Submitted:
12 August 2024
Posted:
13 August 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Methodology
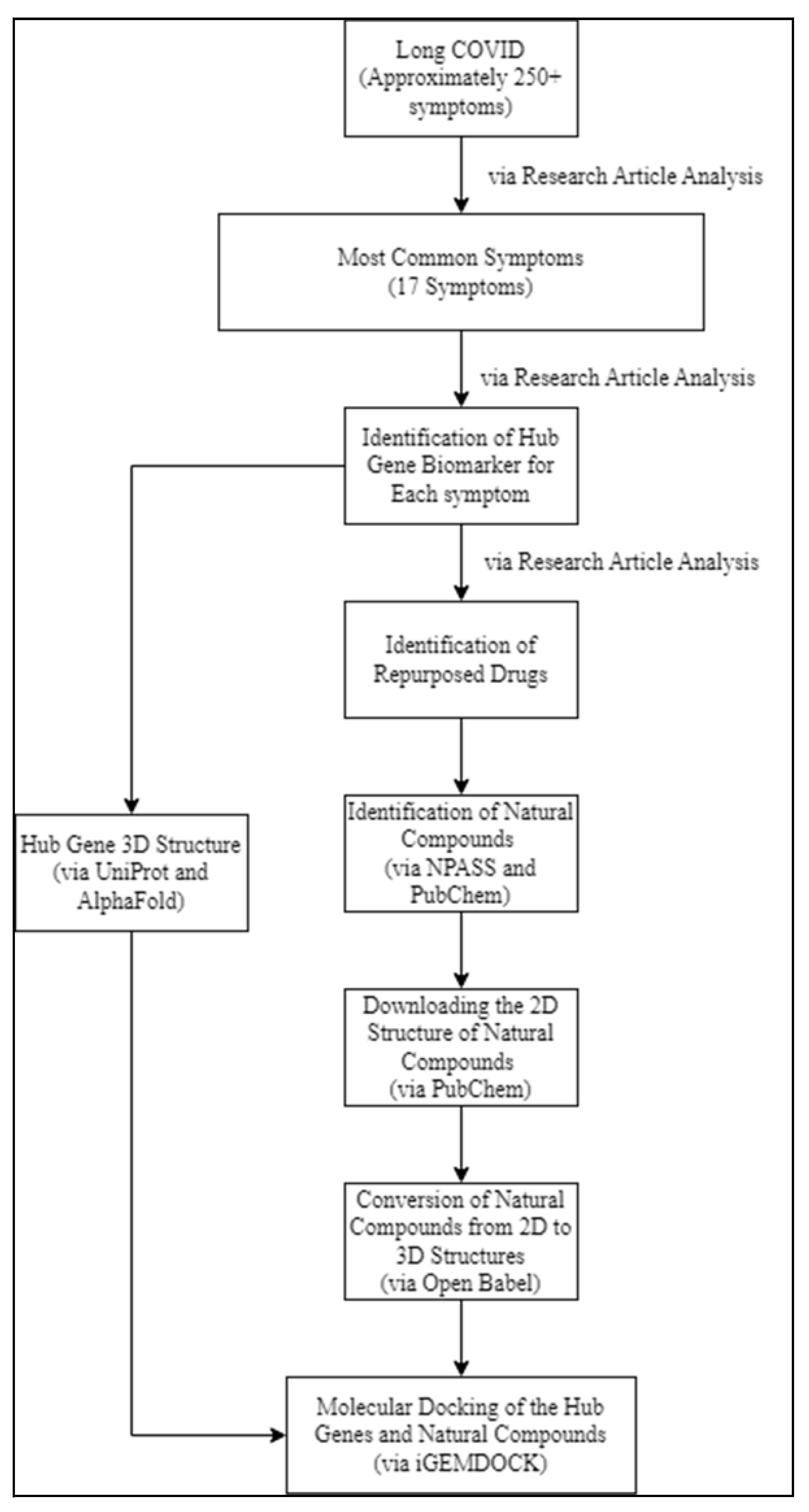
Data Retrieval
Identification of Natural Compounds
Hub Genes Data Collection (3D Structure)
2D Structure of Natural Compounds
Conversion of 2D structures to 3D structures (Natural Compounds)
Molecular Docking of the Hub Genes and Natural Compounds
Results
Discussion
| Hub Gene & Natural Component | Energy | VDW | HBond | Elec |
|---|---|---|---|---|
| RET-NPC56271 | -94.65 | -85.16 | -9.49 | 0.00 |
| ERBB4-NPC56271 | -94.29 | -85.58 | -8.71 | 0.00 |
| NF1-NPC117032 | -83.94 | -80.65 | -2.74 | -0.55 |
| SMAD4-NPC189301 | -73.85 | -53.31 | -21.34 | 0.80 |
| SMAD4-NPC176164 | -73.26 | -47.26 | -26.00 | 0.00 |
| SMAD4-NPC226027 | -70.85 | -56.50 | -13.59 | -0.75 |
| SMAD4-NPC174246 | -69.07 | -55.26 | -15.09 | 1.28 |
| SMAD4-NPC183845 | -67.90 | -31.13 | -36.77 | 0.00 |
| SMAD4-NPC112890 | -65.48 | -31.35 | -32.29 | -1.84 |
| SMAD4-NPC140872 | -64.27 | -46.55 | -17.72 | 0.00 |
| SMAD4-NPC118459 | -59.74 | -38.78 | -20.96 | 0.00 |
| SMAD4-NPC245027 | -55.96 | -44.00 | -10.50 | -1.46 |
| SMAD4-NPC162620 | -54.80 | -47.63 | -7.17 | 0.00 |
|
Table 1.2: Molecular Docking results for Shortness of Breath. Table showing hub genes and natural compounds derived from NPASS database | ||||
|
Hub Gene & Natural Component |
Energy |
VDW |
HBond |
Elec |
| CHRNE-NPC108434 | -106.33 | -93.35 | -12.99 | 0.00 |
| CHRNB1-NPC10908 | -104.70 | -92.98 | -11.72 | 0.00 |
| CHNRD-NPC11296 | -103.00 | -83.34 | -19.66 | 0.00 |
| CHRNB1-NPC115284 | -102.98 | -94.69 | -8.29 | 0.00 |
| CHNRD-NPC10871 | -101.86 | -78.73 | -23.43 | 0.29 |
| CHRNB1-NPC108434 | -98.10 | -89.39 | -8.72 | 0.00 |
| CHRND-NPC108434 | -97.35 | -78.84 | -18.50 | 0.00 |
| CHRNE-NPC10871 | -96.64 | -84.54 | -12.10 | 0.00 |
| CHRNB1-NPC116465 | -94.42 | -81.36 | -13.06 | 0.00 |
| CHNRD-NPC115284 | -94.09 | -93.60 | -0.49 | 0.00 |
| CHRNE-NPC11296 | -93.10 | -74.49 | -18.61 | 0.00 |
| CHRNB1-NPC10871 | -93.05 | -90.87 | -2.90 | 0.73 |
| CHRNB1-NPC11296 | -93.02 | -88.30 | -4.72 | 0.00 |
| CHNRD-NPC13916 | -92.25 | -74.21 | -18.04 | 0.00 |
| CHRNB1-NPC13916 | -92.22 | -81.32 | -10.89 | 0.00 |
| CHRNB1-NPC104196 | -91.30 | -87.10 | -4.20 | 0.00 |
| CHRND-NPC104196 | -88.75 | -71.78 | -16.98 | 0.00 |
| CHRNE-NPC106295 | -86.51 | -84.75 | -1.76 | 0.00 |
| CHNRD-NPC106295 | -84.77 | -83.12 | -1.65 | 0.00 |
| CHRNE-NPC10908 | -84.70 | -74.68 | -10.02 | 0.00 |
| CHNRD-NPC116465 | -83.01 | -68.36 | -14.65 | 0.00 |
| CHRNE-NPC115284 | -82.07 | -79.57 | -2.50 | 0.00 |
| CHRNB1-NPC106295 | -81.97 | -81.97 | 0.00 | 0.00 |
| CHRNB1-NPC114124 | -81.78 | -65.48 | -16.30 | 0.00 |
| CHRNB1-NPC103379 | -81.66 | -60.92 | -20.74 | 0.00 |
| CHRNE-NPC13916 | -81.21 | -65.30 | -15.91 | 0.00 |
| CHRND-NPC10908 | -79.41 | -79.41 | 0.00 | 0.00 |
| CHRNE-NPC116465 | -79.33 | -73.68 | -5.65 | 0.00 |
| CHRNE-NPC114124 | -78.43 | -60.67 | -17.77 | 0.00 |
| CHNRD-NPC103379 | -78.37 | -70.02 | -8.35 | 0.00 |
| CHRNE-NPC104196 | -78.19 | -73.19 | -5.00 | 0.00 |
| CHNRD-NPC114124 | -77.32 | -65.32 | -12.00 | 0.00 |
| CHNRD-NPC123323 | -76.01 | -73.71 | -2.30 | 0.00 |
| CHRNB1-NPC123323 | -74.20 | -66.93 | -7.27 | 0.00 |
| CHRNE-NPC123323 | -73.07 | -73.07 | 0.00 | 0.00 |
| CHRNE-NPC103379 | -72.46 | -59.26 | -13.20 | 0.00 |
|
Table 1.3: Molecular Docking results for Loss of Smell. Table showing hub genes and natural compounds derived from NPASS database | ||||
| Hub Gene & Natural Component | Energy | VDW | HBond | Elec |
| GNRH1-NPC69843 | -105.95 | -91.90 | -12.60 | -1.46 |
| GNRH1-NPC322594 | -100.52 | -82.39 | -18.12 | 0.00 |
| GNRH1-NPC328779 | -95.88 | -46.03 | -49.85 | 0.00 |
| GNRH1-NPC120887 | -92.41 | -70.14 | -22.27 | 0.00 |
| GNRH1-NPC17892 | -88.11 | -63.65 | -24.47 | 0.00 |
| GNRH1-NPC320249 | -82.83 | -55.55 | -27.28 | 0.00 |
| GNRH1-NPC324390 | -77.60 | -56.91 | -20.69 | 0.00 |
| GNRH1-NPC226769 | -73.68 | -44.94 | -28.74 | 0.00 |
| TAC3-NPC17760 | -70.87 | -64.87 | -6.00 | 0.00 |
| GNRH1-NPC474926 | -70.68 | -48.60 | -22.08 | 0.00 |
| GNRH1-NPC229249 | -70.02 | -70.02 | 0.00 | 0.00 |
| TAC3-NPC241086 | -69.42 | -59.93 | -9.49 | 0.00 |
| TAC3-NPC197239 | -68.37 | -61.77 | -6.61 | 0.00 |
| TAC3-NPC282087 | -67.22 | -51.77 | -15.45 | 0.00 |
| GNRH1-NPC106780 | -66.35 | -66.35 | 0.00 | 0.00 |
| TAC3-NPC259800 | -66.01 | -50.62 | -15.39 | 0.00 |
| GNRH1-4-NPC107135 | -58.84 | -36.30 | -22.54 | 0.00 |
| TAC3-NPC142638 | -58.49 | -38.96 | -19.54 | 0.00 |
| TAC3-NPC106551 | -57.96 | -45.84 | -12.78 | 0.66 |
| TAC3-NPC281686 | -57.96 | -44.77 | -13.83 | 0.64 |
| GNRH1-NPC43655 | -55.76 | -47.40 | -8.36 | 0.00 |
| TAC3-NPC188867 | -54.67 | -33.17 | -21.51 | 0.00 |
| TAC3-NPC239697 | -52.34 | -39.72 | -12.62 | 0.00 |
|
Table 1.4: Molecular Docking results for Headache. Table showing hub genes and natural compounds derived from NPASS database | ||||
| Hub Gene & Natural Component | Energy | VDW | HBond | Elec |
| TERT-NPC230098 | -97.62 | -85.53 | -12.09 | 0.00 |
| ESR1-NPC136948 | -96.97 | -78.32 | -18.65 | 0.00 |
| RET-NPC56271 | -94.65 | -85.16 | -9.49 | 0.00 |
| TERT-NPC474324 | -92.79 | -82.29 | -10.50 | 0.00 |
| ESR1-NPC123319 | -92.16 | -77.78 | -14.38 | 0.00 |
| TERT-NPC319549 | -91.83 | -71.07 | -20.76 | 0.00 |
| TERT-NPC33256 | -85.58 | -69.20 | -16.38 | 0.00 |
| ESR1-NPC1015 | -85.42 | -70.77 | -14.64 | 0.00 |
| TERT-NPC237044 | -85.10 | -75.60 | -9.50 | 0.00 |
| ESR1-NPC139397 | -84.00 | -80.50 | -3.50 | 0.00 |
| NF1-NPC117032 | -83.94 | -80.65 | -2.74 | -0.55 |
| TERT-NPC474325 | -83.26 | -62.39 | -20.87 | 0.00 |
| TERT-NPC298186 | -83.22 | -72.58 | -10.94 | 0.30 |
| ESR1-NPC255253 | -82.47 | -71.97 | -10.50 | 0.00 |
| TERT-NPC304675 | -81.06 | -65.27 | -15.79 | 0.00 |
| ESR1-NPC144258 | -80.38 | -74.17 | -6.21 | 0.00 |
| TERT-NPC301189 | -80.12 | -69.50 | -10.61 | 0.00 |
| TERT-NPC165797 | -78.17 | -69.69 | -8.47 | 0.00 |
| ESR1-NPC126993 | -76.15 | -73.83 | -2.33 | 0.00 |
| ESR1-NPC136548 | -74.01 | -61.01 | -13.00 | 0.00 |
| SMAD4-NPC189301 | -73.85 | -53.31 | -21.34 | 0.80 |
| SMAD4-NPC176164 | -73.26 | -47.26 | -26.00 | 0.00 |
| ESR1-NPC129913 | -73.01 | -64.51 | -8.50 | 0.00 |
| SMAD4-NPC226027 | -70.85 | -56.50 | -13.59 | -0.75 |
| SMAD4-NPC174246 | -69.07 | -55.26 | -15.09 | 1.28 |
| SMAD4-NPC183845 | -67.90 | -31.13 | -36.77 | 0.00 |
| SMAD4-NPC112890 | -65.48 | -31.35 | -32.29 | -1.84 |
| SMAD4-NPC140872 | -64.27 | -46.55 | -17.72 | 0.00 |
| SMAD4-NPC118459 | -59.74 | -38.78 | -20.96 | 0.00 |
| SMAD4-NPC245027 | -55.96 | -44.00 | -10.50 | -1.46 |
| SMAD4-NPC162620 | -54.80 | -47.63 | -7.17 | 0.00 |
|
Table 1.5: Molecular Docking results for Brain Fog. Table showing hub genes and natural compounds derived from NPASS database | ||||
| Hub Gene & Natural Component | Energy | VDW | HBond | Elec |
| CACNA1B-NPC198254 | -119.29 | -90.52 | -28.77 | 0.00 |
| CACNA1B-NPC473404 | -115.68 | -86.02 | -33.71 | 4.05 |
| CACNA1B-NPC153554 | -109.71 | -85.83 | -23.88 | 0.00 |
| CACNA1B-NPC240130 | -104.61 | -70.44 | -31.19 | -2.99 |
| CACNA1B-NPC274198 | -100.59 | -76.19 | -24.98 | 0.58 |
| KCNT1-NPC329708 | -85.81 | -77.31 | -8.50 | 0.00 |
| KCNT1-NPC193238 | -85.17 | -70.08 | -15.09 | 0.00 |
| KCNT1-NPC274291 | -84.64 | -70.82 | -13.82 | 0.00 |
| GABRA1-NPC317054 | -83.96 | -77.67 | -6.30 | 0.00 |
| KCNT1-NPC47059 | -80.67 | -73.67 | -7.00 | 0.00 |
| KCNT1-NPC203754 | -78.36 | -72.36 | -6.00 | 0.00 |
| KCNT1-NPC165349 | -76.72 | -72.59 | -4.13 | 0.00 |
| KCNT1-NPC264166 | -74.93 | -72.43 | -2.50 | 0.00 |
| KCNT1-NPC118832 | -70.83 | -62.29 | -8.54 | 0.00 |
| KCNT1-NPC150048 | -70.42 | -65.85 | -4.57 | 0.00 |
| KCNT1-NPC231986 | -69.20 | -65.70 | -3.50 | 0.00 |
|
Table 1.6: Molecular Docking results for Chest Pain. Table showing hub genes and natural compounds derived from NPASS database | ||||
| Hub Gene & Natural Component | Energy | VDW | HBond | Elec |
| RET-NPC56271 | -94.65 | -85.16 | -9.49 | 0.00 |
| NF1-NPC117032 | -83.94 | -80.65 | -2.74 | -0.55 |
| SMAD4-NPC189301 | -73.85 | -53.31 | -21.34 | 0.80 |
| SMAD4-NPC176164 | -73.26 | -47.26 | -26.00 | 0.00 |
| SMAD4-NPC226027 | -70.85 | -56.50 | -13.59 | -0.75 |
| SMAD4-NPC174246 | -69.07 | -55.26 | -15.09 | 1.28 |
| SMAD4-NPC183845 | -67.90 | -31.13 | -36.77 | 0.00 |
| SMAD4-NPC112890 | -65.48 | -31.35 | -32.29 | -1.84 |
| SMAD4-NPC140872 | -64.27 | -46.55 | -17.72 | 0.00 |
| SMAD4-NPC118459 | -59.74 | -38.78 | -20.96 | 0.00 |
| SMAD4-NPC245027 | -55.96 | -44.00 | -10.50 | -1.46 |
| SMAD4-NPC162620 | -54.80 | -47.63 | -7.17 | 0.00 |
|
Table 1.7: Molecular Docking results for Insomnia. Table showing hub genes and natural compounds derived from NPASS database | ||||
| Hub Gene & Natural Component | Energy | VDW | HBond | Elec |
| ABCB11-NPC100366 | -107.98 | -85.27 | -22.72 | 0.00 |
| NR1H4-NPC100366 | -97.79 | -87.37 | -10.42 | 0.00 |
| ABCB4-NPC213206 | -96.30 | -79.94 | -16.35 | 0.00 |
| ABCB4-NPC187022 | -96.10 | -85.42 | -10.68 | 0.00 |
| ABCB4-NPC208890 | -94.01 | -79.52 | -14.49 | 0.00 |
| ABCB4-NPC169387 | -92.86 | -90.86 | -2.00 | 0.00 |
| ABCB4-NPC136860 | -88.28 | -79.85 | -8.43 | 0.00 |
| ABCB4-NPC222524 | -86.16 | -78.11 | -8.05 | 0.00 |
| SLC12A3-NPC321053 | -81.71 | -47.62 | -34.09 | 0.00 |
| ABCB4-NPC188163 | -81.18 | -71.92 | -9.26 | 0.00 |
| ABCB4-NPC128019 | -71.50 | -61.79 | -9.71 | 0.00 |
|
Table 1.8: Molecular Docking results for Heart Palpitations. Table showing hub genes and natural compounds derived from NPASS database | ||||
| Hub Gene & Natural Component | Energy | VDW | HBond | Elec |
| GATA4-NPC101636 | -107.59 | -90.62 | -16.97 | 0.00 |
| GATA4-NPC100818 | -103.25 | -67.84 | -35.40 | 0.00 |
| GATA4-NPC100887 | -98.92 | -67.75 | -31.18 | 0.00 |
| GATA4-NPC10097 | -89.52 | -56.87 | -32.65 | 0.00 |
| GATA4-NPC100985 | -84.82 | -62.35 | -22.47 | 0.00 |
| SCN5A-NPC162417 | -83.58 | -73.08 | -10.50 | 0.00 |
| SCN5A-NPC64436 | -82.21 | -74.38 | -7.83 | 0.00 |
| SCN5A-NPC470971 | -81.67 | -71.48 | -10.19 | 0.00 |
| GATA4-NPC101366 | -79.21 | -54.46 | -24.75 | 0.00 |
| SCN5A-NPC322433 | -78.26 | -60.08 | -18.18 | 0.00 |
| SCN5A-NPC265100 | -77.86 | -70.08 | -7.78 | 0.00 |
| SCN5A-NPC136112 | -77.71 | -66.44 | -11.27 | 0.00 |
| GATA4-NPC101294 | -76.70 | -69.70 | -7.00 | 0.00 |
| SCN5A-NPC26285 | -76.27 | -73.59 | -2.68 | 0.00 |
| SCN5A-NPC242933 | -75.49 | -60.87 | -14.62 | 0.00 |
| GATA4-NPC10027 | -74.31 | -65.03 | -9.28 | 0.00 |
| SCN5A-NPC319645 | -72.42 | -64.05 | -8.37 | 0.00 |
| SCN5A-NPC226143 | -72.40 | -68.95 | -3.45 | 0.00 |
| GATA4-NPC100986 | -71.88 | -55.38 | -16.50 | 0.00 |
| SCN5A-NPC248462 | -71.41 | -58.66 | -12.75 | 0.00 |
| SCN5A-NPC70406 | -70.84 | -52.41 | -18.43 | 0.00 |
| SCN5A-NPC141739 | -70.80 | -65.80 | -5.00 | 0.00 |
| SCN5A-NPC26524 | -68.43 | -63.12 | -5.30 | 0.00 |
| SCN5A-NPC173295 | -63.42 | -55.17 | -8.26 | 0.00 |
| SCN5A-NPC57051 | -63.00 | -60.62 | -2.38 | 0.00 |
| SCN5A-NPC42383 | -61.93 | -56.37 | -5.56 | 0.00 |
| SCN5A-NPC65408 | -61.57 | -55.44 | -6.13 | 0.00 |
| SCN5A-NPC97811 | -58.36 | -55.41 | -2.94 | 0.00 |
| SCN5A-NPC24777 | -57.83 | -50.23 | -7.60 | 0.00 |
|
Table 1.9: Molecular Docking results for Dizziness. Table showing hub genes and natural compounds derived from NPASS database | ||||
| Hub Gene & Natural Component | Energy | VDW | HBond | Elec |
| RET-NPC56271 | -94.65 | -85.16 | -9.49 | 0.00 |
| RYR2-NPC122235 | -85.94 | -78.79 | -7.15 | 0.00 |
| NF1-NPC117032 | -83.94 | -80.65 | -2.74 | -0.55 |
| SCN5A-NPC162417 | -83.58 | -73.08 | -10.50 | 0.00 |
| SCN5A-NPC64436 | -82.21 | -74.38 | -7.83 | 0.00 |
| SCN5A-NPC470971 | -81.67 | -71.48 | -10.19 | 0.00 |
| SCN5A-NPC322433 | -78.26 | -60.08 | -18.18 | 0.00 |
| SCN5A-NPC265100 | -77.86 | -70.08 | -7.78 | 0.00 |
| SCN5A-NPC136112 | -77.71 | -66.44 | -11.27 | 0.00 |
| SCN5A-NPC26285 | -76.27 | -73.59 | -2.68 | 0.00 |
| SCN5A-NPC242933 | -75.49 | -60.87 | -14.62 | 0.00 |
| CACNA1G-NPC264400 | -74.95 | -58.77 | -16.18 | 0.00 |
| CACNA1G-NPC55529 | -73.98 | -62.68 | -11.30 | 0.00 |
| RYR2-NPC150323 | -73.75 | -47.76 | -26.00 | 0.00 |
| CACNA1G-NPC470926 | -73.59 | -71.09 | -2.50 | 0.00 |
| SCN5A-NPC319645 | -72.42 | -64.05 | -8.37 | 0.00 |
| SCN5A-NPC226143 | -72.40 | -68.95 | -3.45 | 0.00 |
| CACNA1G-NPC256452 | -71.53 | -61.21 | -10.32 | 0.00 |
| SCN5A-NPC248462 | -71.41 | -58.66 | -12.75 | 0.00 |
| SCN5A-NPC70406 | -70.84 | -52.41 | -18.43 | 0.00 |
| SCN5A-NPC141739 | -70.80 | -65.80 | -5.00 | 0.00 |
| SCN5A-NPC26524 | -68.43 | -63.12 | -5.30 | 0.00 |
| RYR2-NPC319645 | -68.19 | -57.69 | -10.50 | 0.00 |
| CACNA1G-NPC71140 | -66.84 | -59.10 | -7.75 | 0.00 |
| SCN5A-NPC173295 | -63.42 | -55.17 | -8.26 | 0.00 |
| RYR2-NPC226794 | -63.12 | -47.87 | -15.24 | 0.00 |
| SCN5A-NPC57051 | -63.00 | -60.62 | -2.38 | 0.00 |
| SCN5A-NPC42383 | -61.93 | -56.37 | -5.56 | 0.00 |
| SCN1B-NPC264400 | -61.81 | -41.42 | -20.39 | 0.00 |
| SCN5A-NPC65408 | -61.57 | -55.44 | -6.13 | 0.00 |
| SCN5A-NPC97811 | -58.36 | -55.41 | -2.94 | 0.00 |
| SCN5A-NPC24777 | -57.83 | -50.23 | -7.60 | 0.00 |
|
Table 1.10: Molecular Docking results for Joint Pain. Table showing hub genes and natural compounds derived from NPASS database | ||||
| Hub Gene & Natural Component | Energy | VDW | HBond | Elec |
| FAS-NPC115624 | -115.52 | -84.27 | -31.25 | 0.00 |
| FAS-NPC116759 | -115.52 | -77.79 | -37.72 | 0.00 |
| FAS-NPC103197 | -110.38 | -97.35 | -13.03 | 0.00 |
| FAS-NPC152424 | -108.06 | -81.95 | -26.11 | 0.00 |
| FAS-NPC100465 | -103.29 | -71.46 | -31.83 | 0.00 |
| FAS-NPC119910 | -103.26 | -89.55 | -13.71 | 0.00 |
| FAS-NPC14294 | -102.40 | -73.18 | -29.22 | 0.00 |
| FAS-NPC150943 | -97.35 | -71.67 | -25.68 | 0.00 |
| FAS-NPC115281 | -95.30 | -76.80 | -18.51 | 0.00 |
| FAS-NPC163527 | -91.19 | -70.10 | -21.08 | 0.00 |
| CR2-NPC139397 | -70.48 | -63.87 | -6.61 | 0.00 |
| Hub Gene & Natural Component | Energy | VDW | HBond | Elec |
|---|---|---|---|---|
| CDH23-NPC67043 | -53.12 | -42.68 | -10.44 | 0.00 |
| Hub Gene & Natural Component | Energy | VDW | HBond | Elec |
|---|---|---|---|---|
| CACNA1D-NPC36836 | -109.75 | -94.82 | -16.40 | 1.47 |
| TERT-NPC230098 | -97.62 | -85.53 | -12.09 | 0.00 |
| RET-NPC56271 | -94.65 | -85.16 | -9.49 | 0.00 |
| TERT-NPC474324 | -92.79 | -82.29 | -10.50 | 0.00 |
| CACNA1D-NPC63370 | -92.73 | -85.36 | -8.25 | 0.88 |
| TERT-NPC319549 | -91.83 | -71.07 | -20.76 | 0.00 |
| TERT-NPC33256 | -85.58 | -69.20 | -16.38 | 0.00 |
| TERT-NPC237044 | -85.10 | -75.60 | -9.50 | 0.00 |
| NF1-NPC117032 | -83.94 | -80.65 | -2.74 | -0.55 |
| TERT-NPC474325 | -83.26 | -62.39 | -20.87 | 0.00 |
| TERT-NPC298186 | -83.22 | -72.58 | -10.94 | 0.30 |
| TERT-NPC304675 | -81.06 | -65.27 | -15.79 | 0.00 |
| TERT-NPC301189 | -80.12 | -69.50 | -10.61 | 0.00 |
| CACNA1D-NPC190945 | -79.50 | -61.18 | -18.10 | -0.22 |
| TERT-NPC165797 | -78.17 | -69.69 | -8.47 | 0.00 |
| Hub Gene & Natural Component | Energy | VDW | HBond | Elec | |||
|---|---|---|---|---|---|---|---|
| MEN1-NPC474814 | -114.41 | -96.24 | -18.17 | 0.00 | |||
| MEN1-NPC199737 | -95.91 | -61.68 | -34.24 | 0.00 | |||
| MEN1-NPC471778 | -94.89 | -77.62 | -17.27 | 0.00 | |||
| MEN1-NPC112336 | -90.02 | -69.56 | -20.45 | 0.00 | |||
| MEN1-NPC301702 | -82.55 | -47.69 | -34.86 | 0.00 | |||
| MEN1-NPC181526 | -74.30 | -46.46 | -27.84 | 0.00 | |||
| SMAD4-NPC189301 | -73.85 | -53.31 | -21.34 | 0.80 | |||
| MEN1-NPC323798 | -73.31 | -50.23 | -23.08 | 0.00 | |||
| SMAD4-NPC176164 | -73.26 | -47.26 | -26.00 | 0.00 | |||
| SMAD4-NPC226027 | -70.85 | -56.50 | -13.59 | -0.75 | |||
| SMAD4-NPC174246 | -69.07 | -55.26 | -15.09 | 1.28 | |||
| SMAD4-NPC183845 | -67.90 | -31.13 | -36.77 | 0.00 | |||
| MEN1-NPC316574 | -67.29 | -39.65 | -27.64 | 0.00 | |||
| SMAD4-NPC112890 | -65.48 | -31.35 | -32.29 | -1.84 | |||
| SMAD4-NPC140872 | -64.27 | -46.55 | -17.72 | 0.00 | |||
| SMAD4-NPC118459 | -59.74 | -38.78 | -20.96 | 0.00 | |||
| SMAD4-NPC245027 | -55.96 | -44.00 | -10.50 | -1.46 | |||
| SMAD4-NPC162620 | -54.80 | -47.63 | -7.17 | 0.00 | |||
Conclusion
Supplementary Materials
Acknowledgements
References
- Akbarialiabad, H., Taghrir, M. H., Abdollahi, A., Ghahramani, N., Kumar, M., Paydar, S., Razani, B., Mwangi, J., Asadi-Pooya, A. A., Malekmakan, L., & Bastani, B. (2021). Long COVID, a comprehensive systematic scoping review. Infection, 49(6), 1163–1186. [CrossRef]
- Amaravathi, A., Oblinger, J. L., Welling, D. B., Kinghorn, A. D., & Chang, L. (2021). Neurofibromatosis: Molecular Pathogenesis and Natural Compounds as Potential Treatments. Frontiers in Oncology, 11. [CrossRef]
- Atanasov, A. G., Waltenberger, B., Pferschy-Wenzig, E., Linder, T., Wawrosch, C., Uhrin, P., Temml, V., Wang, L., Schwaiger, S., Heiss, E. H., Rollinger, J. M., Schuster, D., Breuss, J. M., Bochkov, V. N., Mihovilovic, M. D., Kopp, B., Bauer, R., Dirsch, V. M., & Stuppner, H. (2015). Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnology Advances, 33(8), 1582–1614. [CrossRef]
- Atanasov, A. G., Zotchev, S. B., Dirsch, V. M., & Supuran, C. T. (2021). Natural products in drug discovery: advances and opportunities. Nature Reviews Drug Discovery, 20(3), 200–216. [CrossRef]
- Castanares-Zapatero, D., Chalon, P., Kohn, L., Dauvrin, M., Detollenaere, J., De Noordhout, C. M., Jong, C., Cleemput, I., & Van Den Heede, K. (2022). Pathophysiology and mechanism of long COVID: a comprehensive review. Annals of Medicine, 54(1), 1473–1487. [CrossRef]
- Crook, H., Raza, S., Nowell, J., Young, M. K., & Edison, P. (2021). Long covid—mechanisms, risk factors, and management. BMJ, n1648. [CrossRef]
- Das, S., & Kumar, S. (2022). Long COVID: G Protein-Coupled Receptors (GPCRs) responsible for persistent post-COVID symptoms. bioRxiv (Cold Spring Harbor Laboratory). [CrossRef]
- Davis, H. E., McCorkell, L., Vogel, J., & Topol, E. J. (2023). Long COVID: major findings, mechanisms and recommendations. Nature Reviews Microbiology. [CrossRef]
- Deer, R. R., Rock, M. A., Vasilevsky, N., Carmody, L. C., Rando, H. M., Anzalone, A. J., Basson, M. D., Bennett, T. D., Bergquist, T., Boudreau, E. A., Bramante, C. T., Byrd, J. B., Callahan, T. J., Chan, L. M., Chu, H., Chute, C. G., Coleman, B., Davis, H. M., Gagnier, J., . . . Robinson, P. N. (2021). Characterizing Long COVID: Deep Phenotype of a Complex Condition. EBioMedicine, 74, 103722. [CrossRef]
- Degen, C. V., Mikuteit, M., Niewolik, J., Schröder, D., Vahldiek, K., Mücke, U., Heinemann, S., Müller, F., Behrens, G. M. N., Klawonn, F., Jablonka, A., & Steffens, S. (2022). Self-reported Tinnitus and Vertigo or Dizziness in a Cohort of Adult Long COVID Patients. Frontiers in Neurology, 13. [CrossRef]
- DePace, N. L., & DePace, N. L. (2022). Long-COVID Syndrome and the Cardiovascular System: A Review of Neurocardiologic Effects on Multiple Systems. Current Cardiology Reports, 24(11), 1711–1726. [CrossRef]
- El-Seedi, H. R. (2007). Antimicrobial Arylcoumarins from Asphodelus microcarpus. Journal of Natural Products, 70(1), 118–120. [CrossRef]
- Gao, Z., Han, B., Wang, H., Shi, C., Xiong, L., & Gu, A. (2012). Clinical observation of gefitinib as a first-line therapy in sixty-eight patients with advanced NSCLC. Oncology Letters, 3(5), 1064–1068. [CrossRef]
- Garg, P., Ahuja, V., Kumar, A., & Wig, N. (2021). The “post-COVID” syndrome: How deep is the damage? Journal of Medical Virology, 93(2), 673–674. [CrossRef]
- Hentsch, L., Cocetta, S., Allali, G., Santana, I., Eason, R., Adam, E., & Janssens, J. (2021). Breathlessness and COVID-19: A Call for Research. Respiration, 100(10), 1016–1026. [CrossRef]
- Huang, B., Chang, P., Yeh, W., Lee, C., Tsai, C., Lin, C., Lin, H., Liu, Y., Wu, C., Ko, P., Huang, S., Hsu, H., & Lu, D. (2014). Anti-Neuroinflammatory Effects of the Calcium Channel Blocker Nicardipine on Microglial Cells: Implications for Neuroprotection. PLOS ONE, 9(3), e91167. [CrossRef]
- Huijiao, Y., Wang, J., & Kong, L. (2014). Cytotoxic Dammarane-Type Triterpenoids from the Stem Bark of Dysoxylum binecteriferum. Journal of Natural Products, 77(2), 234–242. [CrossRef]
- Joli, J., Buck, P., Zipfel, S., & Stengel, A. (2022). Post-COVID-19 fatigue: A systematic review. Frontiers in Psychiatry, 13. [CrossRef]
- Kirk, R. D., He, H., Wahome, P. K., Wu, S., Carter, G. T., & Bertin, M. J. (2021). New Micropeptins with Anti-Neuroinflammatory Activity Isolated from a Cyanobacterial Bloom. ACS Omega, 6(23), 15472–15478. [CrossRef]
- Korres, G. M., Kitsos, D. K., Kaski, D., Tsogka, A., Giannopoulos, S., Giannopapas, V., Sideris, G., Tyrellis, G., & Voumvourakis, K. I. (2022). The Prevalence of Dizziness and Vertigo in COVID-19 Patients: A Systematic Review. Brain Sciences, 12(7), 948. [CrossRef]
- Lopez-Leon, S., Wegman-Ostrosky, T., Perelman, C., Sepulveda, R., Rebolledo, P. A., Cuapio, A., & Villapol, S. (2021). More than 50 long-term effects of COVID-19: a systematic review and meta-analysis. Scientific Reports, 11(1). [CrossRef]
- Membrilla, J. A., Caronna, E., Trigo, J., Gonzalez-Martinez, A., Layos-Romero, A., Pozo-Rosich, P., Ál, G., Gago-Veiga, A., Andrés-López, A., & De Terán, J. D. (2021). Persistent headache after COVID-19: Pathophysioloy, clinic and treatment. Neurology Perspectives, 1, S31–S36. [CrossRef]
- Nakatomi, K., Nakamura, Y., Tetsuya, I., & Kohno, S. (2011). Treatment with gefitinib after erlotinib-induced liver injury: a case report. Journal of Medical Case Reports, 5(1). [CrossRef]
- Natale, R. B. (2004). Effects of ZD1839 (Iressa, gefitinib) treatment on symptoms and quality of life in patients with advanced non-small cell lung cancer. Seminars in Oncology, 31, 23–30. [CrossRef]
- Nouraeinejad, A. (2022). Brain fog as a Long-term Sequela of COVID-19. SN Comprehensive Clinical Medicine, 5(1). [CrossRef]
- Park, J., Wang, X., & Xu, R. (2022). Revealing the mystery of persistent smell loss in long COVID patients. International Journal of Biological Sciences, 18(12), 4795–4808. [CrossRef]
- Pinzi, L., & Rastelli, G. (2019). Molecular Docking: Shifting Paradigms in Drug Discovery. International Journal of Molecular Sciences, 20(18), 4331. [CrossRef]
- Raveendran, A. V., Jayadevan, R., & Sashidharan, S. (2021). Long COVID: An overview. Diabetes and Metabolic Syndrome: Clinical Research and Reviews, 15(3), 869–875. [CrossRef]
- Roca-Fernandez, A., Wamil, M., Telford, A., Carapella, V., Borlotti, A., Monteiro, D., Thomaides-Brears, H., Dennis, A., Banerjee, R., Robson, M. D., Lip, G. Y. H., Bull, S., Heightman, M., Ntusi, N., & Banerjee, A. (2022). Cardiac impairment in Long Covid 1-year post SARS-CoV-2 infection. European Heart Journal, 43(Supplement_2). [CrossRef]
- Sheung, W., MA, Mutka, T. S., Vesley, B., Amsler, M. O., McClintock, J. B., Amsler, C. D., Perman, J. A., Singh, R. K., Maiese, W. M., Zaworotko, M. J., Kyle, D. E., & Baker, B. J. (2009). Norselic Acids A−E, Highly Oxidized Anti-infective Steroids that Deter Mesograzer Predation, from the Antarctic Sponge Crella sp. Journal of Natural Products, 72(10), 1842–1846. [CrossRef]
- Sudre, C. H., Murray, B. J., Varsavsky, T., Graham, M., Penfold, R. S., Bowyer, R. C. E., Pujol, J., Klaser, K., Antonelli, M., Canas, L. S., Molteni, E., Modat, M., Cardoso,.
- M. J., May, A., Ganesh, S., Davies, R. J., Nguyen, L. H., Drew, D. A., Astley, C. M., . . . Steves, C. J. (2021). Attributes and predictors of long COVID. Nature Medicine, 27(4), 626–631. [CrossRef]
- Van Der Voort, P. H. J., Moser, J., Zandstra, D. F., Kobold, A. C. M., Knoester, M., Calkhoven, C. F., Hamming, I., & Van Meurs, M. (2020). Leptin levels in SARS-CoV-2 infection related respiratory failure: A cross-sectional study and a pathophysiological framework on the role of fat tissue. Heliyon, 6(8), e04696. [CrossRef]
- Vijayakumar, B., Tonkin, J. A., Devaraj, A., Philip, K. E. J., Orton, C. R., Desai, S. R., & Shah, P. L. (2022). CT Lung Abnormalities after COVID-19 at 3 Months and 1 Year after Hospital Discharge. Radiology, 303(2), 444–454. [CrossRef]
- Yang, S., Yusoff, K., Ajat, M., Yap, W. S., Lim, S. H., & Lai, K. (2021). Antimicrobial activity and mode of action of terpene linalyl anthranilate against carbapenemase-producing Klebsiella pneumoniae. Journal of Pharmaceutical Analysis, 11(2), 210–219. [CrossRef]
- Yong, S. J. (2021). Persistent Brainstem Dysfunction in Long-COVID: A Hypothesis. ACS Chemical Neuroscience, 12(4), 573–580. [CrossRef]
- Zilla, M. K., Nayak, D., Amin, H., Nalli, Y., Rah, B., Chakraborty, S., Kitchlu, S., Goswami, A., & Ali, A. (2014). 4′-Demethyl-deoxypodophyllotoxin glucoside isolated from Podophyllum hexandrum exhibits potential anticancer activities by altering Chk-2 signaling pathway in MCF-7 breast cancer cells. Chemico-Biological Interactions, 224, 100–107. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




