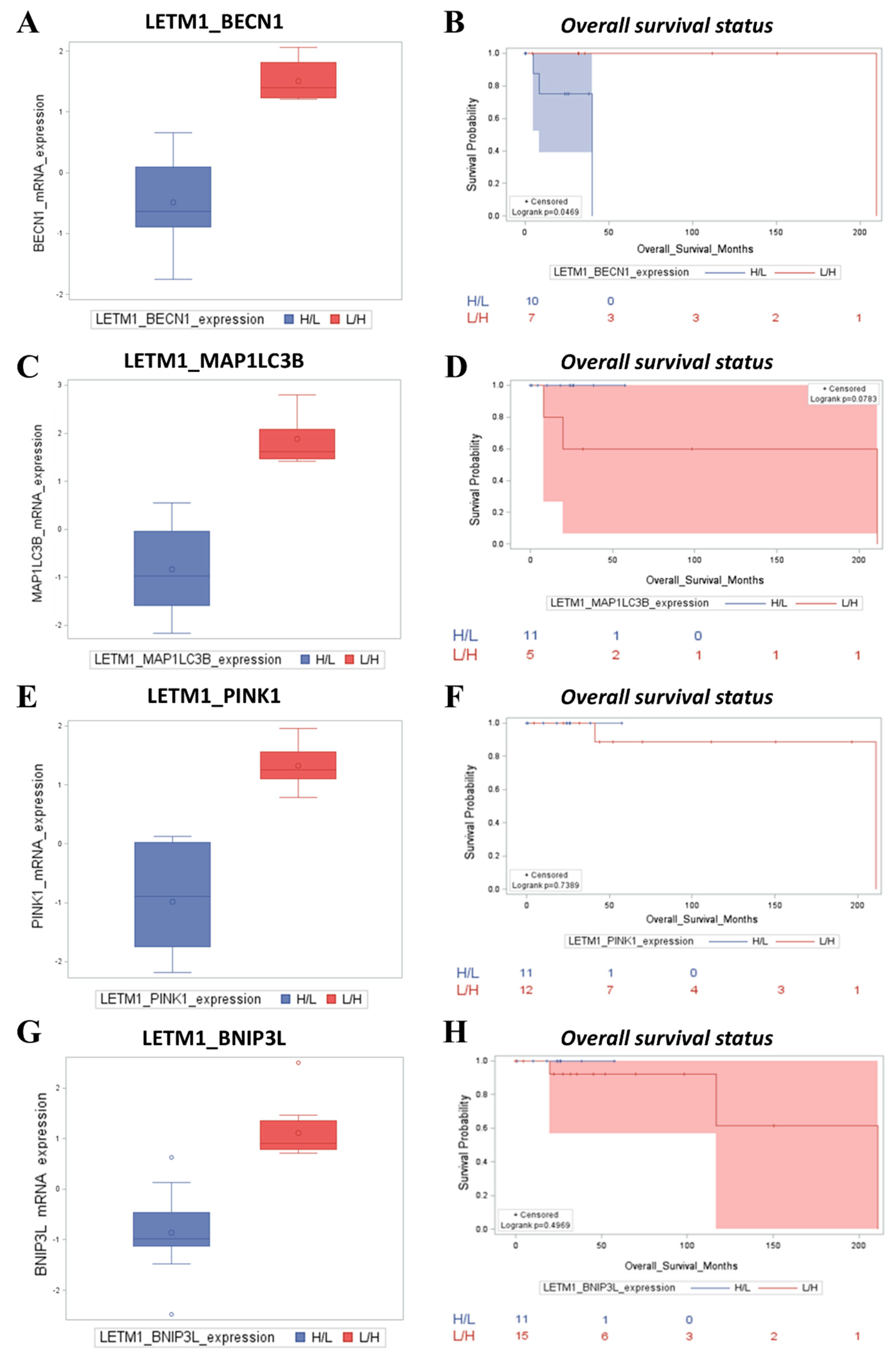
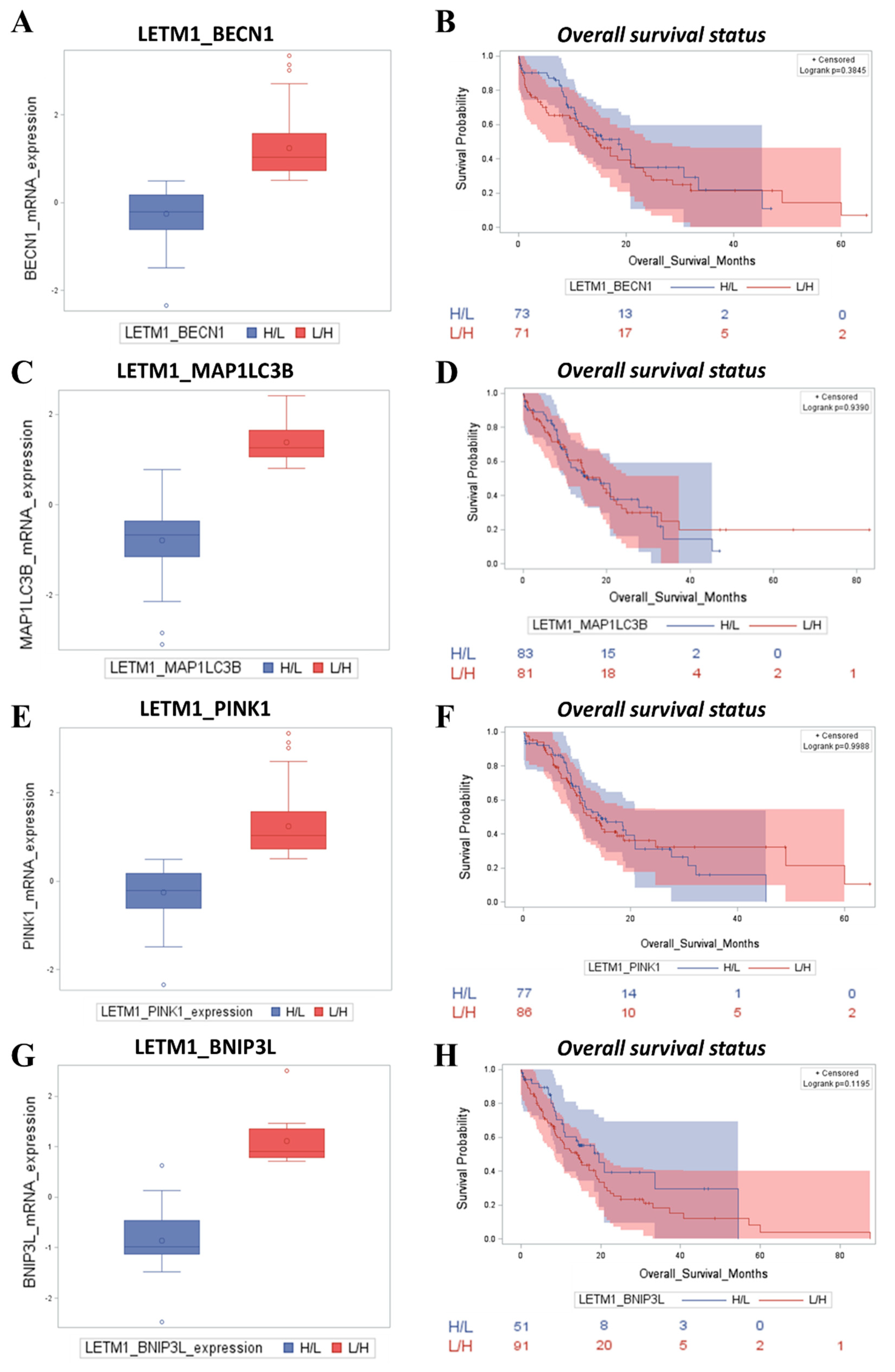
2.3. Patients with Low LETM1 Together with Upregulation of BECN1-Dependent Autophagy/Mitophagy Display a Better Clinical Outcome
3. Discussion
4. Materials and Methods
4.1. TCGA Database
4.2. Correlation Analysis and Screening of LETM1 Differentially Expressed Genes (DEGs)
4.3. Gene Ontology and Pathway Enrichment Analysis of DEGs
4.4. Statistical Analysis of Gene Expression and Clinical Outcomes
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Osellame, L.D.; Blacker, T.S.; Duchen, M.R. Cellular and Molecular Mechanisms of Mitochondrial Function. Best Pract Res Clin Endocrinol Metab 2012, 26, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Vara-Perez, M.; Felipe-Abrio, B.; Agostinis, P. Mitophagy in Cancer: A Tale of Adaptation. Cells 2019, 8, 493. [Google Scholar] [CrossRef] [PubMed]
- Tamai, S.; Iida, H.; Yokota, S.; Sayano, T.; Kiguchiya, S.; Ishihara, N.; Hayashi, J.-I.; Mihara, K.; Oka, T. Characterization of the Mitochondrial Protein LETM1, Which Maintains the Mitochondrial Tubular Shapes and Interacts with the AAA-ATPase BCS1L. J Cell Sci 2008, 121, 2588–2600. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Matsui, A.; Akabane, S.; Tamura, Y.; Hatano, A.; Miyano, Y.; Omote, H.; Kajikawa, M.; Maenaka, K.; Moriyama, Y.; et al. The Mitochondrial Inner Membrane Protein LETM1 Modulates Cristae Organization through Its LETM Domain. Commun Biol 2020, 3, 99. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tran, Q.; Shrestha, R.; Piao, L.; Park, S.; Park, J.; Park, J. LETM1 Is Required for Mitochondrial Homeostasis and Cellular Viability (Review). Mol Med Rep 2019, 19, 3367–3375. [Google Scholar] [CrossRef]
- Hashimi, H.; McDonald, L.; Stríbrná, E.; Lukeš, J. Trypanosome Letm1 Protein Is Essential for Mitochondrial Potassium Homeostasis. J Biol Chem 2013, 288, 26914–26925. [Google Scholar] [CrossRef] [PubMed]
- Waldeck-Weiermair, M.; Jean-Quartier, C.; Rost, R.; Khan, M.J.; Vishnu, N.; Bondarenko, A.I.; Imamura, H.; Malli, R.; Graier, W.F. Leucine Zipper EF Hand-Containing Transmembrane Protein 1 (Letm1) and Uncoupling Proteins 2 and 3 (UCP2/3) Contribute to Two Distinct Mitochondrial Ca2+ Uptake Pathways. J Biol Chem 2011, 286, 28444–28455. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Yang, C.; Yan, X.; Shi, Z.; Xiao, H.; Wei, X.; Jiang, N.; Wu, Z. LETM1 Knockdown Promotes Autophagy and Apoptosis Through AMP-Activated Protein Kinase Phosphorylation-Mediated Beclin-1/Bcl-2 Complex Dissociation in Hepatocellular Carcinoma. Front Oncol 2020, 10, 606790. [Google Scholar] [CrossRef]
- Dong, Z.; Liang, S.; Hu, J.; Jin, W.; Zhan, Q.; Zhao, K. Autophagy as a Target for Hematological Malignancy Therapy. Blood Rev 2016, 30, 369–380. [Google Scholar] [CrossRef]
- Jin, S.; Wei, J.; You, L.; Liu, H.; Qian, W. Autophagy Regulation and Its Dual Role in Blood Cancers: A Novel Target for Therapeutic Development (Review). Oncol Rep 2018, 39, 2473–2481. [Google Scholar] [CrossRef]
- Piya, S.; Kornblau, S.M.; Ruvolo, V.R.; Mu, H.; Ruvolo, P.P.; McQueen, T.; Davis, R.E.; Hail, N.; Kantarjian, H.; Andreeff, M.; et al. Atg7 Suppression Enhances Chemotherapeutic Agent Sensitivity and Overcomes Stroma-Mediated Chemoresistance in Acute Myeloid Leukemia. Blood 2016, 128, 1260–1269. [Google Scholar] [CrossRef] [PubMed]
- Rothe, K.; Porter, V.; Jiang, X. Current Outlook on Autophagy in Human Leukemia: Foe in Cancer Stem Cells and Drug Resistance, Friend in New Therapeutic Interventions. Int J Mol Sci 2019, 20, 461. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. Therapeutic Strategies of Drug Repositioning Targeting Autophagy to Induce Cancer Cell Death: From Pathophysiology to Treatment. J Hematol Oncol 2017, 10, 67. [Google Scholar] [CrossRef]
- Doonan, P.J.; Chandramoorthy, H.C.; Hoffman, N.E.; Zhang, X.; Cárdenas, C.; Shanmughapriya, S.; Rajan, S.; Vallem, S.; Chen, X.; Foskett, J.K.; et al. LETM1-Dependent Mitochondrial Ca2+ Flux Modulates Cellular Bioenergetics and Proliferation. FASEB J 2014, 28, 4936–4949. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zheng, Y.; Xuan, C.; Lin, Z.; Piao, L.; Liu, S. LETM1 Overexpression Is Correlated with the Clinical Features and Survival Outcome of Breast Cancer. Int J Clin Exp Pathol 2015, 8, 12893–12900. [Google Scholar] [PubMed]
- Lin, Q.-T.; Stathopulos, P.B. Molecular Mechanisms of Leucine Zipper EF-Hand Containing Transmembrane Protein-1 Function in Health and Disease. Int J Mol Sci 2019, 20, 286. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Q.; Chen, Y.; Liu, S.; Xu, J.; Cui, X.; Zhang, Y.; Piao, L. Clinical Implication of Leucine Zipper/EF Hand-Containing Transmembrane-1 Overexpression in the Prognosis of Triple-Negative Breast Cancer. Exp Mol Pathol 2015, 98, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Piao, L.; Li, H.; Feng, Y.; Li, X.; Cui, Y.; Xuan, Y. Leucine Zipper-EF-Hand Containing Transmembrane Protein 1 Is a Potential Prognostic Biomarker and Promotes Cell Progression in Prostate Cancer. Cancer Manag Res 2020, 12, 1649–1660. [Google Scholar] [CrossRef] [PubMed]
- Piao, L.; Yang, Z.; Feng, Y.; Zhang, C.; Cui, C.; Xuan, Y. LETM1 Is a Potential Biomarker of Prognosis in Lung Non-Small Cell Carcinoma. BMC Cancer 2019, 19, 898. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy (4th Edition)1. Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Wallace, D.C. A Mitochondrial Paradigm of Metabolic and Degenerative Diseases, Aging, and Cancer: A Dawn for Evolutionary Medicine. Annu Rev Genet 2005, 39, 359–407. [Google Scholar] [CrossRef]
- Moindjie, H.; Rodrigues-Ferreira, S.; Nahmias, C. Mitochondrial Metabolism in Carcinogenesis and Cancer Therapy. Cancers 2021, 13, 3311. [Google Scholar] [CrossRef]
- Wang, S.-F.; Tseng, L.-M.; Lee, H.-C. Role of Mitochondrial Alterations in Human Cancer Progression and Cancer Immunity. J Biomed Sci 2023, 30, 61. [Google Scholar] [CrossRef]
- Doblado, L.; Lueck, C.; Rey, C.; Samhan-Arias, A.K.; Prieto, I.; Stacchiotti, A.; Monsalve, M. Mitophagy in Human Diseases. Int J Mol Sci 2021, 22, 3903. [Google Scholar] [CrossRef] [PubMed]
- Bordi, M.; Nazio, F.; Campello, S. The Close Interconnection between Mitochondrial Dynamics and Mitophagy in Cancer. Front Oncol 2017, 7, 81. [Google Scholar] [CrossRef]
- Stergiou, I.E.; Kapsogeorgou, E.K. Autophagy and Metabolism in Normal and Malignant Hematopoiesis. Int J Mol Sci 2021, 22, 8540. [Google Scholar] [CrossRef]
- Nencioni, A.; Cea, M.; Montecucco, F.; Longo, V.D.; Patrone, F.; Carella, A.M.; Holyoake, T.L.; Helgason, G.V. Autophagy in Blood Cancers: Biological Role and Therapeutic Implications. Haematologica 2013, 98, 1335–1343. [Google Scholar] [CrossRef] [PubMed]
- Salwa, A.; Ferraresi, A.; Secomandi, E.; Vallino, L.; Moia, R.; Patriarca, A.; Garavaglia, B.; Gaidano, G.; Isidoro, C. High BECN1 Expression Negatively Correlates with BCL2 Expression and Predicts Better Prognosis in Diffuse Large B-Cell Lymphoma: Role of Autophagy. Cells 2023, 12, 1924. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, Y.; Liu, S.; Piao, L.; Zhang, Y.; Lin, Z.; Li, Z. High Expression of Leucine Zipper-EF-Hand Containing Transmembrane Protein 1 Predicts Poor Prognosis in Head and Neck Squamous Cell Carcinoma. Biomed Res Int 2014, 2014, 850316. [Google Scholar] [CrossRef]
- Piao, L.; Feng, Y.; Yang, Z.; Qi, W.; Li, H.; Han, H.; Xuan, Y. LETM1 Is a Potential Cancer Stem-like Cell Marker and Predicts Poor Prognosis in Colorectal Adenocarcinoma. Pathol Res Pract 2019, 215, 152437. [Google Scholar] [CrossRef]
- Yang, Z.; Ni, W.; Cui, C.; Qi, W.; Piao, L.; Xuan, Y. Identification of LETM1 as a Marker of Cancer Stem-like Cells and Predictor of Poor Prognosis in Esophageal Squamous Cell Carcinoma. Hum Pathol 2018, 81, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ding, W.; Xu, Y.; Tao, E.; Mo, M.; Xu, W.; Cai, X.; Chen, X.; Yuan, J.; Wu, X. Long Non-Coding RNA RHPN1-AS1 Promotes Tumorigenesis and Metastasis of Ovarian Cancer by Acting as a CeRNA against MiR-596 and Upregulating LETM1. Aging 2020, 12, 4558–4572. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, L.; Cao, Y.; Chen, S.; Xu, C.; Xing, J.; Zhang, K. LETM1 Promotes Gastric Cancer Cell Proliferation, Migration, and Invasion via the PI3K/Akt Signaling Pathway. J Gastric Cancer 2020, 20, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Zhang, J.; Zhang, X.; Huang, C.; Hu, G.; Li, S.; Xie, T.; Liu, M.; Xu, Y. Suppression of LETM1 by SiRNA Inhibits Cell Proliferation and Invasion of Bladder Cancer Cells. Oncol Rep 2017, 38, 2935–2940. [Google Scholar] [CrossRef]
- Xu, J.; Huang, B.; Li, S.; Zhang, X.; Xie, T.; Xu, Y. Knockdown of LETM1 Inhibits Proliferation and Metastasis of Human Renal Cell Carcinoma Cells. Oncol Lett 2018, 16, 6377–6382. [Google Scholar] [CrossRef]
- Che, N.; Yang, Z.; Liu, X.; Li, M.; Feng, Y.; Zhang, C.; Li, C.; Cui, Y.; Xuan, Y. Suppression of LETM1 Inhibits the Proliferation and Stemness of Colorectal Cancer Cells through Reactive Oxygen Species-Induced Autophagy. J Cell Mol Med 2021, 25, 2110–2120. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




