1. Introduction
The rapid emergence of drug resistance in pathogenic bacteria poses a catastrophic threat all around the world. At the same time, the unavailability of new drugs or anti-infective therapies creates an alarming situation concerning global health in the battle against drug resistance [
1]. Overall, there is an urgent need for new antimicrobial or alternative strategies to combat drug-resistant pathogens. Microbial secondary metabolites consist of structurally diverse natural products which are essential phenomena of microbial communities, playing a diverse role in biotic and abiotic interactions [
2]. Bacteriocins are one such secondary metabolites produced by bacterial communities in competitive complex environments such as soil, human gut, or marine sediments [
3,
4,
5]. Bacteriocins are ribosomally-synthesized and posttranslationally modified peptides (RiPPs) and have already been suggested as a viable alternative to conventional antibiotics [
6,
7]. As bacterial communities and their diverse intra or inter-species interactions, bacteriocins are a highly diverse group of antimicrobial peptides (AMPs), and a universal classification to accommodate all is yet to come. Although, there are some proposed classifications available, however, many of the bacteriocins don’t fit into them [
8,
9]. Commonly, there are two types of bacteriocins, one containing an N-terminal leader sequence and another one is leaderless bacteriocins. Leaderless bacteriocins are secreted in matured form, while the leader sequence containing bacteriocins are posttranslationally modified with conserved enzymes such as lanthionine synthetases and the leader sequence is cleaved off during the maturation process for the secretion of matured bacteriocins. Lanthionine-containing bacteriocins are designated as lanthipeptides, characterized by the presence of intramolecular thioether bridges, and turn out into complex polycyclic structures. Lanthipeptides are usually synthesized as a precursor peptide consisting of an N-terminal leader peptide and a C-terminal core peptide that serves as a substrate for the specific lanthionine synthetases. Posttranslational modification of lanthipeptides involves dehydration of selected Ser and Thr residues to dehydroalanines (Dha) and dehydrobutyrines (Dhb)) and subsequently, intramolecular cyclization of nearby Cys thiols to the dehydrated residues (Dha or Dhb), forming lanthionine and methyl-lanthionine bridges, respectively, through a Michael-type addition. Finally, the posttranslationally modified precursor peptide moved to the cell membrane where the N-terminal leader sequence is cleaved off by the C39 protease domain of the ABC transporter and matured modified core peptide is released to the extracellular space [
10]. Interestingly, oligotrophic oceans are the largest ecosystem on Earth estimated to have 3.28 x 10
4 to 2.46 x 10
6 bacteria as amplicon sequence variants, however, little has been explored yet [
11]. Enormous and unexplored bacterial diversity associated with marine ecosystems suggested a huge potential for the availability of novel bacteriocins to fight against rapidly evolving drug-resistant bacteria. Therefore, in the present review, we have summarized the structural and physiochemical diversity of known and fully characterized marine bacteriocins along with their antimicrobial spectrum.
2. Diverse Bacteriocins from Marine Bacteria
2.1. Leaderless Bacteriocins
2.1.1. Piscicocin
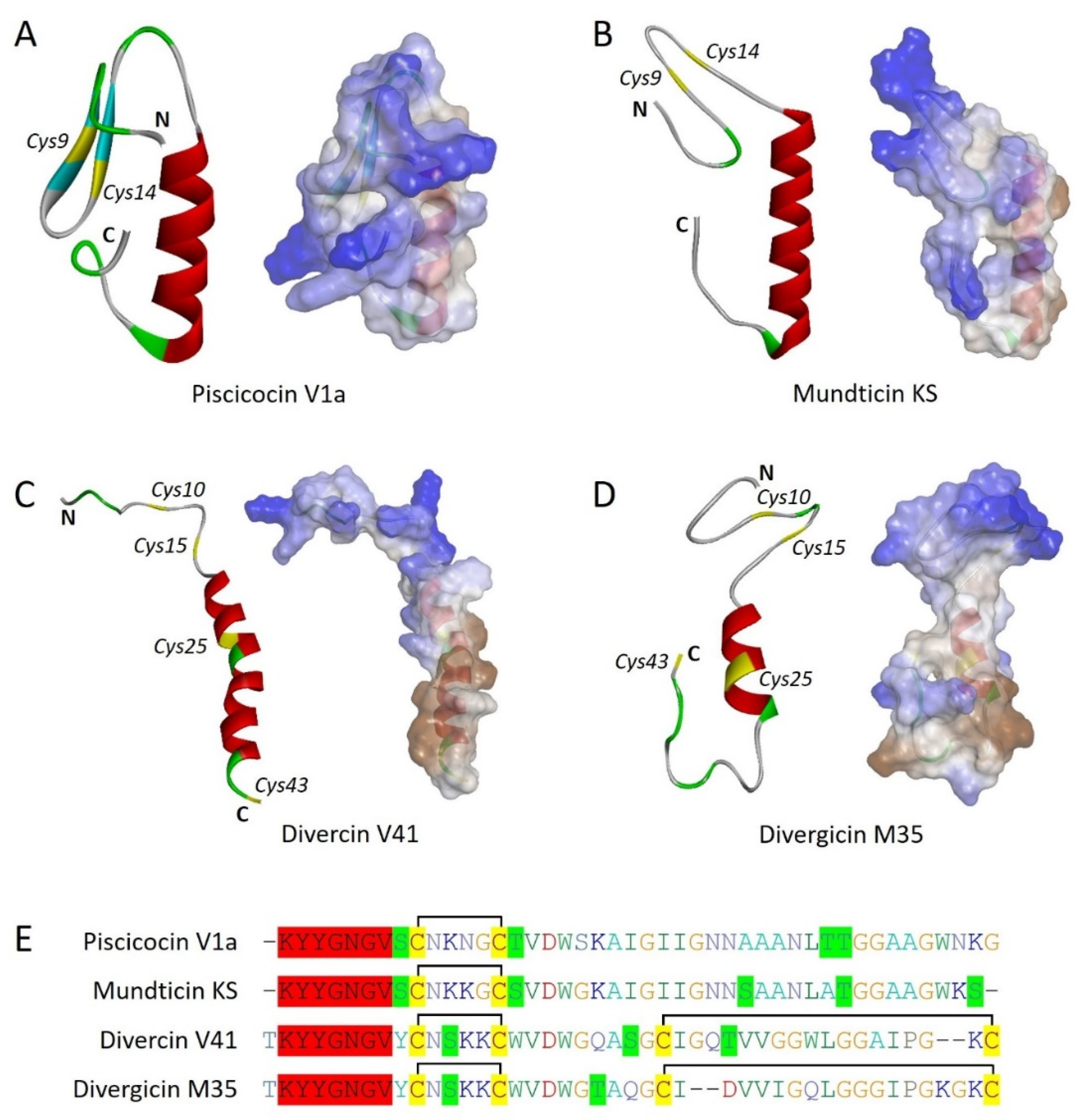
Piscicocins V1a (44 aa) and V1b (43 aa) are leaderless, and non-lanthioninbacteriocins belonging to class IIa. Piscicocins are produced by a lactic acid bacteria (LAB)
Carnobacterium piscicola strain V1 which was originally isolated from fish [
12]. Both V1a and V1b are reported to produce in the cell-free supernatant of the same bacteria, however, their genetic organization is not known. V1a is reported as a novel bacteriocin while V1b was found similar to carnobacteriocin BM1, produced by
Carnobacterium piscicola LV17B, however, both contained the consensus conserved motif YGNGV. Interestingly, V1a contained two cysteine residues at positions 9 and 14 that confirmed to form a disulfide bond (
Figure 1A, and 1E) [
13]. Piscicocins V1a displayed 100 times more potent activity than piscicocin V1b against the Gram-positive bacteria, though the activity spectrum was the same for both. Piscicocin CS526 is another class IIa bacteriocin, produced by
C. piscicola CS526 isolated from surimi (a fish-based product). It showed high similarity to piscicocin while the N-terminal conserved consensus sequence is YGNGL rather than YGNGV where valine is replaced by leucine. It displayed activity against Gram-positive bacteria including
Enterococcus,
Listeria,
Pediococcus, and
Leuconostoc [
14].
2.1.2. Mundticin KS
Mundticin KS is a class IIa bacteriocin that was first reported to produce by
Enterococcus mundtii NFRI 7393, isolated from grass silage [
15]. Later, Schelegueda et. al., isolated and purified the same bacteriocins from
Enterococcus mundtii Tw56, isolated from the intestine of cold water marine fish, silverside (
Odontesthes platensis) [
16]. Munditicin KS is a leaderless, non-lanthionine bacteriocin with the consensus motif YGNGV at the N-terminal, similar to other class IIa bacteriocins. Additionally, muditicin KS contains one di sulfide bond between Cys9 and Cys14, which is well conserved among other class IIa bacteriocins (
Figure 1B, and 1E). Muditicin KS was found active against Gram-positive bacteria including different strains of
Enterococcus faecium,
Lactobacillus plantarum,
L. lactis,
L. curvatus, and
Listeria monocytogenes, however, Schelegueda et. al., reported later that cell-free supernatant of
Enterococcus mundtii Tw56 was also active against
Pediococcus pentosaceus,
Streptococcus thermophiles,
S. pyogenes and some Gram-negative bacteria, including,
Pseudomonas aeruginosa, and
Shewanella putrefaciens [
15,
16].
2.1.3. Divercin V41
Diversin V41 is produced by
Carnobacterium divergens V41, isolated from fish viscera [
12]. Diversin V41 is a non-lanthionine bacteriocin, belonging to class IIa, however, it is reported to have 23 amino acids long N-terminal sequence which has to be cleaved off to generate a 43 amino acid long mature bacteriocin. Interestingly, divercin V41 gene cluster was revealed to have two components of a lantibiotic-type signal transduction system at the same time and also have multiple cysteine residues along with adjacent serine or threonine residues (
Figure 1C, and 1E). This all provides a template for the synthesis of lanthionine bridges, however, lanthionine dehydrates and cyclases are absent in the gene cluster of diversin V41. This suggests an evolutionary position of diversin V41 between lanthipeptides and leaderless bacteriocins. Further, diversin V41 was confirmed to have two disulfide bonds between Cys10-Cys15 and Cys25-Cys43. Interestingly, diversin V41 demonstrates specific activity against Gram-positive bacteria.
2.1.4. Divergicin M35
Divergicin M35 is a class IIa bacteriocin (leaderless, non-lanthionine) produced by
Carnobacterium divergens M35 which is isolated from the frozen smoked mussels. Divergicin M35 was revealed to have a molecular weight of 4518.75 Da (43 amino acids) consisting of 4 cysteine residues that are involved in disulfide bond formation (Cys10-Cys15 and Cys25-Cys43) while consensus sequence YGNGV is present at N-terminal like other bacteriocins of the same class (
Figure 1D, and 1E). It is reported to have specifically strong activity against
L. monocytogenes and could be potentially useful in food preservation [
17].
2.1.5. BaCf3
BaCf3 is a 27 amino acid long, leaderless, non-lanthionine marine bacteriocin produced by
Bacillus amyloliquefaciens BTSS3 isolated from the gut of deep-sea shark (
Centroscyllium fabricii). BaCf3 was confirmed to have a molecular weight of 3028.42 kD with the presence of 3 cysteine residues while Cys6 and Cys13 formed a disulfide bond. Interestingly, the structure prediction of BaCf3 showed a high resemblance with laterosporulin that are highly similar to human beta-defensins [
18]. Furthermore, BaCf3 also showed anticancer activities similar to laterosporulin10 [
19]. BaCf3 was reported to inhibit the biofilm formation by different Gram-positive bacteria including, different
Bacillus sp.,
S. warnie,
Micrococcus luteus,
Geobacillus stearothermophilus. Additionally, it displayed potential activity against food pathogens including,
Salmonella typhimurium,
Clostridium perfringens, and
E. faecalis [
20].
2.1.6. Sonorensin
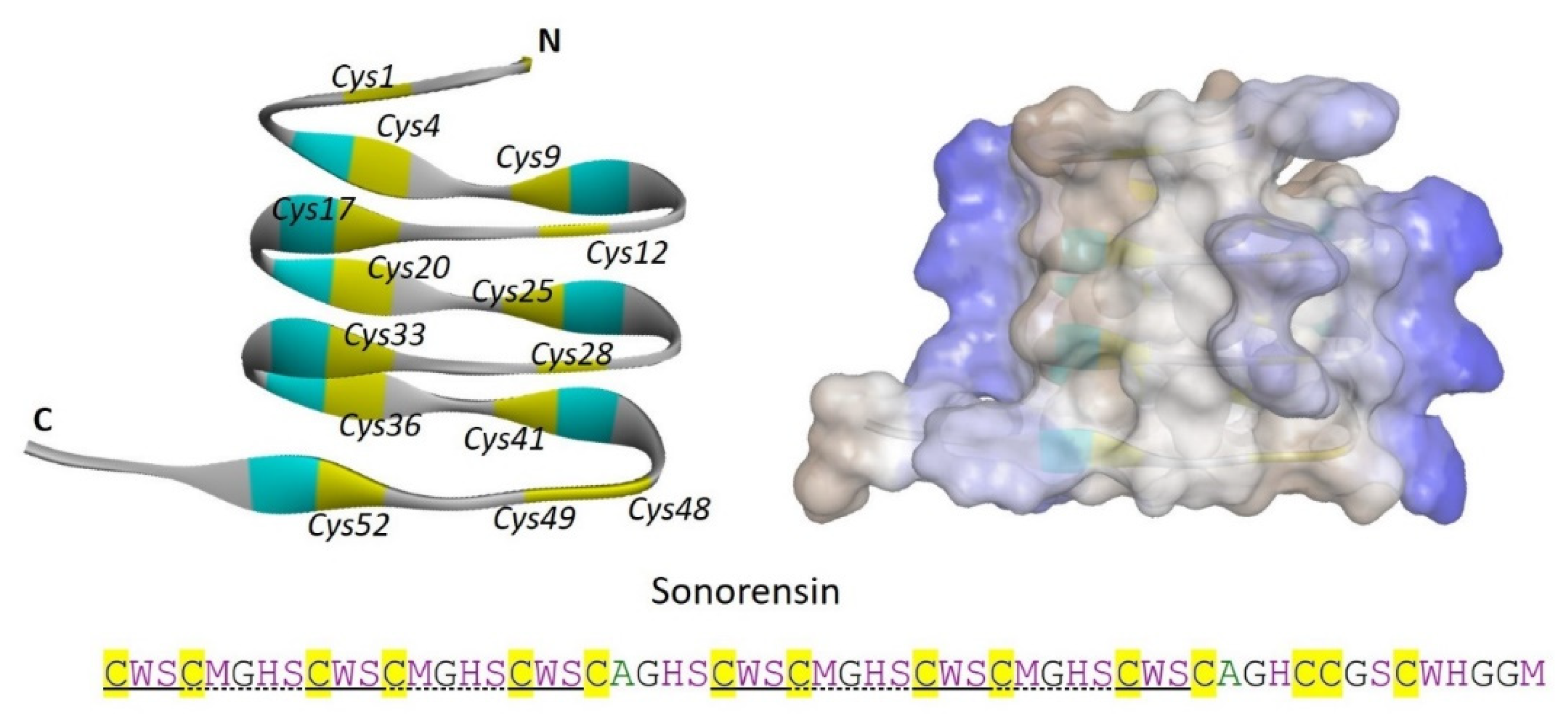
Sonorensin is a 57 amino acid long, non-lanthionine, cysteine-rich bacteriocin produced by a marine bacterial isolate
Bacillus sonorensis MT93. Characterization of sonorensin does not provide sufficient proof to categorize it into any designated class of bacteriocins, while it has having 53 amino acid long leader sequence which is cleaved off during the production of mature peptide. Unusually, sonorensin contains 15 cysteine residues, however, not characterized to be involved in any disulfide bond formation (
Figure 2). Interestingly, sonoresin showed a broad activity spectrum against both Gram-positive and Gram-negative bacteria including,
L. monocytogenes,
V. vulnificus,
B. subtilis,
S. aureus,
P. aeruginosa, and
E. coli [
21].
2.1.7. CAMT6
CAMT6 is a small (12 amino acids), leaderless, non-lanthionine bacteriocin produced by
Enterococcus durans strain YQ-6, isolated from a marine fish,
Larimichthys polyactis. Interestingly, CAMT6 is a unique bacteriocin itself that does not show similarity with other bacteriocins and thus represents a new class of bacteriocins. Additionally, it shows a poor similarity with surfactant-associated anionic peptides from sheep. CAMT6 shows the potential antimicrobial activities against both Gram-positive and Gram-negative bacteria including,
S. aureus,
B. subtilis,
B. equi,
B. cereus,
S. haemolyticus,
Propionibacterium acnes,
Salmonella paratyphi,
V. parahaemolyticus,
P. foulis, and
Enterobacter aerogenes. Interestingly, CAMT6 was also reported to disrupt the biofilm formation by
L. monocytogenes showing its potential applications in the food industry [
22].
2.2. Lanthipeptides
2.2.1. Nisin Z
Nisin Z is (34 amino acids) belongs to class I bacteriocins (Type I lantibiotic), and originally isolated from
Lactococcus lactic NIZO 22186. Nisin Z turned out as a natural variant of nisin A, with a single mutation from histidine to asparagine at position 27 [
23]. In the recent past, nisin Z production has also been reported by the bacterial isolates from the gut of marine fish [
24,
25]. Nisin Z contains 23 amino acid long leader peptides which are cleaved off by peptidase (NisP) while mature nisin Z is released after the formation of lanthionine and methyllanthionine bridges by the respective class-specific lanthionine dehydratase (LanB) and lanthionine cyclases (
Figure 3A) (LanC) [
23]. It is reported that nisin Z produced by a bacterial isolate (
Lactococcus lactis subsp. lactis) from marine fish (olive flounder) showed efficient antimicrobial activity against
Streptococcus iniae when prepared in 3.5% (w/v) NaCl (equivalent to seawater) [
25]. Another study demonstrated that nisin Z produced by
Lactococcus lactis TW34 (isolated from marine fish) effectively kills the fish pathogen
Lactococcus garvieae [
24]. Overall, it suggested that nisin Z is more adapted to its original habitat concerning the displayed antimicrobial activity and thus could be a potential candidate for sea food preservation or aquaculture pathogens.
2.2.2. Subtilomycin
Subtilomycin is a 32 amino acid long, class I bacteriocins (Type I lantibiotic) produced by a marine bacteria
Bacillus subtilis MMA7, isolated from the marine sponge
Haliclona simulans. It possesses a 24 amino acids long N-terminal leader peptide which is cleaved off during the peptide maturation by a peptidase present within the gene cluster. Subtilomycin contained 5 cysteine residues which all are involved in the formation of five lanthionine or mehthylanthionine rings subsequently after dehydration and cyclization by LanB and LanC, respectively (
Figure 3A). Subtilomycin displayed potent activity against both Gram-positive and Gram-negative bacteria including, different species of
Bacillus and
Clostridium,
L.
monocytogenes,
S.
aureus, and
P.
aeruginosa. Interestingly, subtilomycin has also been reported to inhibit the growth of vancomycin-intermediate
S. aureus (VISA), methicillin-resistant
S.
aureus (MRSA), vancomycin-resistant
E. coli, and different pathogenic
Candida species [
26].
2.2.3. Viridisin
Viridisin is a class I bacteriocin (Type I lantibiotic) produced by marine bacteria
Thalassomonas viridans XOM25. Viridisin is an unusual lanthipeptide as it consists of 3 core lanthipeptides in the gene cluster, VdsA1, VdsA2, and VdsA3 which have 4, 2, and 2 cysteine residues respectively (
Figure 3B). While corresponding lanthipeptides for VdsA1 (25 amino acids) and VdsA2 (25 amino acids) are cloned, expressed, and fully characterized for their lanthionine and methlyllanthione ring patterns, their antimicrobial activities are still needed to be explored.
2.2.4. Thalassomonasin
Thalassomonasin is produced by a marine proteobacterium
Thalassomonas actiniarum NBRC 104231 and belongs to class I bacteriocins (Type I lantibiotic). Thalassomonasin is a two-component lanthipeptide consisting of two core lanthipeptide precursor genes,
tln A1 (thalassomonasin A) and
tln A2 (thalassomonasin B) in its gene cluster. Thalassomonasin A (25 amino acids) and Thalassomonasin B (26 amino acids) consist of 3 and 2 cysteine residues, respectively which all are involved in the formation of lanthionine rings. Both thalassomonasin A and B have an N-terminal leader sequence of 29 amino acids that differ from each other, however, having characteristic conserved motifs for type I lanthipeptide leader sequences (
Figure 3B). Interestingly, thalassomonasin A is showing efficient activity against both Gram-positive and Gram-negative bacteria including
B. subtilis,
S. aureus,
M. luteus,
E. coli, and
P. aeruginosa while Thalassomonasin B showed minor activity and thus not explored further [
27].
2.2.5. Formicin
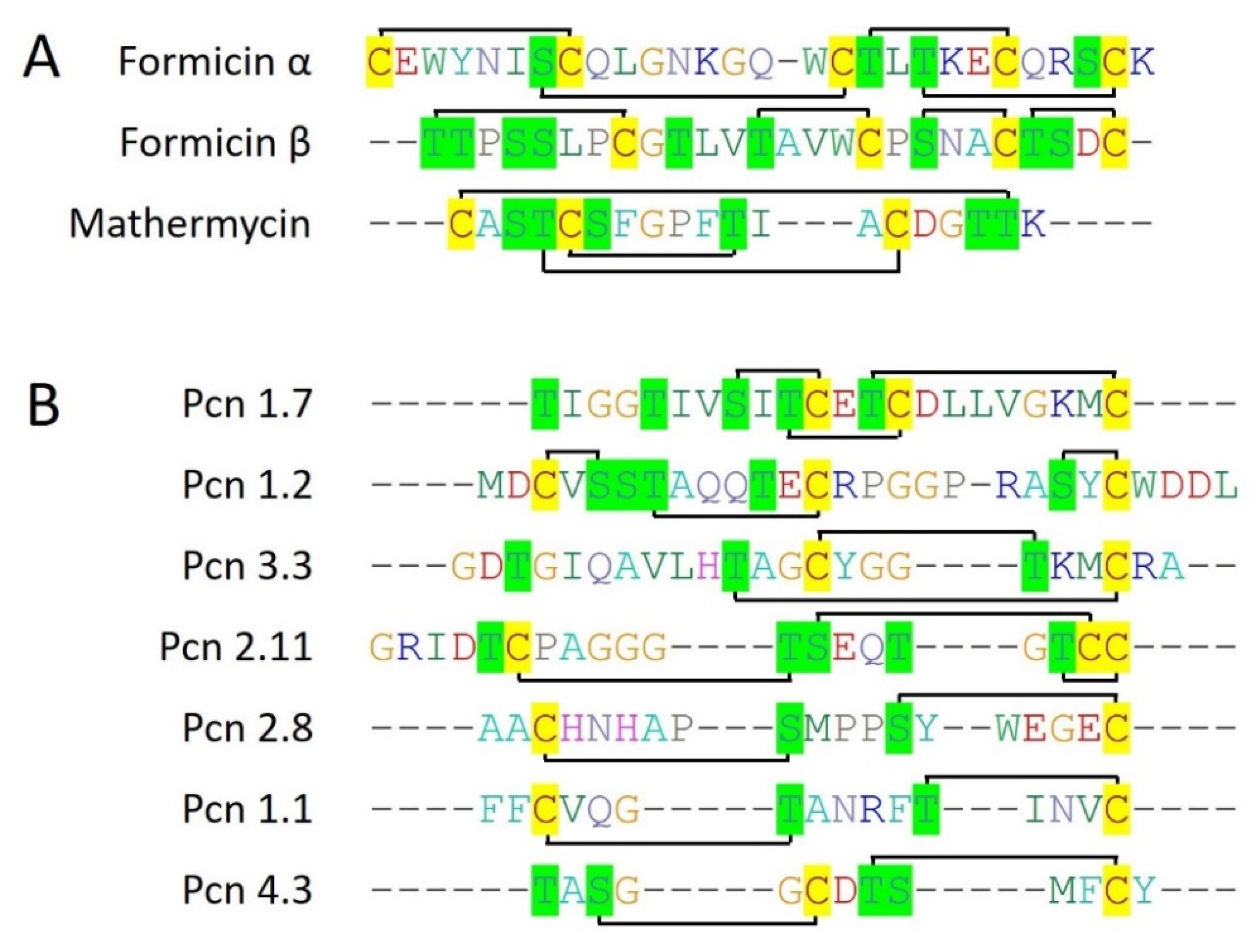
Formicin is a two-component bacteriocin (Type II lantibiotic) that belongs to class I bacteriocins. Formicin is produced by an antimicrobial-producing bacteria,
Bacillus paralicheniformis strain APC 1576, isolated from the intestine of Atlantic mackerel (
Scomber scombrus), a marine fish. Formicin contains a 40 amino acid N-terminal leader sequence which is cleaved by LanP, residing within the gene cluster itself. As formicin is a type II lanthipeptide, its gene cluster consists of a bifunctional enzyme that performs both dehydration and cyclization, subsequently during the maturation process of lanthipeptide before the cleavage of the leader peptide. Additionally, an overall +2 positive charge along with less hydrophobicity make formicin unique among all the type II lanthipeptides (
Figure 4A). Formicin, showed potential antimicrobial activity against Gram-positive pathogenic strains including,
L. monocytogenes,
S. aureus,
S. mutans,
Clostridioides difficile,
Clostridia, and different species of
Enterococcus [
28].
2.2.6. Mathermycin
Mathermycin is produced by marine actinomycete
Marinactinospora thermotolerans SCSIO 00652 (isolated from sea marine sediments), a 19 amino acid long class I bacteriocin (Type II lantibiotic). It contains a 60 amino acid long N-terminal leader sequence and 3 cysteine residues that all are involved in the formation of lanthionine and methyllanthionine rings with nearby Serine and Threonine residues (
Figure 4A). Mathermycine showed a closed structural homology with cinnamycin and duramycin. Also, mathermycin is revealed to have a similar activity spectrum against Gram-positive bacteria such as
B. subtilis, like cinnamycin [
29].
2.2.7. Prochlorosins
Prochlorosins are a structurally diverse set of class I bacteriocins (Type II lantibiotic) produced by marine picocyanobacteria,
Prochlorococcus, and
Synechococcus. Surprisingly, picocyanobacteria employed an unusual mechanism that was able to produce structurally diverse lanthipeptides abundantly using a single lanthionine synthetase. Using a deep sequencing methodology, 50
Prochlorococcus and 26
Synechococcus genomes were analyzed for the presence of lanthionine synthetases and lantibiotic precursor peptides. Out of all,
Prochlorococcus MIT0701,
Prochlorococcus MIT1327,
Prochlorococcus MIT9303, and
Prochlorococcus MIT9313, are found to have 9, 13, 13, 29 while
Synechococcus WH8016,
Synechococcus RS9916,
Synechococcus KORDI100,
Synechococcus MITS9508, and
Synechococcus MITS9504, are found to have 1, 19, 9, 8, 80 core lantibiotic sequences. In total, these 9 genomes were revealed to have 181 diverse core lanthipeptide genes which are almost double the total other lanthipeptides (~90) that have been reported from other different bacterial species [
30]. The most structurally diverse lanthipeptides are observed in
Prochlorococcus MIT9313 and 7 of them have been fully characterized for their ring topologies (
Figure 4B).
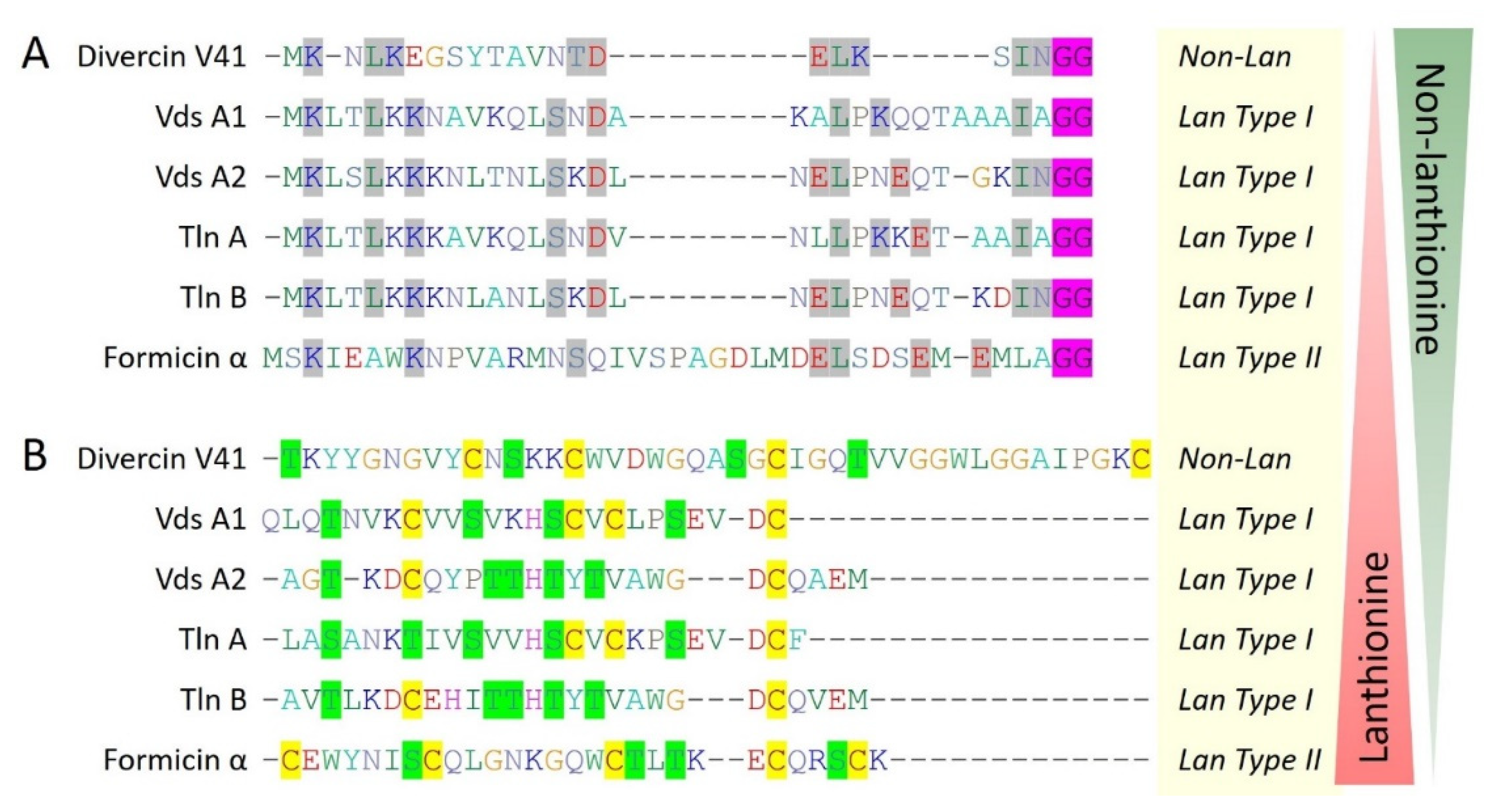
Main Text As the marine ecosystem is enormously diverse, a huge diversity of the marine bacterial communities and thus the marine bacteriocins is also anticipated. However, there are only a few bacteriocins of the defined class of bacteriocins are reported from the marine ecosystem yet, they represent a diverse group of bacteriocins (Figures 1, 2, 3, and 4). Moreover, some of the bacteriocins didn’t fit into any defined class of bacteriocins which suggested the possibility of having novel classes of bacteriocins from the marine environment in the future, which are even not defined yet. For example, BaCf3 is characterized as a leaderless bacteriocin with the unusual presence of 3 cysteine residues [
20]. BaCf3 did not have any consensus sequence or show similarity with a known class of bacteriocins, however, structural resemblance showed some similarities with laterosporulins, class IId bacteriocins produced by different strains of
Brevibacillus laterosporus [
18,
31]. Next, sonorensin contains 15 cysteine residues that make it unusually unique on its own and don’t fit into any existing class of bacteriocins [
21]. CAMT6 is a small marine bacteriocin with no cysteine residues that don’t belong to any class [
22].
Other four marine bacteriocins (piscicocin V1a, mundticin KS, divercin V41, divergicin M35) belongs to class IIa of bacteriocins and all have a disulfide bond followed by characteristic consensus conserved sequence YGNGV at N-terminal. Notably, divercin V41 and divergicin M35 consist of two additional cysteine residues involved in disulfide formation (
Figure 1). It seems there is an evolutionary pressure for the addition of an extra disulfide bond to make more stable bacteriocins. Additionally, the presence of two disulfide bonds, further makes them closer to eukaryotic defensins that have characteristic 3 disulfide bonds. Interestingly, divercin V41 was reported to have an N-terminal leader sequence that cleaved off during the maturation process. An N-terminal leader sequence, double glycine motif at the cleavage site, and presence of 4 cysteine residues along with Ser and Thr residues suggest divercin V41 resemblance with lanthipeptides, where, lanthionine synthetases are absent (
Figure 5).
Other than class IIa bacteriocins, 6 type I lanthipeptides (nisin Z, subtilomycin, viridisin A1, viridisin A2, thalassomonasin A, thalassomonasin B) belonging to class I bacteriocins are characterized from the marine bacteria. They contain 2 to 5 cysteine residues which all are involved in lanthionine ring formation with different topologies, making them diverse (
Figure 3). Next, formicin, and mathermycin (type II lanthipeptides) are reported from the marine bacteria which are consist of 3 to 5 cysteine residues that involved in different ring topologies (
Figure 4). Additionally, 181 type II lanthipeptide genes were identified from gnome mining of the 9 cyanobacteria genomes isolated from the deep sea [
30]. This provides a glimpse of the huge unexplored diversity of marine ecosystems and the possibility of having plenty of novel and diverse bacteriocins from the underexplored marine ecosystem. Out of all,
Prochlorococcus MIT9313 alone was reported to have 29 lanthipeptide gene clusters, out of which 7 most interesting and diverse lanthipeptides are characterized from the genome (
Figure 4). Surprisingly, ring topologies of these 7 peptides were highly dissimilar to all other known lanthipeptides and even with each other as all of the Ser, Thr, or Cys (2 to 3) residues follow unique patterns while the leader is highly conserved. This suggested an advanced evolutionary mechanism in marine cyanobacteria that can generate such a high extent of structural diversity of lanthipeptides. Overall, this indicates marine bacterial communities are residing under an ongoing evolutionary pressure that guides a unique mechanism of structural diversification of bacteriocins. It can be hypothesized that with this huge diversity of different life domains and associated diverse bacterial communities under different physiological and physical conditions or marine ecosystems, what would be the extent of bacteriocins novelty and diversity which has been only a little bit explored yet?
Figure 5.
Multiple sequence alignment (CLUSTALW) based hypothetical model for the evolution of marine bacteriocins from non-lanthionine to lanthionine containing lanthipeptides and vice-versa. (A) Divercin V41 belongs to class IIa while unusually having an N-terminal leader sequence containing a double glycine motif (GG, highlighted in pink color), similar to lanthionine containing type I and type II lanthipeptides. Leader sequence alignment showing conserved residues (highlighted in grey color) suggesting divercin V41 is under evolutionary transition towards lanthipeptides or vice-versa. (B) Core peptide sequence alignment showing diverse structural ring topologies. Cysteine residues are highlighted in yellow color while serine and threonine residues are highlighted in green color.
Figure 5.
Multiple sequence alignment (CLUSTALW) based hypothetical model for the evolution of marine bacteriocins from non-lanthionine to lanthionine containing lanthipeptides and vice-versa. (A) Divercin V41 belongs to class IIa while unusually having an N-terminal leader sequence containing a double glycine motif (GG, highlighted in pink color), similar to lanthionine containing type I and type II lanthipeptides. Leader sequence alignment showing conserved residues (highlighted in grey color) suggesting divercin V41 is under evolutionary transition towards lanthipeptides or vice-versa. (B) Core peptide sequence alignment showing diverse structural ring topologies. Cysteine residues are highlighted in yellow color while serine and threonine residues are highlighted in green color.
4. Challenges and Future Directions
Marine habitats are the largest and one of the most diverse ecosystems on the Earth which has only a little explored yet and much is still far away from human interference. One of the biggest challenges is to access or analyze the vast marine diversity and thus biodiscovery of its associated bacterial communities. Also, marine bacterial communities represent a dilute habitat which is an unusual place for the action of bacteriocins. Next, whatever, the marine bacterial communities we have explored yet are incomplete as many of them are uncultivable in lab conditions unless created in very similar conditions to their native marine habitats. This is why the bacteriocins biosynthetic gene cluster of marine bacteria are fully expressed in vitro lab conditions and remain inaccessible [
32]. Additionally, marine habitats are physically diverse such as sea cost subsurface soil, sea cost sediments, and deep-sea sediments, moreover, the marine ecosystem is divided into five different zones based on different environment pressures, light levels, temperatures, dissolved oxygen concentration, and thus the microbial communities [
33]. Especially low oxygen levels in the deep-sea environment create anaerobic conditions that significantly affect the antimicrobial production by the deep-sea bacteria [
34,
35]. All these various factors play essential roles and affect the secondary metabolites and bacteriocins produced by marine bacterial communities [
36,
37]. Design and development of more sophisticated culture conditions or strategies are required to recover uncultivable marine bacterial communities and thus the diversity of diverse unexplored bacteriocins. Most of the reported marine bacteriocins are from bacterial species isolated either from the gut or in association with the other marine organisms that again suggested the little-known information about the marine bacteriocins. Additionally, each organism has its specific gut microbiome, where bacteriocins-producing bacteria interact with the other prokaryotic organisms within the gut and the eukaryotic host as well. These diverse interactions between different microbial communities from different sea organisms under the diverse conditions of different marine habitats generate a huge diversity of bacteriocins or other secondary metabolites, which still need to be explored. Moreover, lanthipeptide (class I bacteriocins) modifying enzymes lanthionine synthetases are known to play a role as signaling molecules in bacterial communities that suggest their diverse functions [
38].
Next, in the recent past, culture-independent techniques such as genome mining, metagenomics, next-generation sequencing, metagenomic library preparation, and metabolomics have been employed to explore inaccessible microbial communities [
39]. Thanks, to the advancement of technology, however, sample collection itself is a big challenge for marine habitats such as the deep sea. There are a few interesting genome mining tools available including BAGEL and antiSMASH, which can find out bacteriocins gene clusters on the similarity basis of available data sets that include conserved modifying enzymes and motif sequences within the leader or sometimes core peptides [
40,
41]. Additionally, DeepRiPP and RODEO are other machine learning-based bioinformatics tools that can perform high-throughput identification and classification of RiPPs [
42,
43]. Although genome mining tool have proven their potential in finding novel bacteriocins, the search is based on the similarities of the earlier identified RiPPs and might fail to detect novel posttranslational modification in the RiPPs, which are still need to be studied in detail and not available in datasets [
44,
45].
Even after, the identification and mining of bacteriocins-producing gene clusters using in silico genome mining, the gene cluster should be expressed in the native host for the subsequent production and purification of the respective bacteriocins, which is challenging. Moreover, it is reported that many of the bacteriocin gene clusters are “silently” present in the genomes, while the gene can be identified by metagenomic approaches but the biological activity is missing during experimental culture conditions [
46,
47]. Heterologous gene expression is one of the reliable and economical strategies for the controlled expression of these silent bacteriocins gene clusters [
48]. Another strategy is the chemical synthesis of bacteriocins as peptidic in nature, however, chemical synthesis is expensive and not suitable to achieve desired posttranslational modification such as lanthionine rings and disulfide bonds. As of the current scenario in the field of bacteriocins identification and characterization, a combination of various in silico and in vitro experiments and strategies are the most acceptable approach for the marine bacteriocins from free marine sediments as well as from the gut of marine organisms.
5. Conclusions
In the current scenario of rapidly evolving drug resistance and scarcity of new alternative drugs or strategies to fight against, there is a pressing need for novel bioactive molecules with diverse and unique mechanisms of action. The marine bacteriocins are one of the exciting and enormous groups for such biologically active molecules. Interestingly, due to its huge diversity, physical conditions, and several other factors, marine ecology is profoundly diverse, and thus their bioactive compounds including bacteriocins, however, little is explored. Other than the free marine bacterial communities, marine organism’s gut-microbiome is supposed to be more diverse due to interactions with marine hosts and then the overall marine ecology that deals with the high salinity, low temperatures, low oxygen or anaerobic conditions, and hydrostatic pressure. Overall, all of these factors create a cumulative evolutionary pressure for the marine bacterial communities that is reflected in the diverse structural and functional diversity of marine bacteriocins that largely remained unexplored. Limited accessibility for the sample collection and technological barriers to analyzing the huge diversity are major factors in the way of true exploration of marine bacteriocins. In conclusion, marine bacteriocins could be a promising alternative to fight against drug-resistant pathogens, however, the true diversity and potential of marine bacteriocins are yet to be explored. Marine bacterial diversity and bacteriocins might be more.
Author Contributions
PB and DR wrote the manuscript. SMM proofread and edited the manuscript. PB conceived the idea, analyzed the data, prepared the illustrations, and led the whole project.
Funding
There is no funding source associated with the present work.
Data Availability Statement
There is no additional data associated with this work.
Acknowledgments
PB acknowledges the Somatic Cell Genome Editing Center, ASRC, Division of Animal Sciences, University of Missouri, Columbia, United States for providing the space necessary facilities for this work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Walsh, T.R.; Gales, A.C.; Laxminarayan, R.; Dodd, P.C. Antimicrobial Resistance: Addressing a Global Threat to Humanity. PLoS Med. 2023, 20. [CrossRef]
- Traxler, M.F.; Kolter, R. Natural products in soil microbe interactions and evolution. Nat. Prod. Rep. 2015, 32, 956–970. [CrossRef]
- Baindara, P.; Chaudhry, V.; Mittal, G.; Liao, L.M.; Matos, C.O.; Khatri, N.; Franco, O.L.; Patil, P.B.; Korpole, S. Characterization of the antimicrobial peptide penisin, a class Ia novel lantibiotic from Paenibacillus sp. strain A3. Antimicrob. Agents Chemother. 2016. [CrossRef]
- Donia, M.S.; Cimermancic, P.; Schulze, C.J.; Wieland Brown, L.C.; Martin, J.; Mitreva, M.; Clardy, J.; Linington, R.G.; Fischbach, M.A. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 2014, 158, 1402–1414. [CrossRef]
- Ziemert, N.; Lechner, A.; Wietz, M.; Millań-Aguiñaga, N.; Chavarria, K.L.; Jensen, P.R. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc. Natl. Acad. Sci. U. S. A. 2014, 111. [CrossRef]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins-a viable alternative to antibiotics? Nat. Rev. Microbiol. 2013.
- Ghosh, C.; Sarkar, P.; Issa, R.; Haldar, J. Alternatives to Conventional Antibiotics in the Era of Antimicrobial Resistance. Trends Microbiol. 2019, 27, 323–338. [CrossRef]
- Zimina, M.; Babich, O.; Prosekov, A.; Sukhikh, S.; Ivanova, S.; Shevchenko, M.; Noskova, S. Overview of global trends in classification, methods of preparation and application of bacteriocins. Antibiotics 2020, 9, 1–21. [CrossRef]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [CrossRef]
- Willey, J.M.; van der Donk, W. a Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 2007, 61, 477–501. [CrossRef]
- Hoshino, T.; Doi, H.; Uramoto, G.I.; Wörmer, L.; Adhikari, R.R.; Xiao, N.; Morono, Y.; D’Hondt, S.; Hinrichs, K.U.; Inagaki, F. Global diversity of microbial communities in marine sediment. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 27587–27597. [CrossRef]
- Pilet, M.F.; Xavier, D.; Rachel, B.; Georges, N.; Michel, D.; Piard, J.C. Evidence for two bacteriocins produced by carnobacterium piscicola and carnobacterium divergens isolated from fish and active against Listeria monocytogenes. J. Food Prot. 1995, 58, 256–262. [CrossRef]
- Bhugaloo-Vial, P.; Dousset, X.; Metivier, A.; Sorokine, O.; Anglade, P.; Boyaval, P.; Marion, D. Purification and amino acid sequences of piscicocins V1a and V1b, two class IIa bacteriocins secreted by Carnobacterium piscicola VI that display significantly different levels of specific inhibitory activity. Appl. Environ. Microbiol. 1996, 62, 4410–4416. [CrossRef]
- Yamazaki, K.; Suzuki, M.; Kawai, Y.; Inoue, N.; Montville, T.J. Purification and characterization of a novel class IIa bacteriocin, piscicocin CS526, from surimi-associated Carnobacterium piscicola CS526. Appl. Environ. Microbiol. 2005, 71, 554–557. [CrossRef]
- Kawamoto, S.; Shima, J.; Sato, R.; Eguchi, T.; Ohmomo, S.; Shibato, J.; Horikoshi, N.; Takeshita, K.; Sameshima, T. Biochemical and genetic characterization of mundticin KS, an antilisterial peptide produced by Enterococcus mundtii NFRI 7393. Appl. Environ. Microbiol. 2002, 68, 3830–3840. [CrossRef]
- Schelegueda, L.I.; Vallejo, M.; Gliemmo, M.F.; Marguet, E.R.; Campos, C.A. Synergistic antimicrobial action and potential application for fish preservation of a bacteriocin produced by Enterococcus mundtii isolated from Odontesthes platensis. LWT 2015, 64, 794–801. [CrossRef]
- Tahiri, I.; Desbiens, M.; Benech, R.; Kheadr, E.; Lacroix, C.; Thibault, S.; Ouellet, D.; Fliss, I. Purification, characterization and amino acid sequencing of divergicin M35: A novel class IIa bacteriocin produced by Carnobacterium divergens M35. Int. J. Food Microbiol. 2004, 97, 123–136. [CrossRef]
- Dinata, R.; Baindara, P. Laterosporulin25: A probiotically produced, novel defensin-like bacteriocin and its immunogenic properties. Int. Immunopharmacol. 2023, 121, 110500. [CrossRef]
- Baindara, P.; Gautam, A.; Raghava, G.P.S.; Korpole, S. Anticancer properties of a defensin like class IId bacteriocin Laterosporulin10. Sci. Rep. 2017. [CrossRef]
- Bindiya, E.S.; Tina, K.J.; Sasidharan, R.S.; Bhat, S.G. BaCf3: highly thermostable bacteriocin from Bacillus amyloliquefaciens BTSS3 antagonistic on food-borne pathogens. 3 Biotech 2019, 9. [CrossRef]
- Chopra, L.; Singh, G.; Choudhary, V.; Sahoo, D.K. Sonorensin: An antimicrobial peptide, belonging to the heterocycloanthracin subfamily of bacteriocins, from a new marine isolate, Bacillus sonorensis MT93. Appl. Environ. Microbiol. 2014, 80, 2981–2990. [CrossRef]
- Li, Q.; Chen, Q.; Wu, Y.; Chen, Z.; Liu, Y.; Fang, Z.; Deng, Q. Purification, characterization and structural identification of a novel bacteriocin produced by marine original Enterococcus durans YQ-6, and its inhibition of Listeria monocytogenes. LWT 2023, 173. [CrossRef]
- MULDERS, J.W.M.; BOERRIGTER, I.J.; ROLLEMA, H.S.; SIEZEN, R.J.; de VOS, W.M. Identification and characterization of the lantibiotic nisin Z, a natural nisin variant. Eur. J. Biochem. 1991, 201, 581–584. [CrossRef]
- Sequeiros, C.; Garcés, M.E.; Vallejo, M.; Marguet, E.R.; Olivera, N.L. Potential aquaculture probiont Lactococcus lactis TW34 produces nisin Z and inhibits the fish pathogen Lactococcus garvieae. Arch. Microbiol. 2015, 197, 449–458. [CrossRef]
- Heo, W.S.; Kim, E.Y.; Kim, Y.R.; Hossain, M.T.; Kong, I.S. Salt effect of nisin Z isolated from a marine fish on the growth inhibition of Streptococcus iniae, a pathogen of streptococcosis. Biotechnol. Lett. 2012, 34, 315–320. [CrossRef]
- Phelan, R.W.; Barret, M.; Cotter, P.D.; Connor, P.M.O.; Chen, R.; Morrissey, J.P.; Dobson, A.D.W.; Gara, F.O.; Barbosa, T.M. Subtilomycin : A New Lantibiotic from Bacillus subtilis Strain MMA7 Isolated from the Marine Sponge Haliclona simulans. 2013, 1878–1898. [CrossRef]
- Thetsana, C.; Ijichi, S.; Kaweewan, I.; Nakagawa, H.; Kodani, S. Heterologous expression of a cryptic gene cluster from a marine proteobacterium Thalassomonas actiniarum affords new lanthipeptides thalassomonasins A and B. J. Appl. Microbiol. 2022, 132, 3629–3639. [CrossRef]
- Collins, F.W.J.; O’Connor, P.M.; O’Sullivan, O.; Rea, M.C.; Hill, C.; Ross, R.P. Formicin – a novel broad-spectrum twocomponent lantibiotic produced by Bacillus paralicheniformis APC 1576. Microbiol. (United Kingdom) 2016, 162, 1662–1671.
- Chen, E.; Chen, Q.; Chen, S.; Xu, B.; Ju, J.; Wang, H. Mathermycin, a lantibiotic from the marine actinomycete Marinactinospora thermotolerans SCSIO 00652. Appl. Environ. Microbiol. 2017, 83. [CrossRef]
- Cubillos-Ruiz, A.; Berta-Thompson, J.W.; Becker, J.W.; Van Der Donk, W.A.; Chisholm, S.W. Evolutionary radiation of lanthipeptides in marine cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E5424–E5433. [CrossRef]
- Singh, P.K.; Solanki, V.; Sharma, S.; Thakur, K.G.; Krishnan, B.; Korpole, S. The intramolecular disulfide-stapled structure of laterosporulin, a class IId bacteriocin, conceals a human defensin-like structural module. FEBS J. 2015.
- Deming, J.W.; Baross, J.A. Survival, Dormancy, and Nonculturable Cells in Extreme Deep-Sea Environments. In Nonculturable Microorganisms in the Environment; 2000; pp. 147–197.
- Costello, M.J.; Chaudhary, C. Marine Biodiversity, Biogeography, Deep-Sea Gradients, and Conservation. Curr. Biol. 2017, 27, R511–R527.
- Zeng, X.; Alain, K.; Shao, Z. Microorganisms from deep-sea hydrothermal vents. Mar. Life Sci. Technol. 2021, 3, 204–230.
- Yadav, S.; Koenen, M.; Bale, N.J.; Reitsma, W.; Engelmann, J.C.; Stefanova, K.; Damsté, J.S.S.; Villanueva, L. Organic matter degradation in the deep, sulfidic waters of the Black Sea: insights into the ecophysiology of novel anaerobic bacteria. Microbiome 2024, 12, 98. [CrossRef]
- Srinivasan, R.; Kannappan, A.; Shi, C.; Lin, X. Marine bacterial secondary metabolites: A treasure house for structurally unique and effective antimicrobial compounds. Mar. Drugs 2021, 19.
- Petersen, L.-E.; Kellermann, M.Y.; Schupp, P.J. Secondary Metabolites of Marine Microbes: From Natural Products Chemistry to Chemical Ecology. In YOUMARES 9 - The Oceans: Our Research, Our Future; 2020; pp. 159–180.
- Goto, Y.; Li, B.; Claesen, J.; Shi, Y.; Bibb, M.J.; Donk, W.A. Van Der Discovery of Unique Lanthionine Synthetases Reveals New Mechanistic and Evolutionary Insights. 2010, 8, 4–13. [CrossRef]
- Zhang, L.; Chen, F.X.; Zeng, Z.; Xu, M.; Sun, F.; Yang, L.; Bi, X.; Lin, Y.; Gao, Y.J.; Hao, H.X.; et al. Advances in Metagenomics and Its Application in Environmental Microorganisms. Front. Microbiol. 2021, 12. [CrossRef]
- Van Heel, A.J.; De Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M. a.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, 339–346. [CrossRef]
- Tietz, J.I.; Schwalen, C.J.; Patel, P.S.; Maxson, T.; Blair, P.M.; Tai, H.C.; Zakai, U.I.; Mitchell, D.A. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 2017, 13, 470–478. [CrossRef]
- Merwin, N.J.; Mousa, W.K.; Dejong, C.A.; Skinnider, M.A.; Cannon, M.J.; Li, H.; Dial, K.; Gunabalasingam, M.; Johnston, C.; Magarvey, N.A. DeepRiPP integrates multiomics data to automate discovery of novel ribosomally synthesized natural products. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 371–380. [CrossRef]
- Zhong, Z.; He, B.; Li, J.; Li, Y.X. Challenges and advances in genome mining of ribosomally synthesized and post-translationally modified peptides (RiPPs). Synth. Syst. Biotechnol. 2020, 5, 155–172. [CrossRef]
- Russell, A.H.; Truman, A.W. Genome mining strategies for ribosomally synthesised and post-translationally modified peptides. Comput. Struct. Biotechnol. J. 2020, 18, 1838–1851. [CrossRef]
- Seyedsayamdost, M.R. High-Throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 7266–7271. [CrossRef]
- Baindara, P.; Nayudu, N.; Korpole, S. Whole genome mining reveals a diverse repertoire of lanthionine synthetases and lanthipeptides among the genus Paenibacillus. J. Appl. Microbiol. 2020. [CrossRef]
- Jain, P.M.; Nellikka, A.; Kammara, R. Understanding bacteriocin heterologous expression: A review. Int. J. Biol. Macromol. 2024, 133916. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).