Submitted:
31 July 2024
Posted:
01 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Literature Search
2.1.1. Search Strategy
2.1.2. Eligibility Criteria
2.1.3. Selection Procedure
2.2. Categorization by Quality Enhancement Aspects
| Quality enhancement aspect | Definition |
|---|---|
| 1. Spatial resolution | The ability of differentiating two adjacent structures as being distinct from one another, either parallel (axial resolution) or perpendicular (lateral resolution) to the direction of the ultrasound beam [29]. |
| 2. Contrast resolution | The ability to distinguish between different echo amplitudes of adjacent structures through image intensity variations [29]. |
| 3. Detail enhancement of structures | Enhancement of texture, edges, or boundaries between structures. |
| 4. Noise | Minimization of random variability that is not part of the desired signal. |
| 5. General quality improvement | Mapping low-quality images to high-quality reference images, where the quality disparities are inherent to differences in the capture process and not artificially induced. |
2.3. Data Extraction
2.4. Statistical Analysis
3. Results
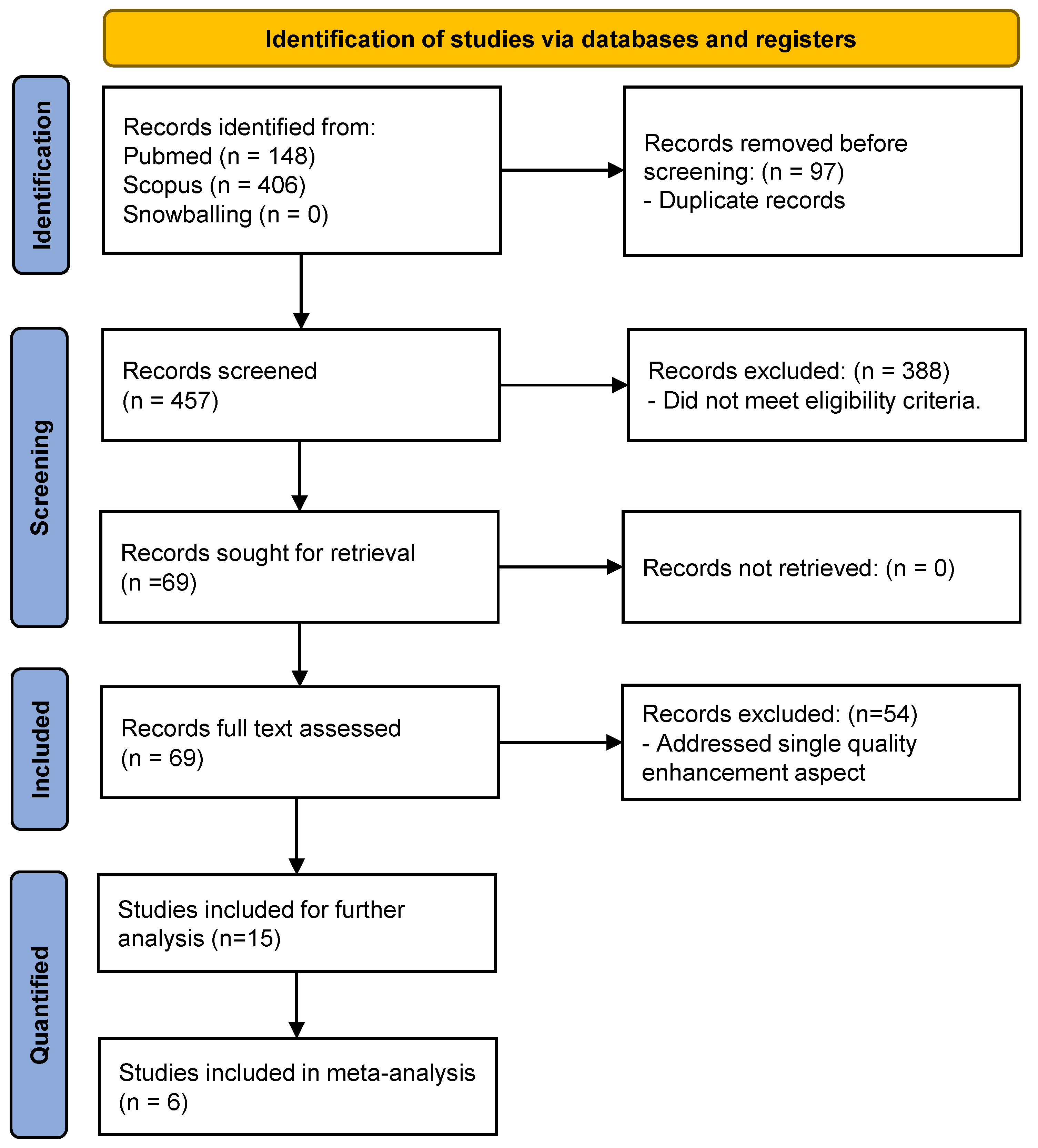
3.1. Study Selection
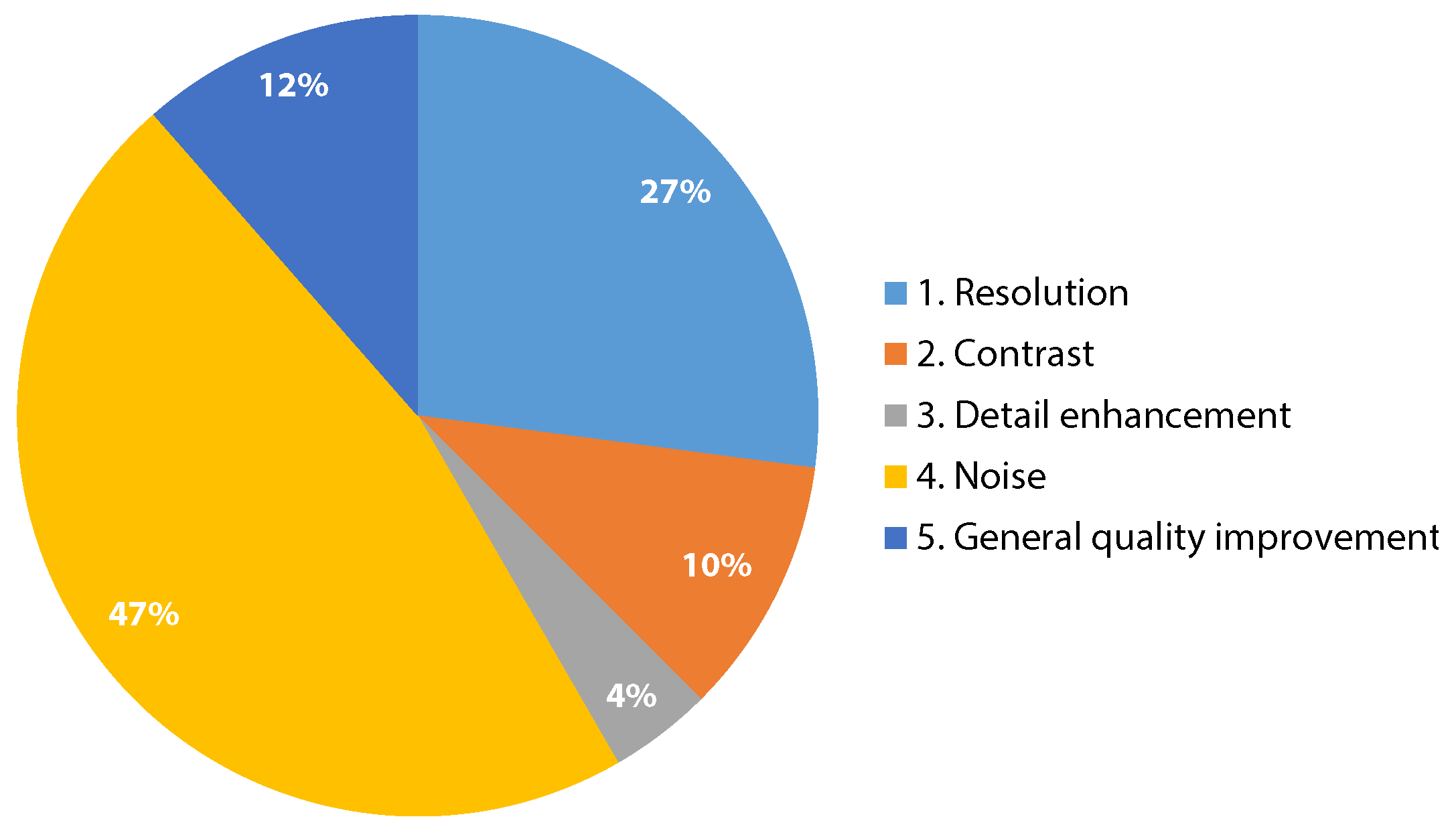
3.2. Quality Enhancement Aspects
3.3. Study Characteristics
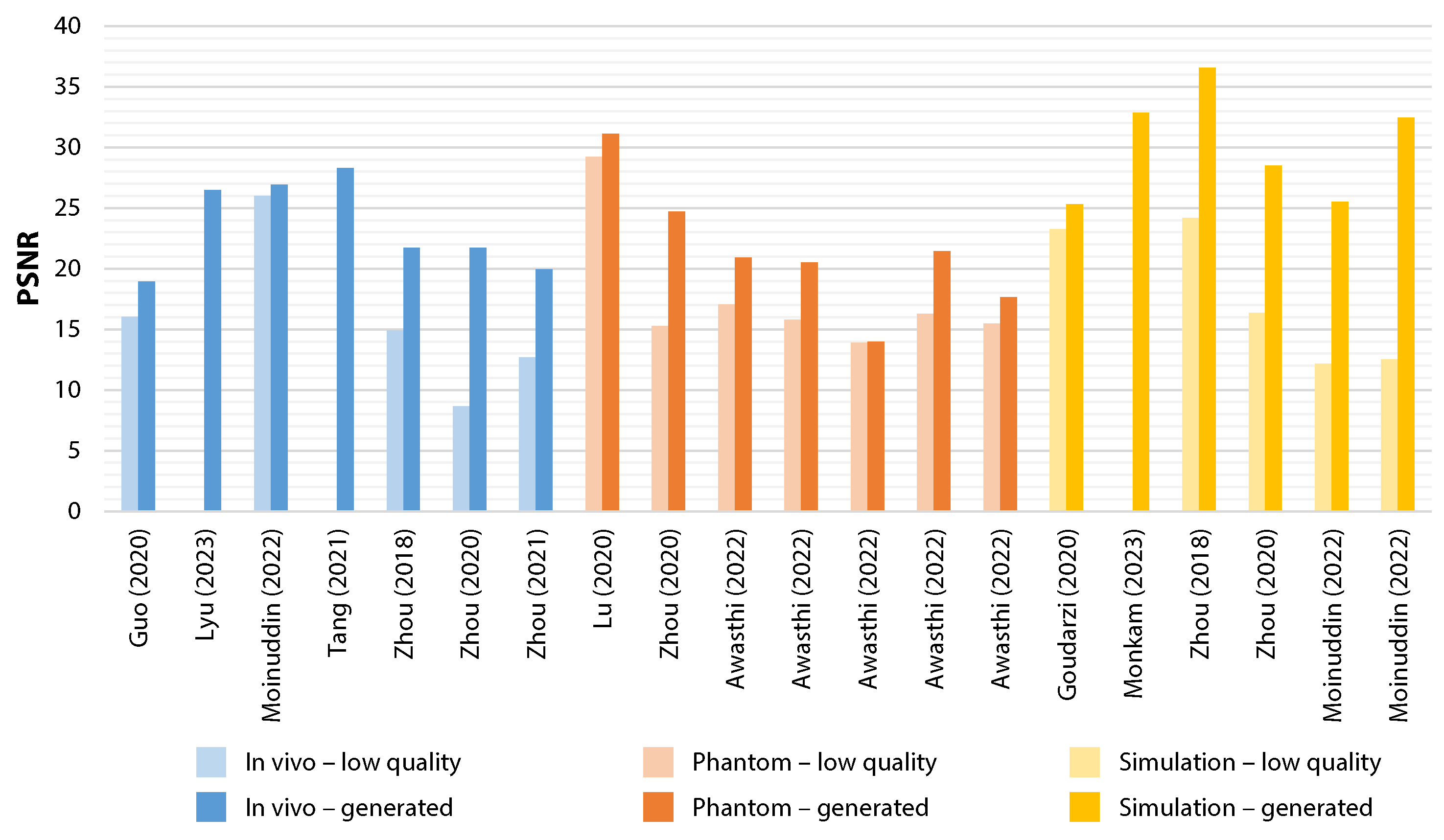
3.4. Study Outcomes
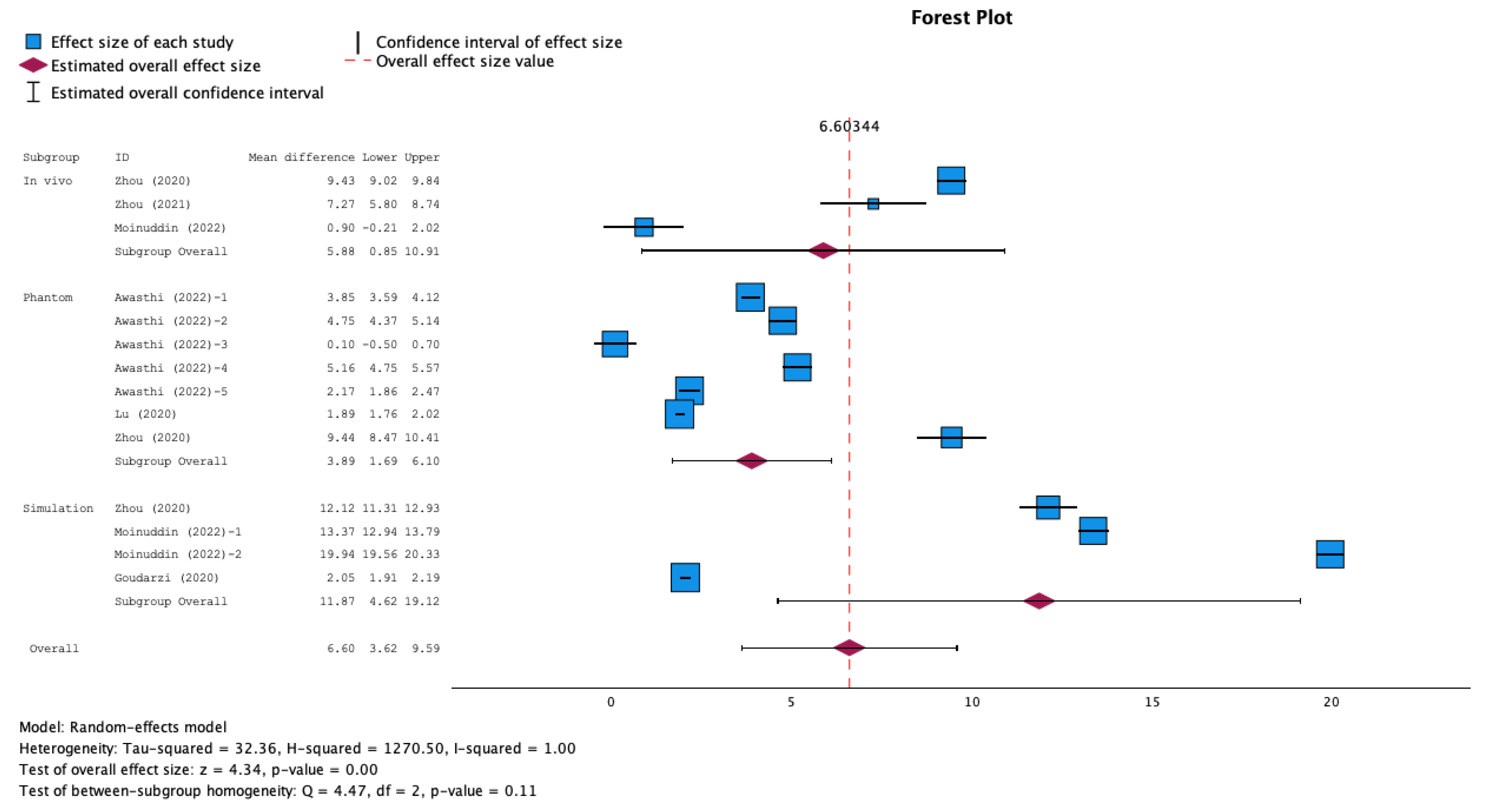
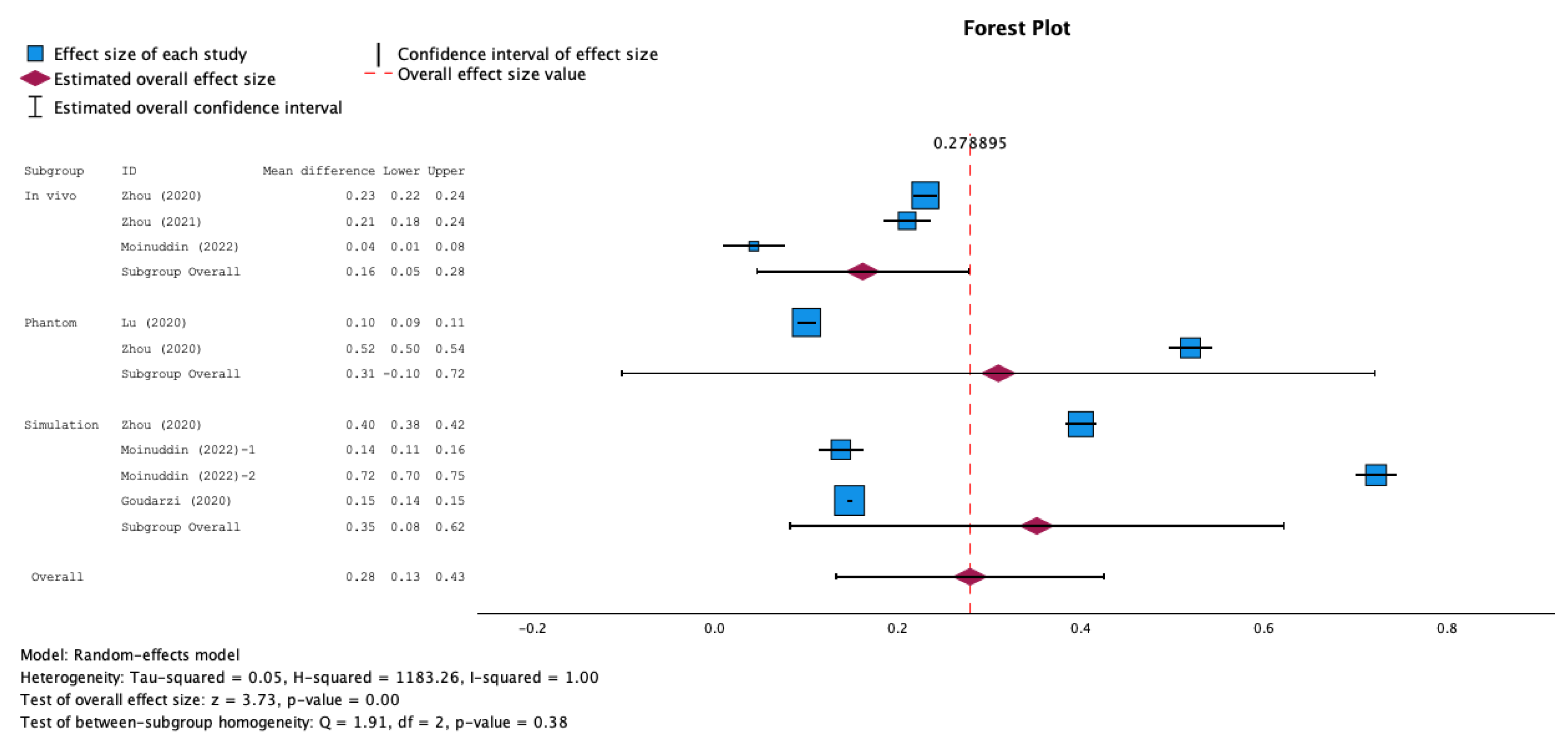
3.5. Meta-Analysis Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Search Strings
References
- Hashim, A.; Tahir, M.J.; Ullah, I.; Asghar, M.S.; Siddiqi, H.; Yousaf, Z. The utility of point of care ultrasonography (POCUS). Ann Med Surg (Lond) 2021, 71, 102982. [Google Scholar] [CrossRef] [PubMed]
- Riley, A.; Sable, C.; Prasad, A.; Spurney, C.; Harahsheh, A.; Clauss, S.; Colyer, J.; Gierdalski, M.; Johnson, A.; Pearson, G.D.; et al. Utility of hand-held echocardiography in outpatient pediatric cardiology management. Pediatr Cardiol 2014, 35, 1379–86. [Google Scholar] [CrossRef] [PubMed]
- Gilbertson, E.A.; Hatton, N.D.; Ryan, J.J. Point of care ultrasound: the next evolution of medical education. Ann Transl Med 2020, 8, 846. [Google Scholar] [CrossRef] [PubMed]
- Stock, K.F.; Klein, B.; Steubl, D.; Lersch, C.; Heemann, U.; Wagenpfeil, S.; Eyer, F.; Clevert, D.A. Comparison of a pocket-size ultrasound device with a premium ultrasound machine: diagnostic value and time required in bedside ultrasound examination. Abdom Imaging 2015, 40, 2861–6. [Google Scholar] [CrossRef] [PubMed]
- Han, P.J.; Tsai, B.T.; Martin, J.W.; Keen, W.D.; Waalen, J.; Kimura, B.J. Evidence basis for a point-of-care ultrasound examination to refine referral for outpatient echocardiography. The American Journal of Medicine 2019, 132, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wang, Y.; Guo, Y.; Qi, Y.; Yu, J. Image Quality Improvement of Hand-Held Ultrasound Devices With a Two-Stage Generative Adversarial Network. IEEE Trans Biomed Eng 2020, 67, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.P.; Sanghvi, A. Out of hospital point of care ultrasound: current use models and future directions. Eur J Trauma Emerg Surg 2016, 42, 139–50. [Google Scholar] [CrossRef] [PubMed]
- Kolbe, N.; Killu, K.; Coba, V.; Neri, L.; Garcia, K.M.; McCulloch, M.; Spreafico, A.; Dulchavsky, S. Point of care ultrasound (POCUS) telemedicine project in rural Nicaragua and its impact on patient management. J Ultrasound 2015, 18, 179–85. [Google Scholar] [CrossRef] [PubMed]
- Stewart, K.A.; Navarro, S.M.; Kambala, S.; Tan, G.; Poondla, R.; Lederman, S.; Barbour, K.; Lavy, C. Trends in Ultrasound Use in Low and Middle Income Countries: A Systematic Review. Int J MCH AIDS 2020, 9, 103–120. [Google Scholar] [CrossRef]
- Becker, D.M.; Tafoya, C.A.; Becker, S.L.; Kruger, G.H.; Tafoya, M.J.; Becker, T.K. The use of portable ultrasound devices in low- and middle-income countries: a systematic review of the literature. Trop Med Int Health 2016, 21, 294–311. [Google Scholar] [CrossRef]
- McBeth, P.B.; Hamilton, T.; Kirkpatrick, A.W. Cost-effective remote iPhone-teathered telementored trauma telesonography. Journal of Trauma and Acute Care Surgery 2010, 69, 1597–1599. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, A.; Galuppo, V.; Méndez, J.; Evangelista, L.; Arpal, L.; Rubio, C.; Vergara, M.; Liceran, M.; López, F.; Sales, C. Hand-held cardiac ultrasound screening performed by family doctors with remote expert support interpretation. Heart 2016, 102, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Salimi, N.; Gonzalez-Fiol, A.; Yanez, N.D.; Fardelmann, K.L.; Harmon, E.; Kohari, K.; Abdel-Razeq, S.; Magriples, U.; Alian, A. Ultrasound Image Quality Comparison Between a Handheld Ultrasound Transducer and Mid-Range Ultrasound Machine. Pocus j 2022, 7, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Guo, Y.; Wang, Y. Handheld Ultrasound Video High-Quality Reconstruction Using a Low-Rank Representation Multipathway Generative Adversarial Network. IEEE Transactions on Neural Networks and Learning Systems 2020, 32, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Huh, J.; Ye, J.C. Contrast and resolution improvement of pocus using self-consistent cyclegan. In Proceedings of the MICCAI Workshop on Domain Adaptation and Representation Transfer. Springer; pp. 158–167.
- Jafari, M.H.; Girgis, H.; Van Woudenberg, N.; Moulson, N.; Luong, C.; Fung, A.; Balthazaar, S.; Jue, J.; Tsang, M.; Nair, P. Cardiac point-of-care to cart-based ultrasound translation using constrained CycleGAN. International journal of computer assisted radiology and surgery 2020, 15, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Henderson, R.; Murphy, S. Portability enhancing hardware for a portable ultrasound system, 2017. US Patent No. 9,629, 606.
- Lockwood, G.R.; Talman, J.R.; Brunke, S.S. Real-time 3-D ultrasound imaging using sparse synthetic aperture beamforming. IEEE transactions on ultrasonics, ferroelectrics, and frequency control 1998, 45, 980–988. [Google Scholar] [CrossRef] [PubMed]
- Matrone, G.; Savoia, A.S.; Caliano, G.; Magenes, G. The delay multiply and sum beamforming algorithm in ultrasound B-mode medical imaging. IEEE transactions on medical imaging 2014, 34, 940–949. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, S.H.C.; Chiu, T.; Fox, M.D. Ultrasound image enhancement: A review. Biomedical Signal Processing and Control 2012, 7, 419–428. [Google Scholar] [CrossRef]
- Anaya-Isaza, A.; Mera-Jiménez, L.; Zequera-Diaz, M. An overview of deep learning in medical imaging. Informatics in medicine unlocked 2021, 26, 100723. [Google Scholar] [CrossRef]
- Zhang, H.M.; Dong, B. A review on deep learning in medical image reconstruction. Journal of the Operations Research Society of China 2020, 8, 311–340. [Google Scholar] [CrossRef]
- Liu, J.; Li, K.; Dong, H.; Han, Y.; Li, R. Medical Image Processing based on Generative Adversarial Networks: A Systematic Review. Curr Med Imaging 2023. [Google Scholar] [CrossRef] [PubMed]
- Lepcha, D.C.; Goyal, B.; Dogra, A.; Sharma, K.P.; Gupta, D.N. A deep journey into image enhancement: A survey of current and emerging trends. Information Fusion 2023, 93, 36–76. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Bmj 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Bhattacharya, M.; Pal, S.; Lee, S.S. From machine learning to deep learning: An advances of the recent data-driven paradigm shift in medicine and healthcare. Current Research in Biotechnology 2023, 100164. [Google Scholar] [CrossRef]
- Makwana, G.; Yadav, R.N.; Gupta, L. , 2022; pp. 303–313.Enhancement. In Internet of Things and Its Applications: Select Proceedings of ICIA 2020; pp. 2022303–313.
- Michailovich, O.V.; Tannenbaum, A. Despeckling of medical ultrasound images. ieee transactions on ultrasonics, ferroelectrics, and frequency control 2006, 53, 64–78. [Google Scholar] [CrossRef]
- Ng, A.; Swanevelder, J. Resolution in ultrasound imaging. Continuing Education in Anaesthesia, Critical Care & Pain 2011, 11, 186–192. [Google Scholar]
- Sara, U.; Akter, M.; Uddin, M.S. Image quality assessment through FSIM, SSIM, MSE and PSNR—a comparative study. Journal of Computer and Communications 2019, 7, 8–18. [Google Scholar] [CrossRef]
- Hore, A.; Ziou, D. Image quality metrics: PSNR vs. In SSIM. In Proceedings of the 2010 20th international conference on pattern recognition. IEEE; pp. 2366–2369.
- Awasthi, N.; van Anrooij, L.; Jansen, G.; Schwab, H.M.; Pluim, J.P.; Lopata, R.G. Bandwidth Improvement in Ultrasound Image Reconstruction Using Deep Learning Techniques. In Proceedings of the Healthcare. MDPI, Vol. 11; p. 123.
- Gasse, M.; Millioz, F.; Roux, E.; Garcia, D.; Liebgott, H.; Friboulet, D. High-quality plane wave compounding using convolutional neural networks. IEEE transactions on ultrasonics, ferroelectrics, and frequency control 2017, 64, 1637–1639. [Google Scholar] [CrossRef]
- Goudarzi, S.; Asif, A.; Rivaz, H. Fast multi-focus ultrasound image recovery using generative adversarial networks. IEEE Transactions on Computational Imaging 2020, 6, 1272–1284. [Google Scholar] [CrossRef]
- Guo, B.; Zhang, B.; Ma, Z.; Li, N.; Bao, Y.; Yu, D. High-quality plane wave compounding using deep learning for hand-held ultrasound devices. In Proceedings of the Advanced Data Mining and Applications: 16th International Conference, ADMA 2020, Foshan, China, 2020, Proceedings 16. Springer, November 12–14; pp. 547–559.
- Huang, C.Y.; Chen, O.T.C.; Wu, G.Z.; Chang, C.C.; Hu, C.L. Ultrasound imaging improved by the context encoder reconstruction generative adversarial network. In Proceedings of the 2018 IEEE International Ultrasonics Symposium (IUS). IEEE; pp. 1–4.
- Lu, J.; Millioz, F.; Garcia, D.; Salles, S.; Liu, W.; Friboulet, D. Reconstruction for diverging-wave imaging using deep convolutional neural networks. IEEE transactions on ultrasonics, ferroelectrics, and frequency control 2020, 67, 2481–2492. [Google Scholar] [CrossRef]
- Lyu, Y.; Jiang, X.; Xu, Y.; Hou, J.; Zhao, X.; Zhu, X. ARU-GAN: U-shaped GAN based on Attention and Residual connection for super-resolution reconstruction. Computers in Biology and Medicine 2023, 164, 107316. [Google Scholar] [CrossRef] [PubMed]
- Moinuddin, M.; Khan, S.; Alsaggaf, A.U.; Abdulaal, M.J.; Al-Saggaf, U.M.; Ye, J.C. Medical ultrasound image speckle reduction and resolution enhancement using texture compensated multi-resolution convolution neural network. Frontiers in Physiology 2022, 2326. [Google Scholar] [CrossRef] [PubMed]
- Monkam, P.; Lu, W.; Jin, S.; Shan, W.; Wu, J.; Zhou, X.; Tang, B.; Zhao, H.; Zhang, H.; Ding, X. US-Net: A lightweight network for simultaneous speckle suppression and texture enhancement in ultrasound images. Computers in Biology and Medicine 2023, 152, 106385. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Zou, B.; Li, C.; Feng, S.; Peng, H. Plane-Wave Image Reconstruction via Generative Adversarial Network and Attention Mechanism. IEEE Transactions on Instrumentation and Measurement 2021, 70. [Google Scholar] [CrossRef]
- Zhang, X.; Li, J.; He, Q.; Zhang, H.; Luo, J. High-quality reconstruction of plane-wave imaging using generative adversarial network. In Proceedings of the 2018 IEEE International Ultrasonics Symposium (IUS). IEEE; pp. 1–4.
- Zhou, Z.; Wang, Y.; Yu, J.; Guo, W.; Fang, Z. Super-resolution reconstruction of plane-wave ultrasound imaging based on the improved CNN method. In Proceedings of the VipIMAGE 2017: Proceedings of the VI ECCOMAS Thematic Conference on Computational Vision and Medical Image Processing Porto, Portugal, October 18-20, 2017.; pp. 111–120.
- Zhou, Z.; Wang, Y.; Yu, J.; Guo, Y.; Guo, W.; Qi, Y. High Spatial-Temporal Resolution Reconstruction of Plane-Wave Ultrasound Images With a Multichannel Multiscale Convolutional Neural Network. IEEE Trans Ultrason Ferroelectr Freq Control 2018, 65, 1983–1996. [Google Scholar] [CrossRef] [PubMed]
- Goudarzi, S.; Asif, A.; Rivaz, H. High Frequency Ultrasound Image Recovery Using Tight Frame Generative Adversarial Networks. Annu Int Conf IEEE Eng Med Biol Soc 2020, 2020, 2035–2038. [Google Scholar] [CrossRef] [PubMed]
- Goudarzi, S.; Asif, A.; Rivaz, H. Multi-focus ultrasound imaging using generative adversarial networks. In Proceedings of the 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019). IEEE; pp. 1118–1121.
- Lu, J.; Liu, W. Unsupervised super-resolution framework for medical ultrasound images using dilated convolutional neural networks. In Proceedings of the 2018 IEEE 3rd International Conference on Image, Vision and Computing (ICIVC). IEEE; pp. 739–744.
- Li, Y.; Lu, W.; Monkam, P.; Wang, Y. IA-Noise2Noise: An Image Alignment Strategy for Echocardiography Despeckling. In Proceedings of the 2023 IEEE International Ultrasonics Symposium (IUS). IEEE; pp. 1–3.
- Mansouri, N.J.; Khaissidi, G.; Despaux, G.; Mrabti, M.; Clézio, E.L. Attention gated encoder-decoder for ultrasonic signal denoising. IAES International Journal of Artificial Intelligence 2023, 12, 1695–1703. [Google Scholar] [CrossRef]
- Basile, M.; Gibiino, F.; Cavazza, J.; Semplici, P.; Bechini, A.; Vanello, N. Blind Approach Using Convolutional Neural Networks to a New Ultrasound Image Denoising Task. In Proceedings of the 2023 IEEE International Workshop on Biomedical Applications, Technologies and Sensors, BATS 2023 - Proceedings; pp. 68–73. [Google Scholar] [CrossRef]
- Shen, Z.; Tang, C.; Xu, M.; Lei, Z. Removal of Speckle Noises from Ultrasound Images Using Parallel Convolutional Neural Network. Circuits, Systems, and Signal Processing 2023, 42, 5041–5064. [Google Scholar] [CrossRef]
- Gan, J.; Wang, L.; Liu, Z.; Wang, J. Multi-scale ultrasound image denoising algorithm based on deep learning model for super-resolution reconstruction. In Proceedings of the ACM International Conference Proceeding Series; pp. 6–11. [CrossRef]
- Asgariandehkordi, H.; Goudarzi, S.; Basarab, A.; Rivaz, H. Deep Ultrasound Denoising Using Diffusion Probabilistic Models. In Proceedings of the IEEE International Ultrasonics Symposium, IUS. [CrossRef]
- Liu, J.; Li, C.; Liu, L.; Chen, H.; Han, H.; Zhang, B.; Zhang, Q. Speckle noise reduction for medical ultrasound images based on cycle-consistent generative adversarial network. Biomedical Signal Processing and Control 2023, 86. [Google Scholar] [CrossRef]
- Mahmoudi Mehr, O.; Mohammadi, M.R.; Soryani, M. Deep Learning-Based Ultrasound Image Despeckling by Noise Model Estimation. Iranian Journal of Electrical and Electronic Engineering 2023, 19. [Google Scholar] [CrossRef]
- Senthamizh Selvi, R.; Suruthi, S.; Samyuktha Shrruthi, K.R.; Varsha, B.; Saranya, S.; Babu, B. Ultrasound Image Denoising Using Cascaded Median Filter and Autoencoder. In Proceedings of the Proceedings of the 4th International Conference on Smart Electronics and Communication, ICOSEC 2023, pp.; pp. 296–302. [CrossRef]
- Mikaeili, M.; Bilge, H.S. Evaluating Deep Neural Network Models on Ultrasound Single Image Super Resolution. In Proceedings of the TIPTEKNO 2023 - Medical Technologies Congress, Proceedings. [CrossRef]
- Liu, H.; Liu, J.; Hou, S.; Tao, T.; Han, J. Perception consistency ultrasound image super-resolution via self-supervised CycleGAN. Neural Computing and Applications 2023, 35, 12331–12341. [Google Scholar] [CrossRef]
- Vetriselvi, D.; Thenmozhi, R. Advanced Image Processing Techniques for Ultrasound Images using Multiscale Self Attention CNN. Neural Processing Letters 2023, 55, 11945–11973. [Google Scholar] [CrossRef]
- Gomez, Y.Z.O.; Costa, E.T. Ultrasound Speckle Filtering Using Deep Learning. In Proceedings of the IFMBE Proceedings, Vol. 99; pp. 283–289. [CrossRef]
- Li, Y.; Zeng, X.; Dong, Q.; Wang, X. RED-MAM: A residual encoder-decoder network based on multi-attention fusion for ultrasound image denoising. Biomedical Signal Processing and Control 2023, 79. [Google Scholar] [CrossRef]
- Yang, T.; Wang, W.; Cheng, G.; Wei, M.; Xie, H.; Wang, F.L. FDDL-Net: frequency domain decomposition learning for speckle reduction in ultrasound images. Multimedia Tools and Applications 2022, 81, 42769–42781. [Google Scholar] [CrossRef]
- Karaoğlu, O.; Bilge, H.S.; Uluer, I. Removal of speckle noises from ultrasound images using five different deep learning networks. Engineering Science and Technology, an International Journal 2022, 29. [Google Scholar] [CrossRef]
- Markco, M.; Kannan, S. Texture-driven super-resolution of ultrasound images using optimized deep learning model. Imaging Science Journal 2023. [Google Scholar] [CrossRef]
- Karthiha, G.; Allwin, S. Speckle Noise Suppression in Ultrasound Images Using Modular Neural Networks. Intelligent Automation and Soft Computing 2023, 35, 1753–1765. [Google Scholar] [CrossRef]
- Kalaiyarasi, M.; Janaki, R.; Sampath, A.; Ganage, D.; Chincholkar, Y.D.; Budaraju, S. Non-additive noise reduction in medical images using bilateral filtering and modular neural networks. Soft Computing 2023. [Google Scholar] [CrossRef]
- Sawant, A.; Kasar, M.; Saha, A.; Gore, S.; Birwadkar, P.; Kulkarni, S. Medical Image De-Speckling Using Fusion of Diffusion-Based Filters And CNN. In Proceedings of the 8th International Conference on Advanced Computing and Communication Systems, ICACCS 2022; pp. 1197–1203. [Google Scholar] [CrossRef]
- Dutta, S.; Georgeot, B.; Kouame, D.; Garcia, D.; Basarab, A. Adaptive Contrast Enhancement of Cardiac Ultrasound Images using a Deep Unfolded Many-Body Quantum Algorithm. In Proceedings of the IEEE International Ultrasonics Symposium, IUS, Vol. 2022-October. [Google Scholar] [CrossRef]
- Sanjeevi, G.; Krishnan Pathinarupothi, R.; Uma, G.; Madathil, T. Deep Learning Pipeline for Echocardiogram Noise Reduction. In Proceedings of the 2022 IEEE 7th International conference for Convergence in Technology, I2CT 2022. [Google Scholar] [CrossRef]
- Makwana, G.; Yadav, R.N.; Gupta, L. Analysis of Various Noise Reduction Techniques for Breast Ultrasound Image Enhancement. In Proceedings of the Lecture Notes in Electrical Engineering, Vol. 825; pp. 303–313. [CrossRef]
- Suseela, K.; Kalimuthu, K. An efficient transfer learning-based Super-Resolution model for Medical Ultrasound Image. In Proceedings of the Journal of Physics: Conference Series, Vol. 1964. [Google Scholar] [CrossRef]
- Chennakeshava, N.; Luijten, B.; Drori, O.; Mischi, M.; Eldar, Y.C.; Van Sloun, R.J.G. High resolution plane wave compounding through deep proximal learning. In Proceedings of the IEEE International Ultrasonics Symposium, IUS, Vol. 2020-September. [Google Scholar] [CrossRef]
- Dong, G.; Ma, Y.; Basu, A. Feature-Guided CNN for Denoising Images from Portable Ultrasound Devices. IEEE Access 2021, 9, 28272–28281. [Google Scholar] [CrossRef]
- Jarosik, P.; Lewandowski, M.; Klimonda, Z.; Byra, M. Pixel-Wise Deep Reinforcement Learning Approach for Ultrasound Image Denoising. In Proceedings of the IEEE International Ultrasonics Symposium, IUS. [CrossRef]
- Kumar, M.; Mishra, S.K.; Joseph, J.; Jangir, S.K.; Goyal, D. Adaptive comprehensive particle swarm optimisation-based functional-link neural network filtre model for denoising ultrasound images. IET Image Processing 2021, 15, 1232–1246. [Google Scholar] [CrossRef]
- Shen, Z.; Li, W.; Han, H. Deep Learning-Based Wavelet Threshold Function Optimization on Noise Reduction in Ultrasound Images. Scientific Programming 2021, 2021. [Google Scholar] [CrossRef]
- Kokil, P.; Sudharson, S. Despeckling of clinical ultrasound images using deep residual learning. Computer Methods and Programs in Biomedicine 2020, 194. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Huang, Q.; Li, X. Ultrasound image de-speckling by a hybrid deep network with transferred filtering and structural prior. Neurocomputing 2020, 414, 346–355. [Google Scholar] [CrossRef]
- Ma, Y.; Yang, F.; Basu, A. Edge-guided CNN for denoising images from portable ultrasound devices. In Proceedings of the Proceedings - International Conference on Pattern Recognition; pp. 6826–6833. [CrossRef]
- Lan, Y.; Zhang, X. Real-time ultrasound image despeckling using mixed-attention mechanism based residual UNet. IEEE Access 2020, 8, 195327–195340. [Google Scholar] [CrossRef]
- Vasavi, G.; Jyothi, S. Noise Reduction Using OBNLM Filter and Deep Learning for Polycystic Ovary Syndrome Ultrasound Images. In Proceedings of the Learning and Analytics in Intelligent Systems, Vol. 16; pp. 203–212. [CrossRef]
- Shelgaonkar, S.L.; Nandgaonkar, A.B. Deep Belief Network for the Enhancement of Ultrasound Images with Pelvic Lesions. Journal of Intelligent Systems 2018, 27, 507–522. [Google Scholar] [CrossRef]
- Singh, P.; Mukundan, R.; De Ryke, R. Feature Enhancement of Medical Ultrasound Scans Using Multifractal Measures. In Proceedings of the Proceedings - 2019 IEEE International Conference on Signals and Systems, ICSigSys 2019; pp. 85–91. [Google Scholar] [CrossRef]
- Choi, W.; Kim, M.; Haklee, J.; Kim, J.; Beomra, J. Deep CNN-Based Ultrasound Super-Resolution for High-Speed High-Resolution B-Mode Imaging. In Proceedings of the IEEE International Ultrasonics Symposium, IUS, Vol. 2018-January. [Google Scholar] [CrossRef]
- Ando, K.; Nagaoka, R.; Hasegawa, H. Speckle reduction of medical ultrasound images using deep learning with fully convolutional network. Japanese Journal of Applied Physics 2020, 59. [Google Scholar] [CrossRef]
- Temiz, H.; Bilge, H.S. Super Resolution of B-Mode Ultrasound Images with Deep Learning. IEEE Access 2020, 8, 78808–78820. [Google Scholar] [CrossRef]
- Liu, J.; Liu, H.; Zheng, X.; Han, J. Exploring multi-scale deep encoder-decoder and patchgan for perceptual ultrasound image super-resolution. In Proceedings of the Communications in Computer and Information Science, Vol. 1265 CCIS; pp. 47–59. [Google Scholar] [CrossRef]
- Mishra, D.; Chaudhury, S.; Sarkar, M.; Soin, A.S. Ultrasound image enhancement using structure oriented adversarial network. IEEE Signal Processing Letters 2018, 25, 1349–1353. [Google Scholar] [CrossRef]
- Mishra, D.; Tyagi, S.; Chaudhury, S.; Sarkar, M.; Singhsoin, A. Despeckling CNN with Ensembles of Classical Outputs. In Proceedings of the Proceedings - International Conference on Pattern Recognition, Vol. 2018-August; pp. 3802–3807. [Google Scholar] [CrossRef]
- Oliveira-Saraiva, D.; Mendes, J.; Leote, J.; Gonzalez, F.A.; Garcia, N.; Ferreira, H.A.; Matela, N. Make It Less Complex: Autoencoder for Speckle Noise Removal-Application to Breast and Lung Ultrasound. J Imaging 2023, 9. [Google Scholar] [CrossRef]
- Vimala, B.B.; Srinivasan, S.; Mathivanan, S.K.; Muthukumaran, V.; Babu, J.C.; Herencsar, N.; Vilcekova, L. Image Noise Removal in Ultrasound Breast Images Based on Hybrid Deep Learning Technique. Sensors (Basel) 2023, 23. [Google Scholar] [CrossRef]
- Sineesh, A.; Shankar, M.R.; Hareendranathan, A.; Panicker, M.R.; Palanisamy, P. Single Image based Super Resolution Ultrasound Imaging Using Residual Learning of Wavelet Features. Annu Int Conf IEEE Eng Med Biol Soc 2023, 2023, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y.; Zhao, Y.; Wei, Y. Fast Speckle Noise Suppression Algorithm in Breast Ultrasound Image Using Three-Dimensional Deep Learning. Front Physiol 2022, 13, 880966. [Google Scholar] [CrossRef] [PubMed]
- Tamang, L.D.; Kim, B.W. Super-Resolution Ultrasound Imaging Scheme Based on a Symmetric Series Convolutional Neural Network. Sensors (Basel) 2022, 22. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, M.; Chung, K.; Kuppoor, V.; Mahapatra, S.; Pustavoitau, A.; Manbachi, A. USDL: Inexpensive Medical Imaging Using Deep Learning Techniques and Ultrasound Technology. Proc Des Med Devices Conf 2020, 2020. [Google Scholar] [CrossRef]
- Yu, H.; Ding, M.; Zhang, X.; Wu, J. PCANet based nonlocal means method for speckle noise removal in ultrasound images. PLoS One 2018, 13, e0205390. [Google Scholar] [CrossRef] [PubMed]
- S, L.S.; M, S. Bayesian Framework-Based Adaptive Hybrid Filtering for Speckle Noise Reduction in Ultrasound Images Via Lion Plus FireFly Algorithm. J Digit Imaging 2021, 34, 1463–1477. [Google Scholar] [CrossRef]
- Liebgott, H.; Rodriguez-Molares, A.; Cervenansky, F.; Jensen, J.A.; Bernard, O. Plane-wave imaging challenge in medical ultrasound. In Proceedings of the 2016 IEEE International ultrasonics symposium (IUS). IEEE; pp. 1–4.
- Yap, M.H.; Pons, G.; Marti, J.; Ganau, S.; Sentis, M.; Zwiggelaar, R.; Davison, A.K.; Marti, R. Automated breast ultrasound lesions detection using convolutional neural networks. IEEE journal of biomedical and health informatics 2017, 22, 1218–1226. [Google Scholar] [CrossRef]
- Xia, C.; Li, J.; Chen, X.; Zheng, A.; Zhang, Y. What is and what is not a salient object? learning salient object detector by ensembling linear exemplar regressors. In Proceedings of the Proceedings of the IEEE conference on computer vision and pattern recognition, pp.; pp. 4142–4150.
- van den Heuvel, T.L.; de Bruijn, D.; de Korte, C.L.; Ginneken, B.v. Automated measurement of fetal head circumference using 2D ultrasound images. PloS one 2018, 13, e0200412. [Google Scholar] [CrossRef]

| Study | Aim | Dataset (availability) | Ultrasound specifications | Deep learning algorithm | Loss function |
| Awasthi et al., 2022 [32] | Reconstruction of high-quality high-bandwidth images from low-bandwidth images | Phantom: Five separate datasets, tissue-mimicking, commercial and in vitro porcin carotid artery. (private) | Verasonics, L11-5v transducer with PWs at range -25∘ to 25∘. LQ: limited bandwidth down to 20%, HQ: full bandwidth. | Residual encoder decoder Net | Scaled MSE |
| Gasse et al., 2017 [33] | Reconstruct high-quality US images from a small number of PW acquisitions | In-vivo: carotid, thyroid and liver regions of healthy subjects; Phantom: Gammex. (private) | Verasonics, ATL L7-4 probe (5.2 MHz, 128 elements) with range ±15∘. LQ: 3 PWs. HQ: 31 PWs. | CNN | L2 loss |
| Goudarzi et al., 2020 [34] | Achieve the quality of multifocus US images by using a mapping function on a single-focus US image. | Phantom: CIRS phantom and ex vivo lamb liver;Simulation: Field II software. (private) | E-CUBE 12 Alpinion machine, L3-12H transducer (8.5 MHz). LQ: image with single focal point. HQ: multi-focus image with 3 focal points. | Boundary-Seeking GAN | Binary cross entropy (discriminator), MSE + boundary seeking loss (generator) |
| Guo et al., 2020 [35] | Improve the quality of handheld US devices using a small number of plane waves | In vivo: dataset provided by Zhang et al. [42] (carotid artery and brachioradialis images of healthy volunteers);Phantom: PICMUS [98] dataset, CIRS phantom;Simulation: US images from natural images using field-II software (only for pre-training LG_Unet). (private and public) | (Derived from dataset sources)In vivo: Verasonics, L10-5 probe (7.5 MHz). LQ: 3 PWs, HQ: Compounded image of 31 PWs with range -15∘ to 15∘.Phantom data:Verasonics, L11 probe. (5.2 MHz, 128 elements). | Local Global Unet (LG-Unet) + Simplified residual network (S_ResNet) | MSE + SSIM (LG_Unet) and L1 (S_Resnet) |
| Huang et al., 2018 [36] | Improve the quality of ultrasonic B-mode images from 32 to that from 128 channels. | Simulation: Field-II software. (private) | Simulation data set at 5MHz center frequency, 0.308mm pitch, 71% bandwidth. LQ: 32-channel image, HQ: 128-channel image. | Context encoder reconstruction GAN | Not reported |
| Khan et al., 2021 [15] | Contrast and resolution enhancement of handheld POCUS images | In vivo: carotid and thyroid regions;Phantom: ATS-539 phantom;Simulation: intermediate domain images generated by down grading the in vivo and phantom images acquired from high-end system. (private) | LQ: NPUS050 portable US system were used as low-quality input. HQ: E-CUBE 12R US, L3–12 transducer. | Cascade application of unsupervised self-consistent CycleGAN + supervised super-resolution network. | Cycle consistency + adversarial loss (cycleGAN), MAE + SSIM (super-resolution network) |
| Lu et al., 2020 [37] | High-quality reconstruction for DW imaging using a small number (3) of DW transmissions competing with those obtained by compounding with 31 DWS | In vivo: Thigh muscle, finger phalanx, and liver regions;Phantom: CIRS and Gammex. (private) | Verasonics, ATL P4-2 transducer. LQ: 3 DWs, HQ: Compounded image of 31 DWs. | CNN with inception module | MSE |
| Lyu et al., 2023 [38] | Reconstruct super-resolution high-quality images from single-beam plane-wave images | PICMUS 2016 dataset [98] modulated following the guidelines of CUBDL, consisting ofSimulation: generated with Field II software;Phantom: CIRS;In vivo: carotid artery of healthy volunteer. (public) | (Derived from dataset source)Verasonics, L11 probe with range -16∘, 16∘. LQ: single PW image. HQ: PW images synthesized from 75 different angles using CPWC | U-shaped GAN based on Attention and Residual connection (ARU-GAN) | Combination of MS-SSIM, classical adversarial and perceptual loss |
| Moinuddin et al., 2022 [39] | Enhance US images using a network where the task of noise suppression and resolution enhancement are carried out simultaneously. | In vivo: breast US (BUS) dataset [99], for which high resolution and low noise label images are generated using NLLR normal filtration;Simulation: Salient object detection (SOD) dataset [100] augmentated using image formaton physics information, divided in two datasets. (public) | (Derived from dataset source)Siemens ACUSON Sequoia C512, 17L5 HD transducer (8.5 MHz) | Deep CNN | MSE |
| Monkam et al., 2023 [40] | Suppress speckle noise and enhance texture and fine-details. | Simulation: original low-quality US images of HC18 Challenge fetal data set [101], from which high-quality target images and additional low-quality images are generated (for training and testing);In vivo publicly available datasets: HC18 Challenge (fetal) [101], BUSI (breast), CCA (common carotid artery) (for testing). (public) | (Derived from HC18 dataset source)Voluson E8 or the Voluson 730 US device. | U-Net with added feature refinement attention block (US-Net) | L1 loss |
| Tang et al., 2021 [41] | Reconstruct high-resolution, high-quality plane-wave images from low-quality plane-wave images from different angles. | PICMUS 2016 dataset [98] modulated following the guidelines of CUBDL, consisting ofSimulation: generated with Field II software;Phantom: CIRS;In vivo: carotid artery of healthy volunteer. (public) | (Derived from dataset source)Verasonics, L11 probe with range -16∘, 16∘. LQ: PW image using 3 angles. HQ: PW images synthesized from 75 different angles using CPWC | Attention mechanism and Unet-based GAN | cross-entropy + MSE + perceptual loss |
| Zhang et al., 2018 [42] | Reconstruct high-quality US images from small number of PWs (3). | In vivo: carotid artery and brachioradioalis of heathy volunteer;Phantom: CIRS phantom, ex vivo swine muscles. (private) | Verasonics, L10-5 (7.5 MHz) with range -15∘ to 15∘. LQ: 3 PWs, HQ: coherent compounding using 31 PWs. | GAN, with feed-forward CNN as both generator and discriminator network | MSE + adversarial loss (generator), binary cross entropy (discriminator) |
| Zhou et al., 2018 [43] | Improve the image quality of a single angle PW image to that of a PW image synthesized from 75 different angles | PICMUS 2016 dataset [98] synthesized by three different beamforming methods:In vivo: 1) thyroid gland and 2) carotid artery of human volunteers. (public)Phantom: CIRS phantom;Simulation: 1) point images and 2) cyst images generated using Field-II software. | (Derived from dataset sources)Verasonics, L11 probe with range -16∘, 16∘. LQ: single PW image. HQ: PW images synthesized from 75 different angles. | Multi-scaled CNN | MSE |
| Zhou et al., 2020 [6] | Improve quality of portable US, by mapping low-quality images to corresponding high-quality images. | Single-/multiangle PWI simulation, phantom and in vivo data (only used for transfer learning). For training and testing:In vivo: carotid and thyroid images of healthy volunteers;Phantom: CIRS and self-made gelatin and raw pork;Simulation: Field-II software. (private) | LQ: mSonics MU1, L10-5v. transducer. HQ: Verasonics, L11-4v transducer (phantom data) and Toshiba Aplio 500, 7.5 MHz (clinical data). | Two-stage GAN with U-Net and gradual learning strategy. | MSE + SSIM + Conv loss |
| Zhou et al., 2021 [14] | Enhance video quality of handheld US devices. | In vivo: single and multiangle PW videos (only for training). Handheld and high-end images and videos of different bodyparts of healthy volunteers (for training and testing). (private) | PW videos: Verasonics, L11-4v transducer (6.25MHz, 128-element) with range -16∘ to 16∘. High-end US (HQ): Toshiba Aplio 500 device. Handheld US (LQ): mSonics MU1, L10-5 transducer. | Low-rank representation multipathway GAN | adversarial + MSE + ultrasound specific perceptual loss |
| Study | Computation time(source code availability) | Number of images in test set | Performance (±SD) of low-quality input image | Performance (±SD) of generated image |
| Awasthi et al., 2022 [32] | "Light weight" (available) | Phantom:dataset 1: n=134dataset 2: n=90dataset 3: n=31dataset 4: n=70dataset 5: n=239 | Phantom:dataset 1: PSNR=17.049±1.107, RMSE=0.141±0.016, PC=0.788dataset 2: PSNR=15.768±1.376, RMSE=0.165±0.026dataset 3: PSNR=13.885±1.276, RMSE=0.204±0.032dataset 4: PSNR=16.297±1.212, RMSE=0.155±0.021dataset 5: PSNR=15.487±1.876, RMSE=0.172±0.040 | Phantom:dataset 1: PSNR=20.903±1.189, RMSE=0.091±0.012, PC=0.86dataset 2: PSNR=20.523±1.242, RMSE=0.095±0.013dataset 3: PSNR=13.985±1.120, RMSE=0.201±0.025dataset 4: PSNR=21.457±1.238, RMSE=0.085±0.012dataset 5: PSNR=17.654±1.536, RMSE=0.133±0.022 |
| Gasse et al., 2017 [33] | Not reported (not available) | Mixed test set of in vivo and phantom data:n=1000 | Only graphs given, showing CR and LR reached by the proposed model with 3 PWs compared to the standard compounding of an increasingly larger number of PWs. | - |
| Goudarzi et al., 2020 [34] | Not reported (available) | Phantom (CIRS):n=-Simulation:n=360 | Phantom:FWHM=1.52, CNR=9.6Simulation:SSIM=0.622±0.02, PSNR=23.27±1, FWHM=1.3, CNR=7.2 | Phantom:FWHM=1.44, CNR=11.1Simulation:SSIM=0.769±0.017, PSNR=25.32±0.919, FWHM=1.09, CNR=8.02 |
| Guo et al., 2020 [35] | Not reported (not available) | 225 (out of 9225) patch images from the in vivo, phantom and simulation dataset (distribution between datasets not reported) | In vivo:PSNR=16.04Phantom:FWHM=1.8 mm, CR=0.36, CNR=24.93 | In vivo:PSNR=18.94Phantom:FWHM=1.3 mm, CR=0.79, CNR=32.81 |
| Huang et al., 2018 [36] | Not reported (not available) | Simulation:n=1 | Simulation:CNR: 0.939, PICMUS CNR: 2.381, FWHM: 13.34 | Simulation:CNR: 1.508, PICMUS CNR: 6.502, FWHM: 11.15 |
| Khan et al., 2021 [15] | 13.18 ms (not available) | In vivo:n=43Phantom:n=32 | Not reported | Gain compared to simulated intermediate quality images of in vivo and phantom data (only measuring fitness of super-resolution network):PSNR=13.58, SSIM=0.63Non-reference metrics for entire proposed method for in vivo and phantom data:CR=14.96, CNR=2.38, GCNR=0.8604 (which is 21.77%, 30.06%, and 44.42% higher than those of the low-quality input images.) |
| Lu et al., 2020 [37] | 0.75 ± 0.03 ms (not available) | Mixed in vivo and phantom data:n=1000 | Mixed in vivo and phantom data:PSNR=29.24±1.57, SSIM=0.83±0.15, MI=0.51±0.16Non-reference metrics are only shown in graph form for low-quality images. | Mixed in vivo and phantom data:PSNR=31.13±1.47, SSIM=0.93±0.06, MI=0.82±0.20,CR (near field)=19.54, CR (far field)=14.95, CNR (near field)=7.63, CNR (far field)=5.21, LR (near field)=0.90, LR (middle field)=1.64, LR (far field)=2.35 |
| Lyu et al., 2023 [38] | Not reported (not available) | In vivo:n=150Phantom:n=150Simulation:n=150 | No performance metrics available for low-quality images, only for other traditional deep learning methods for comparison. | In vivo:PSNR=26.508, CW-SSIM=0.876, NCC=0.943Phantom:FWHM=0.424, CR=26.900, CNR=3.693Simulation:FWHM=0.277, CR=39.472, CNR=5.141 |
| Moinuddin et al., 2022 [39] | Not reported | In vivo:n=33Simulation:SOD-1: n=200SOD-2: n=200Evaluated with 5-fold cross-validation approach. | In vivo:PSNR=26.0071±2.3083, SSIM= 0.7098 ± 0.0761Simulation:SOD-1: PSNR=12.1587±0.7839, SSIM=0.5570±0.1205SOD-2: PSNR=12.5272±0.8243, SSIM=0.1556±0.1451,GCNR=0.9936±0.0039 | In vivo:PSNR=26.9112±2.3025, SSIM=0.7522±0.0635Simulation:SOD-1: PSNR=25.5275±2.9712, SSIM=0.6946±0.1267SOD-2: PSNR=32.4719±2.6179, SSIM=0.8785±0.0766,GCNR=0.9966±0.0026 |
| Monkam et al., 2023 [40] | 52.16 ms (not available) | In vivo:HC18: n=30BUSI: n=30CCA: n=30Simulation:HC18: n=335 | No performance metrics available for low-quality images, only for other enhancement methods for comparison. | In vivo:HC18: SNR=39.32, CNR=1.10, AGM=27.46, ENL=15.71BUSI: SNR=34.54, CNR=4.20, AGM=39.88, ENL=17.04CCA: SNR=40.87, CNR=2.59, AGM=35.92, ENL=23.03Simulation:HC18: SSIM=0.9155, PSNR=32.87, EPI= 0.6371 |
| Tang et al., 2021 [41] | Not reported (not available) | n=360 (total number of images in test set for the in vivo, phantom and simulation datasets, distribution not reported) | Phantom:FWHM=0.5635, CR=8.718, CNR=1.109, GCNR=0.609Simulation:FWHM=0.2808, CR=13.769, CNR=1.576, GCNR=0.735 | In vivo:PSNR=28.278, SSIM=0.659, MI=0.9980, NCC=0.963Phantom:FWHM=0.3556, CR=24.571, CNR=2.495, GCNR=0.915Simulation:FWHM=0.2695, CR=39.484, CNR=5.617, GCNR=0.998 |
| Zhang et al., 2018 [42] | Not reported (not available) | In vivo:n=500phantom:n=30 | Mixed in vivo and phantom test set:FWHM=0.50, CR=10.23, CNR=1.30 | Mixed in vivo and phantom test set:FWHM=0.53, CR=19.46, CNR=2.25 |
| Zhou et al., 2018 [43] | Not reported (not available) | In vivo:Thyroid dataset: n=30Simulation:Point dataset: n=30Cyst dataset: n=30Evaluated with 5-fold cross-validation approach. | In vivo:Thyroid dataset: PSNR=14.9235, SSIM=0.0291, MI=0.3474Simulation: Point dataset: PSNR=24.1708, SSIM=0.1962, MI=0.4124,FWHM=0.49Cyst dataset: PSNR=15.8860, SSIM=0.5537, MI=1.1976,CR=137.0473 | In vivo: Thyroid dataset: PSNR=21.7248, SSIM=0.3034, MI=0.8856Simulation: Point dataset: PSNR=36.5884, SSIM=0.9216, MI=0.4483,FWHM=0.196 Cyst dataset: PSNR=24.0167, SSIM=0.6135, MI=1.5622,CR=184.0432 |
| Zhou et al., 2020 [6] | Not reported (not available) | In vivo:n=94Phantom:n=40Simulation:n=56Evaluated with 5-fold cross validation approach. | In vivo: PSNR=8.65±1.32, SSIM=0.18±0.04, MI=0.22±0.13,BRISQUE=38.91±4.99Phantom: PSNR=15.26±2.91, SSIM=0.12±0.03, MI=0.20±0.11,BRISQUE=24.61±4.50Simulation: PSNR=16.38±2.35, SSIM=0.19±0.06, MI=0.22±0.16,BRISQUE=29.08±3.45 | In vivo: PSNR=18.08±1.57, SSIM=0.41±0.05, MI=0.68±0.18,BRISQUE=35.25±4.13Phantom: PSNR=24.70±1.11, SSIM=0.64±0.07, MI=0.26±0.09,BRISQUE=21.68±3.36Simulation: PSNR=28.50±2.01, SSIM=0.59±0.02, MI=0.42±0.04,BRISQUE=23.30±3.09 |
| Zhou et al., 2021 [14] | Not reported (not available) | In vivo:n=40 videosFor full-reference methods, a single frame in handheld video was used and most similar frame in high-end video was selected. | In vivo:PSNR=12.68±3.45, SSIM=0.24±0.06, MI=0.71±0.09,NIQE=19.48±4.66, ultrasound quality score=0.06±0.03 | In vivo:PSNR=19.95±3.24, SSIM=0.45±0.06, MI=1.05±0.07,NIQE=6.95±1.97, ultrasound quality score=0.89±0.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





