1. Introduction
Influenza A viruses are members of the
Orthomyxoviridae family and contain a segmented, negative-sense RNA genome. They are commonly described by their combinations of two surface proteins, hemagglutinin (H, H1 to H18) and neuraminidase (NA, N1 to N11). While the viruses replicate in numerous terrestrial avian and mammalian species, wild aquatic birds are considered the natural reservoirs of avian influenza viruses (AIVs) [
1].
Globally, there has been a substantial increase in detections of diverse highly pathogenic avian influenza (HPAI) and low pathogenicity avian influenza (LPAI) viruses in wild birds and poultry in the past two decades, including in Africa [
2,
3]. The evolution of new strains and the dispersal and establishment of these strains in new areas across the world have been associated with seasonal wild bird migration along flyways [
2,
4]. One such migratory pathway, the East Africa-West Asia flyway, encompasses portions of southern and eastern Africa[
4]. Likewise, poultry trade and, specifically, live bird markets (LBM), are recognized as key nodes for the transmission of influenza A viruses and act as key sites to monitor for the introduction and circulation of AIVs in poultry populations.
Globally, human infections with different AIV H5, H7, and H9 influenza A subtypes with varying N proteins have occurred, commonly as spill-over events during AIV outbreaks in poultry [
5,
6]. The earliest human infections with H9N2 were reported in China in 1998 [
6]. By 2019, through ongoing targeted surveillance, 59 H9N2 human cases had been reported in China, Hongkong, Egypt, Bangladesh, Pakistan, and Oman. The majority of AIV human cases have reported exposure to poultry, underscoring an ongoing public health risk given the intrinsic potential for influenza virus reassortment [
5,
6,
7,
8]. In 2021–2023, the WHO reported 20 human cases of HPAI H5N1 virus [
9]. All but one case in India had confirmed exposure to poultry [
10].
Low pathogenicity avian influenza (LPAI) H9N2 viruses are the most prevalent subtype of AIVs in poultry globally and have been reported throughout Asia, the Middle East, Europe, and Africa, including in Kenya [
6,
7,
11,
12]. Since introduction in 1966, H9N2 has frequently been observed with high prevalence rates, exceeding 10% reported among poultry in LBMs in Vietnam, China, and Egypt [
6]. H5N2 viruses are less prevalent in Africa and while HPAI H5N2 outbreaks have been reported in farmed ostriches in South Africa and the LPAI H5N2 viruses have been reported in West Africa, the H5N2 viruses have not been reported in Eastern Africa [
13,
14] . Between 2016 and 2018, an Asian origin HPAI H5N8 rapidly spread in a variety of avian species including wild birds and poultry in sub-Saharan Africa [
11,
15,
16,
17]. The pan-African spread of H5N8 demonstrated the potential for large scale transmission of AIVs in different ecosystems across the continent, highlighting knowledge gaps on the contribution of environmental conditions on AIV transmission and underscoring the need for more robust continental surveillance for monitoring and mitigation measures [
15]. In January 2017, Uganda confirmed infection with HPAI H5N8 among wild birds in Lutembe Bay along the shores of Lake Victoria and in poultry in two districts of its southeastern region [
16]. In response to the outbreak risk assessment considered the spread of H5N8 across Eastern African countries as likely [
16]. On phylogenetic analysis, theH5N8 viruses in the Uganda outbreak clustered with viruses collected in 2017 in the Democratic Republic of the Congo and in West Africa suggesting migratory birds could have played a role in transmission in the region.
In Kenya, AIVs surveillance among poultry and wild birds has not been routinely conducted, particularly following surveillance conducted from 2009-2011 which detected AIV in only 0.8% of chickens [
18]. In response to the 2018 outbreaks of H5N8 virus in Uganda, we initiated surveillance for AIVs among poultry traded in LBMs and among wild birds to assess possible introduction and circulation of AIVs in these populations in Kenya. In addition, given the potential of human infections with AIVs, we conducted surveillance for influenza viruses among poultry workers in the LBMs.
2. Materials and Methods
LBM surveillance
Site Selection
We targeted LBMs for surveillance of poultry and poultry workers from March 2019 through February 2020 within five counties of Kenya, including Nairobi, Nakuru, Kisumu, Kiambu, and Busia. Kisumu and Busia were selected because of their proximity to poultry trade flow with Uganda, where an HPAI H5N8 outbreak had previously been reported [
16]. Nakuru and Nairobi were selected to represent areas that receive poultry from different parts of Kenya. The market in Kiambu was selected because it primarily traded in non-chicken poultry. Overall, seven LBMs were selected including three in Nairobi and one market in each of the other counties.
Sample Size Estimation
We calculated the number of poultry specimens collected based on an estimated seroprevalence of 0.8% (95% confidence interval [CI] 0.6–1.1)[
18] reported among Kenyan poultry in LBMs in 2009 with an error margin of 1.3% at the 95% confidence level using formulas described by Thompson[
19]. A design effect of 1.5 was applied to account for clustering resulting in a sample size of 5,380.
Specimen Collection
Poultry: LBMs were visited monthly for the duration of the survey. Up to 20 stalls were selected in each market and 3–5 birds sampled in each stall. In Nyambari market in Kiambu County, where poultry were not caged, approximately 30% of the birds available at the market on any day of sample collection were sampled by species. For each sampled poultry, one oropharyngeal and one cloacal specimen were collected.
Poultry workers: All poultry workers aged >13 years were eligible for enrollment after providing consent/assent. A questionnaire collecting demographic characteristics of the enrolled participant was completed during the first contact. On subsequent monthly visits, a brief questionnaire on respiratory illness during the visit or the preceding 14 days was administered. Those who met the case definition for acute respiratory infection (ARI), defined as history of fever, cough, or red eyes, and with onset within the last seven days, had an oropharyngeal (OP) and nasal (NS) swab collected for influenza testing. Sample size calculations were not performed for poultry worker surveillance.
Wild Bird Surveillance
Site Selection
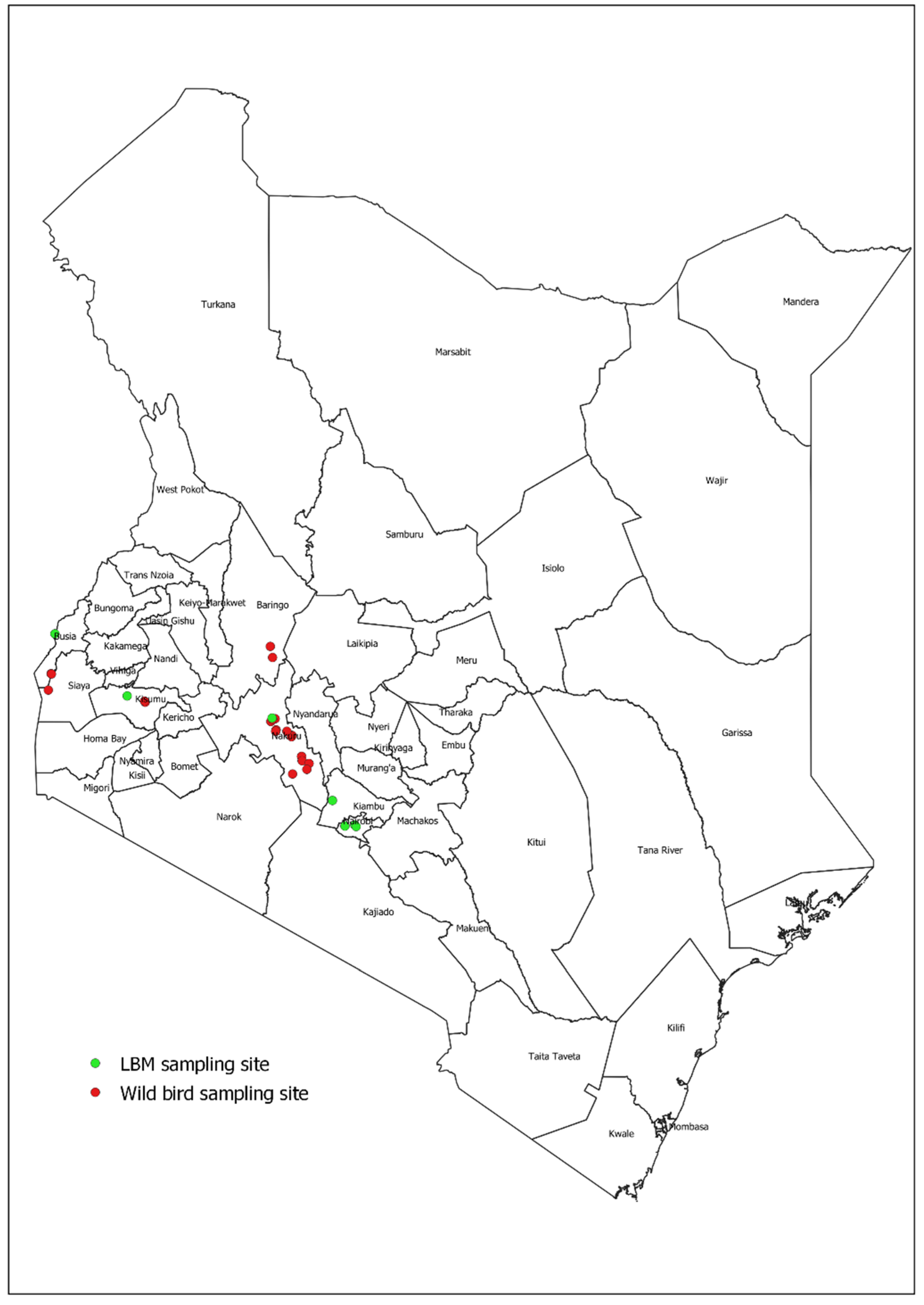
During January 2018, we collected freshly voided fecal specimens from wild birds on the shores of Lake Victoria, along its edges from Homabay to Busia County. From February–March 2020, sampling was conducted along the shorelines of Lake Bogoria, Lake Elementaita, Lake Nakuru, Lake Naivasha, and Lake Victoria (
Figure 1).
Sample Size Estimation
Prevalence of AIVs among wild birds in Kenya is not known but prevalence of AIVs in fecal specimen pools was reported at 2.3%, where 12/504 pools (of 5 specimens each) tested positive (and 3.7% in fecal specimens collected from fresh water lakes)[
20]. Using the lower prevalence and targeting surveillance strategies to detect at least 15 positive pools, we set out to collect 3,260 specimens to detect a prevalence as low as 1.2% at a 95% confidence level.
Sample Collection
At each site, an ornithologist on the team recorded data on the bird species observed including the dominant species, their migratory status (Palearctic, Afrotropical, or local), presence of dead birds, and any wild bird interaction with humans or poultry. Geographic coordinates for each site were also recorded.
Fresh environmental fecal specimens were collected using plastic-shafted polyester-tipped swabs and placed individually in cryovials containing 2 mL of freshly prepared viral transport media (VTM) containing bovine serum albumin and veal infusion broth supplemented with amphotericin B and gentamycin (
www.who.int/csr/resources/publications/surveillance/Annex8.pdf).
Virus Testing and Sequencing Analysis
Specimens from LBMs were labeled and maintained at 2–8 0C upon collection and subsequently frozen at -80 0C at the CDC-supported Kenya Medical Research Institute (KEMRI) Center for Global Health Research (CGHR) laboratory in Kisumu until testing. Wild bird fecal specimens were placed in cool boxes after collection and then frozen in liquid nitrogen within four hours of collection and subsequently frozen at -80 0C until testing. Not all collected specimens were screened for influenza A because of the high cost of reagents. Poultry worker specimens were maintained at 2–8 0C upon collection and subsequently frozen at -80 0C at the CDC-supported KEMRI- CGHR laboratory in Kisumu until testing.
Specimens were pooled in groups of five according to species and site of collection prior to RNA extraction. Pooled specimens were tested for influenza A virus RNA by real-time reverse transcription-polymerase chain reaction (RT-PCR) using primers and probes that target the matrix gene of influenza A viruses (Spackman et al, 2002). Cut-off for positivity was a cycle threshold (CT) value ≤40. Specimens from any positive pool were retested individually. Viral subtype was determined using primers and probes targeting the hemagglutinin genes of three avian influenza A subtypes (H5, H7, H9). Human specimens were screened individually for influenza A virus by RT-PCR.
Full genome sequencing was performed at the Influenza Division laboratories (CDC, Atlanta) on a random sample of positive LBM specimens (n=34), and all wild bird specimens (n=2) that tested positive for influenza A virus and had a CT value of <30. Total RNA was extracted from 100 μL of sample using the MagNA Pure 96 DNA and Viral NA Small Volume Kit (catalog no. 06542588001; Roche Diagnostics) following the manufacturer's protocol. Purified RNA was eluted into 50 μL nuclease-free water and screened for influenza A virus and Newcastle Disease Virus (NDV) using TaqMan real-time RT-PCR assays) [
21,
22]. Full genome sequencing was performed using Illumina MiSeq technology, as previously described [
23]. Sequences were deposited in GISAID.
1
HA sequences from Kenyan H9N2 isolates were used to construct a phylogenetic tree. Data were aligned using MUSCLE [
24] and sequences were trimmed to the beginning of the mature HA1 protein gene sequence using BioEdit v7.0. The neighbor-joining method [
25] with the Jukes-Cantor model in the Mega 7.0 software package [
26] were used to construct the final tree. The percentage of replicate trees in which the associated taxa clustered together was measured by the bootstrap test (1000 replicates) and bootstrap values are shown next to the branches. Nucleotide sequence alignments used for the HA trees were translated to amino acid protein sequences for comparison to the closest A(H9N2) World Health Organization (WHO) candidate vaccine virus (CVV), A/Oman/2747/2019. Amino acid changes are annotated on each branch of the tree.
Antigenic Characterization of AIVs
Specimens received at the CDC Influenza Division laboratories were inoculated into 10-day old embryonated eggs, incubated for 48 hours at 37°C, and harvested[
27]. Antigenic relatedness of the six isolates was determined using the hemagglutination inhibition (HI) assay. Ferret antisera used in the HI test were created by inoculating naïve ferrets intranasally with selected reference virus using a 50% egg infectious dose (EID50) of 10^
6. Antisera were harvested 14 days post-infection. Each ferret antiserum was treated with a receptor-destroying enzyme (RDE: Denka Seiken) at a 1:4 dilution for 18 hours at 37°C and further diluted with physiological saline for a total dilution of 1:10. The diluted antiserum was adsorbed with packed turkey red blood cells. Antiserum was serially diluted 2-fold in a 96-well v-bottom plate and all antigens were standardized to 8 HAU/50µL. Antisera and antigens were incubated together for 30 minutes at room temperature. Standardization and hemagglutination inhibition were determined using turkey red blood cells (0.5%)[
27].
1 GISAID accession numbers EPI_ISL_18856481 – EPI_ISL_18856545.
3. Results
Between March 2019 and February 2020, 10,340 poultry specimens were collected from seven LBMs in five counties in Kenya (
Table 1). Most of the specimens were collected from chickens (8,176; 79%) followed by ducks/geese (1,166; 11%), turkeys (590; 6%), pigeons (270; 3%), domesticated guinea fowl (114; 1%), and doves (24; 0.2%). The median stay of poultry at the market was 1 day for chickens [interquartile range (IQR), 1–2 days], 9 days [IQR 5–17 days] for guinea fowls, 12 days [IQR 10–21 days] for pigeons, 12 days [IQR 9–22 days] for doves, 16 days [IQR 10–21 days] for ducks/geese, and 17 days [IQR 10–21 days] for turkeys,.
During the LBM surveillance period, 155 poultry workers at the market were enrolled for follow-up from Burma (37; 24%), Busia (19; 12%), Kariokor (14; 9%), Kawangware (16; 10%), Kisumu (28; 18%), Nakuru (23; 15%), and Nyambari (18; 12%). The median age was 38 years [IQR 30–46 years] and 91 (59%) were males. Three quarters of the participants (n=120) were present during all 12 follow-up visits. A total of 18 OP/NS swabs from poultry workers with ARI (12%) were collected; none were positive for influenza A virus.
Across five lakes in Kenya, a total of 6,531 fecal specimens from wild birds were collected, including 3,351 (51.3%) in January 2018 (Lake Victoria only) and 3,180 (48.7%) in February–March of 2020 (
Figure 1,
Table 1). A total of 580 different bird species were observed in the sampling sites including 26% (n=149) Palearctic migrants, 19% (n=109) afrotropical migrants, and 56% (n=322) local types (
Supplementary table 1).
Detection of Influenza A Virus in LBM Poultry Specimens
From the specimens collected at LBMs (n=10,340), we randomly selected and tested 7,464 (72.2%), including 3,737 cloacal swabs and 3,727 OP swabs in 1,507 pools. A total of 135/1507 (9.0%) pools were positive for influenza A segregated as: 19/754 (2.5%) cloacal and 116/753 (15.4%) OP pools. On individual specimen testing, overall, 292 (3.9%) swabs were positive for influenza A, including 246/3727 (6.6%) OP swabs and 46/3737 (1.2%) cloacal swabs. Chickens most frequently tested positive for influenza A viruses (5.0%), followed by turkeys (0.3%), and ducks/geese (0.2%) (
Table 2). None of the specimens collected from pigeons, guinea fowl, and doves tested positive for influenza A. On hemagglutinin subtyping, most of the influenza A positive specimens (274/292, 93.8%) were H9. We randomly selected 34 H9 specimens for further subtyping and all were H9N2.
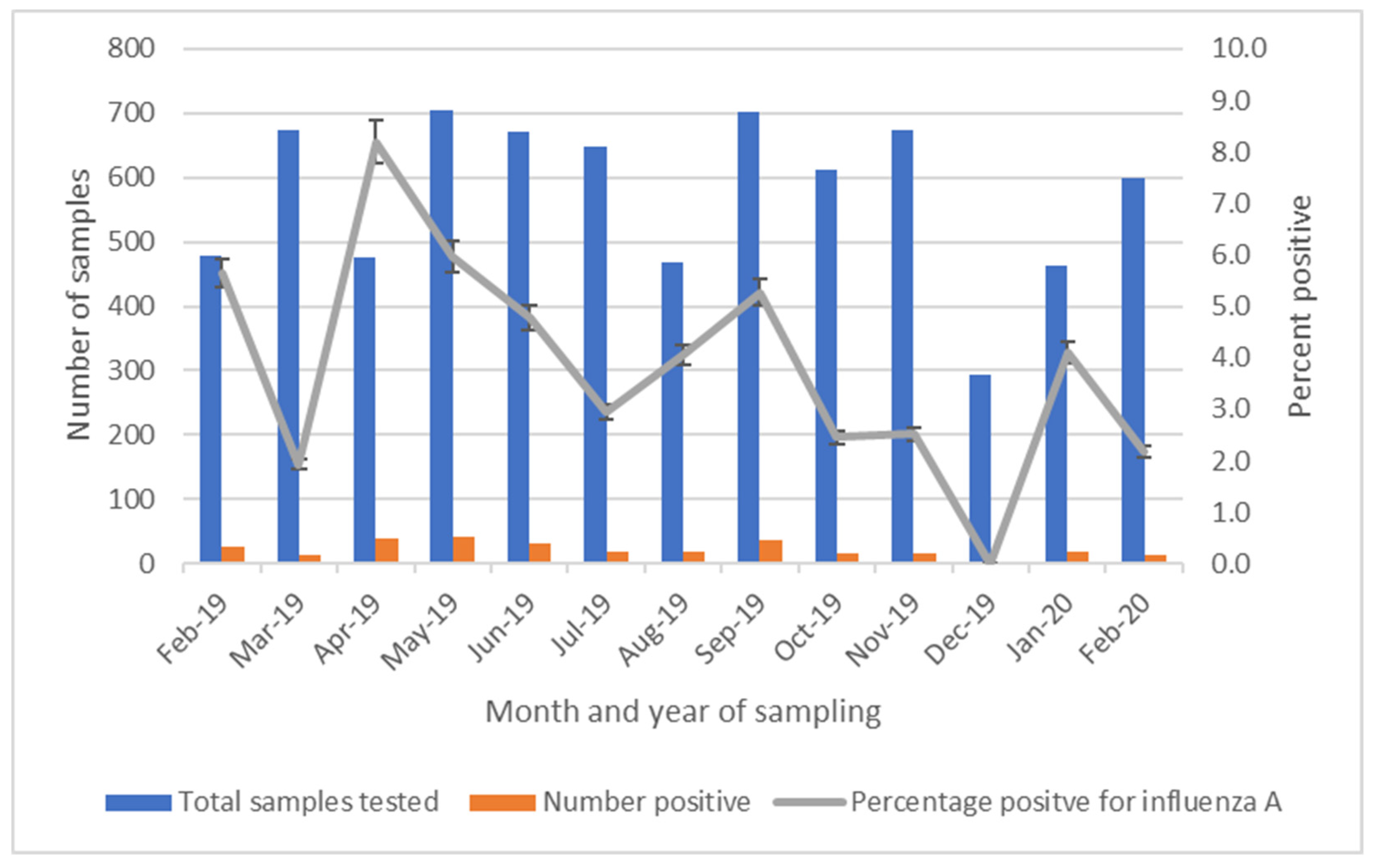
By region, influenza A was most commonly detected among specimens collected in Kawangware (17.5%), followed by Burma (11.1%), and Busia (0.7%). None of the specimens collected from the Nakuru market were positive (
Table 2). Influenza A was detected in 12 of 13 months during LBM surveillance, with detection most common during April 2019 and least common during December 2019 (range 0 – 8.2%) (
Figure 2).
Detection of Influenza A Virus in Wild Bird Fecal Specimens
A total of 6,531 specimens (in 1,307 pools of five) were tested and 10/1307 pools (0.8%) were positive for influenza A. Of individually tested specimens, 10/6531 (0.2%; 95% CI) 0.1–0.3) were positive for influenza A including 1/3351 (0.0%; 95% CI 0.0–0.2) collected in 2018 and 9/3180 (0.3%; 95% CI 0.1–0.5) collected in 2020. Among specimens collected in 2020, influenza A positivity was slightly more frequent in specimens from Lake Nakuru (0.7%; 4/560), followed by 0.4 % (2/484) from Lake Elementaita, and 0.3% (3/904) from Lake Naivasha; fecal specimens collected from Lake Bogoria and Lake Victoria basin tested negative.
Phylogenetic Analyses of the H9N2 Viruses from LBM Poultry
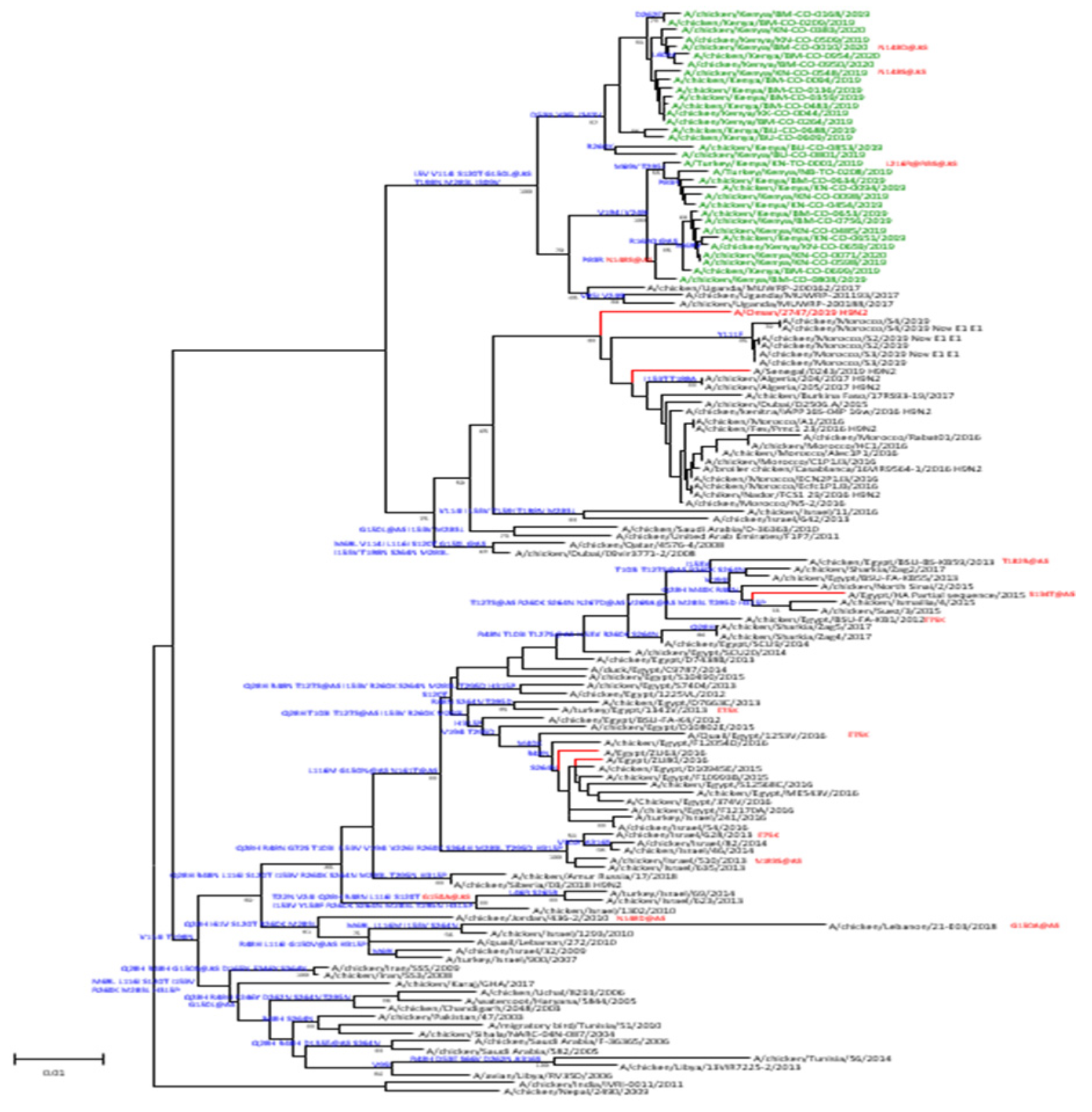
We obtained codon complete nucleotide sequences of all eight segments of 34 AIV isolates, including 32 from chickens and two from turkeys. Phylogenetic analyses of the HA sequences showed that the isolates belonged to the G1-lineage of H9N2 viruses (
Figure 3). Viruses from this study clustered with sequences of specimens collected in Uganda in 2017. On the same phylogenetic tree, sequences from West Africa and North Africa clustered separately from the East Africa viruses indicating evidence of local and regional circulation among poultry. Moreover, amino acid differences relative to the closest candidate vaccine virus (CVV) in the G1-linage, A/Oman/2747/2019, indicate multiple conserved changes in the HA1 portion of the sequenced viruses from Kenya. Viruses sequenced from this study had between 9–13 amino acid changes compared to the reference CVV of which several were identified at putative antigenic or receptor binding sites. All the viruses from this study had amino acid changes G150L at antigenic site B. Other mutations of the antigenic sites included N148S at antigenic site B, and N161S and R162Q at antigenic site D (
Supplementary Table). Of note, viruses collected from turkeys had changes of either L216R or Q compared to viruses from chickens, indicating likely species-specific receptor binding among terrestrial poultry.
Phylogenetic Analysis of the H5N2 Virus from Wild Bird Fecal Specimens
One influenza A positive fecal specimen collected from the Lake Victoria basin in 2018 was H5-positive and, upon phylogenetic analysis, determined to be LPAI A(H5N2) virus of the Eurasian lineage. We obtained whole-genome codon complete sequences of all eight segments of the A/environment/Kenya/Z201801195/2018 A(H5N2) virus. Additionally, one of the nine influenza A positive wild bird fecal specimens collected in 2020 was successfully sequenced and was subtyped as an A(H11) virus. Only the HA could be sequenced.
Antigenic Analysis Results of H9N2 Poultry Specimens
A panel of genetically related A(H9N2) viruses and CVVs were compared by HI assay to six A(H9N2) viruses from Kenya to determine antigenic relatedness. The panel of reference viruses and ferret antisera included three G1-lineage WHO CVVs; A/Hong Kong/1073/1999, IDCDC-RG31 (A/Bangladesh/0994/2011-like), and IDCDC-RG66A (A/Oman/2747/2019-like), a more recent CVV of the G1 lineage [
28].
The viruses isolated from Kenya are given below
2. Amino acid comparison of the hemagglutinin protein of the six isolates versus the closest genetically related WHO CVV IDCDC-RG66A (A/Oman/2747/2019-like) showed between 11 and 13 amino acid changes (
Supplementary Table 2). One amino acid change (N148S), in putative antigenic site B, was identified in multiple of the H9N2 viruses and two that were selected for virus isolation and HI testing, A/chicken/Kenya/BM-CO-0756/2019 and A/chicken/Kenya/BM-CO-0808/2019. Amino acid 148S in combination with 216L in antigenic site B has been shown to increase α2-6 binding [
29].
Antigenic analysis showed that the six virus isolates were inhibited within four-fold of homologous titers of the most recent G1 lineage World Health Organization CVV, IDCDC-RG66A (A/Oman/2747/2019-like). A CVV previously selected in the G1 lineage, IDCDC-RG31 (A/Bangladesh/0994/2011-like), also inhibited viruses within four-fold of the homologous virus titer (
Table 3), indicating that the identified amino acid differences did not result in substantial antigenic variation relative to the G1 lineage CVVs.
4. Discussion
Using multiple strategies and locations to monitor and identify avian influenza viruses in Kenya, we identified avian influenza viruses among traded poultry and wild birds, including two low pathogenicity viruses, H9N2 and H5N2. We found influenza A positivity in 3.9% of specimens collected from poultry traded in Kenyan LBMs between 2019 and 2020. On phylogenetic analysis, these viruses were genetically similar to other H9 viruses detected in East Africa. Concurrently, we found low prevalence circulation of LPAI H5N2 in wild bird populations. While there was no apparent transmission of these viruses to persons working in the LBMs during this monitoring period, related viruses belonging to the G1 lineage have resulted in human infections in Africa and elsewhere underscoring the public health threat and need for continuous surveillance [
6,
7,
30]
The observed prevalence in poultry represents a five-fold increase in influenza A virus detection compared with similar surveillance conducted a decade earlier (0.8% positivity during 2009–2011)[
18]. Virus detection was five times more frequent among tested oropharyngeal swabs (6.6%) compared to cloacal swabs (1.2%), a common observation among poultry [
18,
31]. H9N2 viruses were detected in chicken and turkey specimens during this period, and detection was variable by month of collection (0.0–8.2%), and by markets. Two of three LBMs in Nairobi that primarily traded indigenous poultry sourced from different counties had significantly higher influenza A virus positivity compared to all the LBMs. This suggests widespread transmission of H9N2 viruses in poultry populations across Kenya. High prevalence of H9N2 viruses has been reported in many countries in Asia, the Middle East, and Africa [
6]. The overall H9N2 prevalence observed was higher than previously reported (5.6%) in 2018 in Kenya[
12].
Phylogenetic analyses showed that all 34 H9N2 viruses belonged to the G1 lineage with minimal genetic variation among the cluster detected in Kenya, suggesting circulation of viruses from a single lineage. As previously shown [
12], viruses from this study in Kenya also clustered with those detected in Uganda, a neighboring country, further supporting that the outbreaks in East Africa were likely from a single lineage. This indicates that despite the contemporaneous circulation of virus clusters identified in West, North, and East Africa, distinct clusters were delineated implying that H9N2 viruses in East Africa may have evolved independently with no intermingling of virus gene pools. Analysis of HA protein sequences identified mutations at several antigenic sites (125, 148, 150, 161, 162, 178, 216) compared to the IDCDC-RG66A CVV (A/Oman/2747/2019-like). Despite these changes, the viruses isolated in this study remained antigenically similar to G1 lineage CVVs recommend for development by the WHO.
We found a low influenza prevalence among wild bird populations in Kenya. This prevalence was lower than that observed during the 2008 study (2.3%) in Kenya that detected H4N6 [
20]. On subtyping, we detected LPAI H5N2 in one fecal specimen. Phylogenetic analysis of the hemagglutinin (HA) gene of isolates from wild bird fecal specimen clustered with viruses from the Eurasian lineage. The LPAI H5N2 virus was collected around the Lake Victoria basin, an important bird resting area that hosts a variety of Palearctic and Afro-tropical migrants in addition to resident bird species. LPAI H5N2 viruses in migratory birds and poultry have been documented around the globe including in Nigeria in 2008 among wild birds with Eurasian migratory patterns implicating introduction of the virus to wild birds in African wetlands. Notably, during the 2016–2018 period, there were multiple independent introductions of HPAI H5N8 to different countries on the African continent by migratory birds, and sporadic spill-over to poultry populations, including the first ever recorded introduction of HPAI H5N8 in East Africa, was recorded [
2,
15,
16,
32]. While LPAI H5N2 has not been reported in poultry in this region over that period, it is likely that the virus was introduced during the same period.
These findings have several limitations. Because of limited resources, sampling among wild birds was conducted in only two time periods and in only one site (Lake Victoria basin) in 2018. Hence, we may have failed to detect some AIV introductions among wild bird populations. Similarly, LBM surveillance was limited to a certain number of counties and spanned only one year, which may not provide a comprehensive representation of all AIVs circulating in poultry populations in Kenya. However, the findings of this study highlight the importance of continued AIV surveillance in domestic and wild bird populations.
Since 2020, there have been reports of an increase in detection of AIVs in Europe, Asia, and North America. However, detections in the African continent have been limited to certain countries in Southern and Western Africa. Due to insufficient surveillance of AIVs in poultry and wild bird populations in most African countries, our findings along with recent findings indicating rapid annual expansion of HPAI H5N8 and LPAI H9N2 on the African continent demonstrate significant risk of virus introduction from outside of the continent, likely persistence in local poultry and wild bird populations, and potential spillover to domestic birds and humans. Strategies such as systematic LBM surveillance and targeted surveillance for influenza viruses in wild bird populations aligned to the seasonal migratory patterns could achieve early detection of virus incursion providing the opportunity for intervention measures to mitigate socioeconomic and public health impacts caused by outbreaks of HPAI viruses.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org, Table S1. List of predominant bird species per Lake (Feb/Mar 2020); Table S2. Amino acid difference table of H9N2 Kenya viruses compared with nearest candidate vaccine virus.
Author Contributions
Conceptualization, Peninah Munyua, Eric Osoro and Harry Oyas; Data curation, Eric Osoro, Ruth Njoroge and Doris Marwanga; Formal analysis, Peninah Munyua, Eric Osoro, Genyan Yang, Juliana DaSilva, Yunho Jang, John R Barnes, Clayton Onyango and Charles Davis; Funding acquisition, Peninah Munyua and Eric Osoro; Investigation, Peninah Munyua, Eric Osoro, Joyce Jones, George Njogu, Ruth Njoroge, Romona Ndanyi, Vincent Obanda, Carolyne Nasimiyu and Yunho Jang; Methodology, Peninah Munyua, Eric Osoro, Joyce Jones, Genyan Yang, Elizabeth Hunsperger, Doris Marwanga, Ben Andagalu, Vincent Obanda, Clayton Onyango and Charles Davis; Project administration, Eric Osoro; Resources, Eric Osoro, Harry Oyas, Romona Ndanyi, Nancy Otieno, Carolyne Nasimiyu, Obadiah Njagi, Gideon Emukule and Charles Davis; Software, Doris Marwanga; Supervision, Peninah Munyua, Eric Osoro, Elizabeth Hunsperger, Vincent Obanda, Gideon Emukule, Clayton Onyango and Charles Davis; Validation, Peninah Munyua, Eric Osoro and Charles Davis; Visualization, Peninah Munyua; Writing – original draft, Peninah Munyua, Eric Osoro and Charles Davis; Writing – review & editing, Peninah Munyua, Eric Osoro, Joyce Jones, George Njogu, Genyan Yang, Elizabeth Hunsperger, Christine Szablewski, Ruth Njoroge, Doris Marwanga, Harry Oyas, Ben Andagalu, Romona Ndanyi, Nancy Otieno, Vincent Obanda, Carolyne Nasimiyu, Obadiah Njagi, Juliana DaSilva, Yunho Jang, John R Barnes, Gideon Emukule, Clayton Onyango and Charles Davis.
Funding
Funding for the project was provided by U.S. Centers for Disease Control and Prevention under the research cooperative agreement with Washington State University (CDC 1U01GH002143).
Institutional Review Board Statement
This work was reviewed and received approval from Kenya Medical Research scientific and Ethics Review committee (#SERU/3670), the KEMRI Institutional Animal Care and Use Committee, Washington State University (IRB # 16743 and IACUC # 6186), and the US CDC Institutional Review Board (#7164) and Institutional Animal Care and Use Committee.. Approval for wild bird sampling was authorized by Kenya Wildlife Service (KWS/BRM/5001).
Informed Consent Statement
Written informed consent was obtained from all human participants and poultry owners gave informed consent for poultry sampling
Data Availability Statement
All data underlying this article are available in the article..
Acknowledgments
We thank staff of the Washington State University Global Health Program Kenya including Naomi Kemunto, Directorate of veterinary services at national and participating counties (Nairobi, Nakuru, Kiambu, Busia and Kisumu), Veterinary Department of the Kenya wildlife service and National museums of Kenya for specimen collection and data management and Kenya Medical research institute laboratories staff for specimen testing. We appreciate the poultry traders in all markets who participated in this study.
Conflicts of Interest
The authors declare no conflicts of interest.
Disclaimer
The findings and conclusions in this manuscript are those of the authors and do not necessarily represent the official position of the US Centers for Disease Control and Prevention or the Government of Kenya.
References
- Webster, R.G., et al., Evolution and ecology of influenza A viruses. Microbiological Reviews, 1992. 56(1): p. 152-79. [CrossRef]
- Sims, L. and I.H. Brown, Multicontinental panzootic of H5 highly pathogenic avian influenza (1996–2015). in Avian Influenza, D.E. Swayne, Editor. 2016, Wiley Blackwell. p. 202-247.
- Lee, D.H., et al., Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J Vet Sci, 2017. 18(S1): p. 269-280. [CrossRef]
- Olsen, B., et al., Global patterns of influenza a virus in wild birds. Science, 2006. 312(5772): p. 384-8. [CrossRef] [PubMed]
- Morens, D.M., J.K. Taubenberger, and A.S. Fauci, H7N9 avian influenza A virus and the perpetual challenge of potential human pandemicity. mBio, 2013. 4(4). [CrossRef]
- Peacock, T.H.P., et al., A Global Perspective on H9N2 Avian Influenza Virus. Viruses, 2019. 11(7). [CrossRef] [PubMed]
- Pusch, E.A. and D.L. Suarez, The Multifaceted Zoonotic Risk of H9N2 Avian Influenza. Vet Sci, 2018. 5(4). [CrossRef]
- Li, Y.T., et al., Avian influenza viruses in humans: lessons from past outbreaks. Br Med Bull, 2019. 132(1): p. 81-95. [CrossRef] [PubMed]
- WHO, Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003-2023. 2023, WHO: Geneva.
- WHO, Monthly risk assessment summary. 2024, WHO: Geneva.
- Wade, A., et al., Highly Pathogenic Avian Influenza A(H5N8) Virus, Cameroon, 2017. Emerg Infect Dis, 2018. 24(7): p. 1367-1370.
- Kariithi, H.M., et al., Genetic characterization and pathogenesis of the first H9N2 low pathogenic avian influenza viruses isolated from chickens in Kenyan live bird markets. Infect Genet Evol, 2020. 78: p. 104074. [CrossRef] [PubMed]
- Kalonda, A., et al., Avian Influenza Viruses Detected in Birds in Sub-Saharan Africa: A Systematic Review. Viruses, 2020. 12(9). [CrossRef]
- Venter, M., et al., Risk of Human Infections With Highly Pathogenic H5N2 and Low Pathogenic H7N1 Avian Influenza Strains During Outbreaks in Ostriches in South Africa. J Infect Dis, 2017. 216(suppl_4): p. S512-S519. [CrossRef]
- Sergei Khomenko, C.A., Laura Roberts, Lauren Waller, Kevin Shaw, Isabella Monne, Joanne Taylor, Madhur Dhingra, Claudia Pittiglio, Moon Mugyeom, Xavier Roche, Kivaria Fredrick, Akiko Kamata, Sam Okuthe, Philippe Kone, Lidewij Wiersma, Sophie Von Dobschuetz, Baba Soumare, Yilma Makonnen, Subhash Morzaria, Juan Lubroth., 2016–2018 Spread of H5N8 highly pathogenic avian influenza (HPAI) in sub-Saharan Africa: epidemiological and ecological observations. FOCUS ON, 2018. 12(12).
- FAO, H5N8 HPAI in Uganda Further Spread in Uganda and Neighbouring Countries (February 2017), in FAO Animal Health Risk Analysis – Assessment,, FAO, Editor. 2017, FAO: Rome.
- Ndumu, D., et al., Highly pathogenic avian influenza H5N8 Clade 2.3.4.4B virus in Uganda, 2017. Infect Genet Evol, 2018. 66: p. 269-271. [CrossRef] [PubMed]
- Munyua, P.M., et al., Detection of influenza A virus in live bird markets in Kenya, 2009-2011. Influenza and Other Respiratory Viruses, 2013. 7(2): p. 113-9. [CrossRef] [PubMed]
- Thompson, S., Sample Size in Sampling, I. John Wiley & Sons, Editor. 2012, John Wiley & Sons, Inc. p. 53-56.
- Ofula, V.O., et al., Detection of avian influenza viruses in wild waterbirds in the Rift Valley of Kenya using fecal sampling. Vector Borne Zoonotic Dis, 2013. 13(6): p. 394-400. [CrossRef] [PubMed]
- Miller, P.J. and M.K. Torchetti, Newcastle disease virus detection and differentiation from avian influenza. Methods Mol Biol, 2014. 1161: p. 235-9.
- Prevention, U.C.f.D.C.a., CDC Laboratory Support for Influenza Surveillance (CLSIS). 2013, CDC: Atlanta Geogia, USA.
- Shepard, S.S., et al., Viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler. BMC Genomics, 2016. 17: p. 708. [CrossRef]
- Edgar, R.C., MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res, 2004. 32(5): p. 1792-7. [CrossRef] [PubMed]
- Saitou, N. and M. Nei, The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol, 1987. 4(4): p. 406-25. [CrossRef] [PubMed]
- Tamura, K., et al., MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol, 2011. 28(10): p. 2731-9. [CrossRef] [PubMed]
- WHO, WHO Manual on Animal Influenza Diagnosis and Surveillance. 2002, World Health Organization: Geneva. p. 78.
- Group, S.H.W., Assessing the fitness of distinct clades of influenza A (H9N2) viruses. Emerg Microbes Infect, 2013. Nov;2(11):e75(11).
- Ilyushina, N.A., et al., Human-like receptor specificity does not affect the neuraminidase-inhibitor susceptibility of H5N1 influenza viruses. PLoS Pathog, 2008. 4(4): p. e1000043. [CrossRef]
- Arbani, O., et al., Low Pathogenic Avian Influenza H9N2 Viruses in Morocco: Antigenic and Molecular Evolution from 2021 to 2023. Viruses, 2023. 15(12). [CrossRef]
- Germeraad, E.A., et al., Virus Shedding of Avian Influenza in Poultry: A Systematic Review and Meta-Analysis. Viruses, 2019. 11(9). [CrossRef]
- El-Shesheny, R., et al., Genesis of Influenza A(H5N8) Viruses. Emerg Infect Dis, 2017. 23(8): p. 1368-1371. [CrossRef] [PubMed]
| 1 |
GISAID accession numbers EPI_ISL_18856481 – EPI_ISL_18856545. |
| 2 |
The viruses isolated from Kenya were A/chicken/Kenya/BU-CO-0688/2019, A/chicken/Kenya/BM-CO-0136/2019, A/chicken/Kenya/KN-CO-0454/2019, A/chicken/Kenya/BM-CO-0756/2019, A/chicken/Kenya/BM-CO-0808/2019, and A/chicken/Kenya/BM-CO-0010/2020 H9N2. |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).