1. Introduction
Methane (CH₄) is a potent greenhouse gas pivotal to global climate dynamics, primarily sourced from biogenic emissions driven by microbial activity, which constitute approximately 90% of the global CH₄ budget, estimated at 380-755 Tg annually. Significant contributors include coal beds, seafloor sediments, and subsurface reservoirs, with deep-sea sediments alone contributing around 20% of these emissions [
1,
2,
3].
Archaea are well documented as major producers of CH₄, utilizing various biochemical pathways such as CO₂ reduction with H₂, acetate reduction, and methylotrophic pathways involving methanol and methoxy group-containing substrates [
4]. Recent research has also identified aerobic bacterial pathways contributing to CH₄ production, including methylthio-alkane reductase involved in methionine biosynthesis and C-P lyase in phosphonate ester degradation [
5,
6]. Furthermore, both plant and animal cells have been observed releasing CH₄ under aerobic conditions independently of endosymbionts, although the precise mechanisms remain elusive [
7,
8]. In contrast, fungi, a vital group of eukaryotic organisms, have received comparatively less attention concerning CH₄ production. Lenhart et al. (2012) provided initial evidence that wood-decaying fungi such as
Pleurotus sapidus,
Trametes versicolor,
Lentinula edodes,
Laetiporus sulphureus, and
Hypholoma fasciculare produce CH₄ under aerobic conditions, identifying serine as a precursor for methane synthesis [
9]. Subsequent studies by Ernst et al. (2022) presented contradictory findings, showing that fungi like
Saccharomyces cerevisiae S288C and
Aspergillus niger DSM 821 primarily generate CH₄ via Fenton chemistry rather than enzymatic reactions [
10].
Recent research by Huang et al. (2022), employing biochemical, genetic, and stable isotopic tracer analyses, revealed that strains of
Schizophyllum commune 20R-7-F01, isolated from coal-bearing sediments ~2 km below the seafloor (under 35 MPa hydrostatic pressure), utilized a novel halomethane-dependent pathway for CH₄ production during anaerobic degradation of phenol, benzoic acid mono- and polymers, and cyclic sugars [
11]. Similar anaerobic methanogenic pathways have been confirmed in other wood-rot fungi such as
Agaricus bisporus,
Hypsizygus marmoreus, and
Pleurotus ostreatus [
12,
13,
14]. Nonetheless, uncertainties persist regarding the capability of these fungi to produce CH₄ under in situ high hydrostatic pressure (HHP) conditions, as well as specific mechanism governing CH₄ production in response to HHP.
In this study, we cultured the fungal strain S. commune 20R-7-F01 anaerobically in 1-liter stainless steel vessels at 30 °C to mimic in situ subseafloor environments. Our findings revealed a significant increase in CH₄ production by this subseafloor fungus under elevated hydrostatic pressure (HP), primarily attributed to the induction of reactive oxygen species (ROS) by HHP. These results underscore fungi’s potential role as methane producers in the deep biosphere, an aspect previously underestimated in global methane budgets.
2. Materials and Methods
2.1. High Hydrostatic Pressure Cultivation Experiments
Schizophyllum commune 20R-7-F01 (CGMCC 11604) was isolated from a sediment core collected at a depth of 1966 meters below seafloor (mbsf) from the Western Pacific Ocean [
15]. Mycelial inocula were prepared following the method described by Zain Ul Arifeen et al. (2021). For the high hydrostatic pressure (HHP) cultivation experiments, fresh mycelial inocula (7 g) were introduced into 170 ml sterile culture bottles containing Seawater Medium (SM). The SM composition included CaCl2 (2.99 g/L), MgCl2 (4.17 g/L), KBr (0.10 g/L), NH4Cl (0.16 g/L), KCl (5.05 g/L), NaCl (33.43 g/L), H3BO3(0.02 g/L), Na2SO4 (0.21 g/L), and C6H12O6 (20 g/L). The bottles were then purged with 99.99 % N2 for 15 minutes to remove oxygen from the culture bottles [
16]. Culture bottles, fitted with sterile syringes for pressure equilibration, were incubated at 30°C under HP of 15 MPa and 35 MPa, achieved by manual pumping of water into the vessel. A control culture under standard atmospheric pressure (0.1 MPa) was maintained under identical conditions. Fungal mycelia were harvested after 1, 3, and 5 days of culture, and one vial of mycelia was filtered through sterile gauze, rinsed three times with deionized water, immediately treated with liquid nitrogen, and stored at −80°C for transcriptome analysis. Additionally, three replicates of harvested mycelia underwent the same gauze filtration and rinsing steps before being immediately utilized for biomass and CH
4 quantification. Simultaneously, one vial of mycelia was also employed for assessing cellular activity following the aforementioned procedures.
For the ROS testing experiments, fresh mycelial inocula (7 g) were inoculated into 170 mL culture bottles containing SM supplemented with 0.75 mM, 1.5 mM, and 3 mM concentrations of cadmium chloride or hydrogen peroxide [
17,
18]. Culture bottles were incubated at 30°C under standard atmospheric pressure. Fungal mycelia were harvested after 1, 3, and 5 days of incubation. Subsequently, three replicates of harvested mycelia underwent the same gauze filtration and rinsing steps before being immediately utilized for biomass and CH4 quantification. Simultaneously, one vial of mycelia was also employed for assessing cellular activity following the aforementioned procedures.
2.2. Assessment of Fungal Hyphal Vitality and Biomass Determination
To assess fungal mycelial viability, a 0.4% trypan blue staining technique was employed [
19]. Viable mycelial cells were identified as colorless under optical microscopy, whereas non-viable cells stained blue. Mycelia harvested by filtration were dried in a 65°C oven for one day to determine biomass. Dry weights were measured to quantify biomass production [
20].
2.3. Assessment of ROS, O2-, OH·, H2O2, and CH₄ Levels in Fungal Mycelia
Intracellular levels of ROS in fungal mycelia were assessed following established methodologies [
21,
22]. The 2’,7’-dichlorofluorescein diacetate (DCFH-DA) probe was introduced into cells, and ROS concentrations were determined through fluorescence microscopy examination and subsequent quantification using ImageJ software. The intracellular content of O
2- in fungal mycelia was determined as per established protocols [
23,
24]. The superoxide anion reacts with hydroxylamine hydrochloride to form NO
2-, which, upon reaction with p-aminobenzenesulfonami-de and naphthalene ethylenediamine hydrochloride, produces a red azo compound with a characteristic absorption peak at 530 nm. The content of O
2- can be determined based on the absorbance at 530 nm. The intracellular content of OH· in fungal mycelia was determined as per established protocols [
25]. The hydroxyphenyl fluorescein (HPF) probe was introduced into cells, and OH· concentrations were determined through fluorescence microscopy examination and subsequent quantification using ImageJ software. The intracellular content of H2O2 in fungal mycelia was determined as per established protocols [
26,
27]. Absorbance at 415 nm, resulting from the formation of a titanium peroxide complex (Ti
4+ and H
2O
2), was measured to quantify H
2O
2 concentrations. Intracellular CH₄ levels in fungal mycelia were evaluated following detailed procedures , utilizing gas chromatography (GC) for accurate quantification of CH₄.
2.4. Transcriptomic Analysis
Mycelial samples for transcriptome sequencing were labeled as follows: “d1_01M”, “d3_01M”, “d5_01M”, “d1_15M”, “d3_15M”, “d5_15M”, “d1_35M”, “d3_35M”, and “d5_35M”. Here, “dn” denotes sampling days (1, 3, and 5 days), and “nM” indicates pressure levels (0.1, 15, and 35 MPa) applied to the strain. Total RNA extraction utilized TRIzol reagent (TIANGEN, Beijing, China) per the manufacturer’s instructions, followed by cDNA library construction. Sequencing employed the Illumina HiSeq platform with default RNA protocols (Meiji, Shanghai, China). Clean reads were obtained by removing adapters, sequences with >10% N bases, and low-quality sequences (Phred score Q ≤ 5, >50% of reads) (
Table S1). Clean reads were mapped to the strain 20R-7-F01 assembled genome (BioProject ID: PRJNA544166) using TopHat2 [
28]. Gene expression levels were quantified in Transcripts Per Million (TPM) using Cufflinks software [
29]. Raw RNA-seq data were deposited in the NCBI Sequence Read Archive under BioProject ID PRJNA1101667.
Differential gene expression analysis utilized the DESeq method DESeq2 [
30], applying a threshold of p-value < 0.05 and |Log2 (Fold-change)| ≥ 1 to identify significant DEGs [
31]. Functional analysis of DEGs included Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses using clusterProfiler in R (v4.3.0). Enriched pathways were visualized with the Pathview package, setting the threshold for enriched gene annotations at p-value < 0.05. DEGs related to key pathways underwent hierarchical clustering, with correlation analysis performed using psych and reshape 2 packages in R (v4.3.0). A correlation network of pathway genes was constructed using Cytoscape software version 3.9.1 (
https://cytoscape.org/releasenotes.html).
2.5. Quantitative Real-Time PCR Analysis
Quantitative real-time PCR (qRT-PCR) followed the protocol outlined by Zain Ul Arifeen et al. Fungal cultures were maintained under identical conditions and durations as those for RNA-seq samples. SYBR qPCR Master Mix (Vazyme, Nanjing, China) and specific primer pairs for each gene (
Table S2) were used for qRT-PCR analysis. Thermal cycling conditions included initial denaturation at 95 ℃ for 30 s, followed by 43 cycles of 95 ℃ for 10 s, 58.5 ℃ for 30 s, and 72 ℃ for 30 s. Relative gene expression was calculated using the 2
−ΔΔCT method [
32].
2.6. Statistical Analysis
The data were presented as mean ± standard deviation. One-way analysis of variance (ANOVA) or student’s t-test, performed using GraphPad Prism version 8.0.2, was used to analyze significant differences between treatments (p < 0.05).
3. Results and Discussion
3.1. Impact of High Hydrostatic Pressure on Fungal Methane Productivity
To assess the effect of HHP on fungal CH₄ productivity, strain 20R-7-F01 was cultured in bottles under varying HHP conditions, and CH₄ yield in the headspace was quantified using GC. Results demonstrated a significant enhancement in CH₄ production of strain 20R-7-F01 with increasing HP. Specifically, CH₄ production at 15 MPa was approximately 1.3, 1.7, and 1.9 times higher on days 1, 3, and 5 of culture, respectively, compared to atmospheric conditions. Furthermore, at 35 MPa (equivalent to in situ pressure), CH₄ production increased to approximately 2.0, 2.4, and 2.5 times higher than at atmospheric pressure. This substantial increase suggests a strong influence of HHP in enhancing CH₄ production by strain 20-7-1. The effect of hydrostatic pressure on biological methanogenesis may be a universal phenomenon that affects all methanogens, but similar observations have only been noted in archaea, such as
Thermophilic marburgensis, which exhibited CH₄ production levels approximately 3 times higher than atmospheric levels when cultured at 50 MPa [
33]. The mechanism behind enhanced CH₄ production under HHP in archaea is hypothesized to involve oxidative stress induced by HP. However, it remains unclear whether analogous mechanisms govern the impact of HHP on fungal CH₄ production.
Figure 1.
Methane production by strain 20R-7-F01 under varying hydrostatic pressures. Note: * represents p<0.05; ** represents p<0.01; *** represents p<0.001.
Figure 1.
Methane production by strain 20R-7-F01 under varying hydrostatic pressures. Note: * represents p<0.05; ** represents p<0.01; *** represents p<0.001.
3.2. Transcriptomic Analysis of Methane Synthesis Genes under High Hydrostatic Pressure
Transcriptomic analysis revealed significant upregulation of key genes involved in methane synthesis under high hydrostatic pressure conditions in
S. commune 20R-7-F01. Following cultivation at 15 MPa for 3 days, expression levels of mct1, dh3, and ms increased by 5.9-fold, 2.8-fold, and 2.7-fold, respectively, compared to ambient pressure (0.1 MPa) (
Table 1). Similarly, at 35 MPa, these genes showed increases of 7.9-fold, 4.5-fold, and 3.1-fold, respectively (
Table 1). Quantitative PCR validated these findings, demonstrating significant upregulation of mct1, dh3, and ms under HHP conditions (
Table 1). This enhanced methane production is primarily attributed to the upregulation of genes encoding key enzymes involved in methane synthesis.
Transcriptional correlation analysis identified 2316 differentially expressed genes (DEGs) highly correlated (|p| ≥ 0.9) with mct1 (689 DEGs), dh3 (817 DEGs), and ms (810 DEGs) (
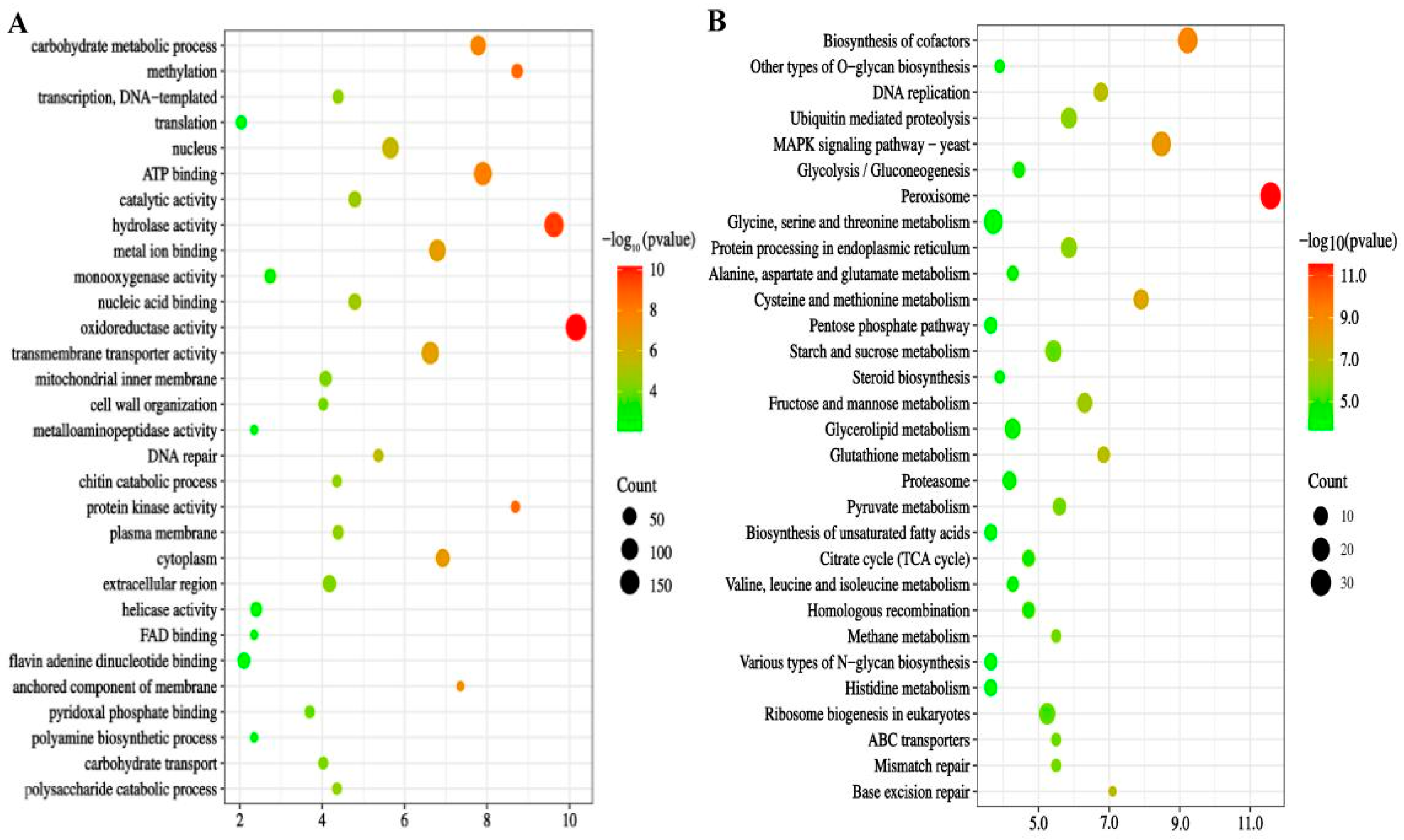
Table S1). Gene ontology (GO) enrichment analysis revealed that these DEGs were enriched in activities associated with oxidoreductase functions, carbohydrate metabolism, methylation, ATP binding, hydrolase activity, metal ion binding, and transmembrane transport. Notably, oxidoreductase activity exhibited the highest enrichment, comprising 8.5% of the total 2316 DEGs (p = 6.95 × 10^
-11) (
Figure 2A,
Table S1). KEGG pathway enrichment analysis further indicated significant enrichment of these DEGs in pathways such as peroxisomes, glycolysis/gluconeogenesis, tricarboxylic acid cycle, and pentose phosphate pathway under HHP conditions (
Figure 2B).
Hierarchical clustering analysis illustrated the upregulation of genes involved in oxidoreductase activities, particularly those implicated in oxidative stress response (e.g., SOD, CAT, and BCP), in S. commune 20R-7-F01 under HHP conditions (
Figure 3,
Table S2). Collectively, these findings suggest that the enhancement of methane synthesis metabolism under HHP conditions may be linked to alterations in oxidoreductase activities. Similar observations in other piezophilic organisms, such as
Sporosarcina psychrophila DSM 6497 and
Shewanella piezotolerans WP3, underscore that
S. commune 20R-7-F01, like other piezophiles, counters oxidative stress induced by HHP through activation of oxidative-reductive pathways [
34,
35].
Furthermore, the investigation of DEGs related to antioxidant genes revealed significant correlations with key genes involved in methane biosynthesis in strain 20R-7-F01. A total of 197 DEGs of antioxidant genes were identified, with 54 (27.4%) showing notable correlations with mct1, 79 (40.1%) with dh3, and 64 (32.5%) with metE. These findings indicate that these antioxidant genes were significantly involved in methane production by
S. commune under HHP conditions. Enhanced methane release in response to oxidative stress induced by HHP may serve as a protective mechanism against biological membrane damage caused by reactive oxygen species (ROS) [
36,
37]. In summary, our study highlights that the increased methane production observed in
S. commune 20R-7-F01 under HHP conditions is a response to oxidative stress induced by HHP, mediated through the upregulation of genes associated with both methane synthesis and antioxidant defense mechanisms. These findings contribute to our understanding of microbial adaptation to extreme environmental conditions, emphasizing the role of methane production in stress response strategies.
3.3. ROS and H2O2 Induced in Fungal Cells by High Hydrostatic Pressure
To explore the enhanced methane production of
S. commune 20R-7-F01 under high hydrostatic pressure (HHP) as a potential response to elevated fungal cell ROS and H
2O
2 levels induced by HHP, we assessed intracellular ROS and H
2O
2 levels, as well as the activities of antioxidative enzymes (SOD, CAT, POD) at 0.1 (control), 15, and 35 MPa pressures. Our results revealed significant increases in ROS and H
2O
2 levels in fungal cells under HHP compared to 0.1 MPa, accompanied by reduced fungal cell viability (
Table 2,
Figure S1). For instance, after 5 days of cultivation, ROS and H
2O
2 levels were 7.04-fold and 6.12-fold higher at 15 MPa, and 10.33-fold and 8.51-fold higher at 35 MPa, respectively, compared to at 0.1 MPa, yet the cells were clearly labeled blue with trypan blue. Further analysis demonstrated that the increase in ROS levels induced by HHP was primarily due to changes in H
2O
2 levels, while the contributions of superoxide anion (O
2-) and hydroxyl radical (OH·) were relatively insignificant (
Table S3). This is similar to findings by Zhe et al. (2018) who reported oxidative damage in deep-sea bacterium
Shewanella piezotolerans WP3 due to elevated intracellular H
2O
2 levels under 20 MPa conditions [
38]. Additionally, antioxidative enzyme activities within fungal hyphae increased with pressure, notably with peroxidase (POD) activity at 35 MPa after 5 days of cultivation, being 5.95 times higher than at 0.1 MPa over the same period, this is consistent with the results shown in
Figure 3. The heightened activities of SOD, CAT, and POD in scavenging ROS and H2O2-induced cellular damage under HHP suggest that
S. commune, in response to HHP stress, employed metabolic mechanisms akin to those observed in deep-sea bacterium
Shewanella piezotolerans WP3 and yeast
Saccharomyces cerevisiae [
39,
40].
After establishing the occurrence of oxidative damage in
S. commune 20R-7-F01 under HHP conditions, we conducted correlation analysis between intracellular ROS and H
2O
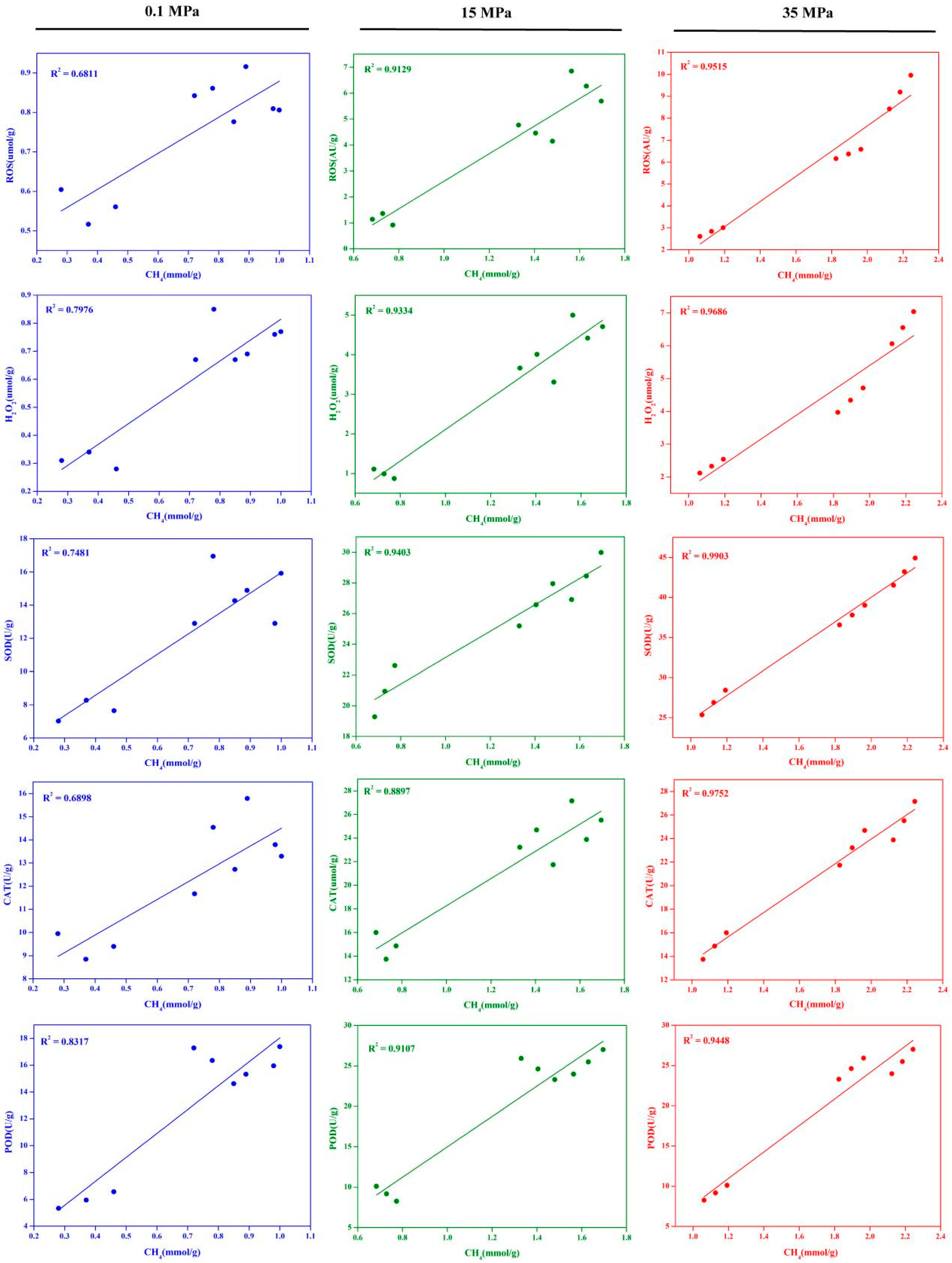
2 levels, antioxidant enzyme activities (SOD, CAT, POD), and methane content across different pressures. The results depicted in
Figure 5 reveal that at 0.1 MPa, there existed a moderate positive correlation between methane production in the strain and intracellular ROS and H
2O
2 levels, as well as antioxidant enzyme activities, though not statistically significant. In contrast, at 15 MPa and 35 MPa, significant positive correlations were observed between methane production and intracellular ROS and H
2O
2 levels, as well as antioxidant enzyme activities. Particularly notable was the pronounced positive correlation between methane production and H
2O
2 levels. For instance, at 15 MPa, the correlation coefficient between methane production and H
2O
2 levels in the strain reached 0.9334. Similarly, at 35 MPa, the correlation coefficient between methane production and H
2O
2 levels also reached 0.9686. These results indicate that alongside the increase in intracellular H
2O
2 levels in
S. commune 20R-7-F01, there is a corresponding increase in methane release. This further corroborates the conclusion from our transcriptome analysis showing that enhancing methane metabolism in
S. commune 20R-7-F01 is a fungal response mechanism to HHP-induced ROS. While Ernst et al. (2022) have demonstrated the existence of an ROS-driven methane production mechanism in general organisms, it is based on the Fenton chemistry rather than a direct ROS-driven pathway specific to methane production in organisms. Similarly, although Mauerhofer et al. (2021) found an increase in methane production by methanogenic archaea with increasing HHP, they did not identify the fundamental reasons behind HHP-induced enhancement of methane production in archaea [
33]. Here, we provide the first evidence that HHP can promote methane production in
S. commune 20R-7-F01 for adapting to oxidative stress.
3.4. Experimental Evidence of ROS Contribution to Increased Methane Production in S. commune 20R-7-F01
To investigate the influence of oxidative stress induced by H
2O
2 or CdCl
2 on methane production by
S. commune 20R-7-F01, we cultured the fungus in liquid mPD medium supplemented with varying concentrations (0.75 mM, 1.5 mM, and 3 mM) of these stressors. Control cultures lacking H
2O
2 or CdCl
2 served as a baseline. Methane production capacity was assessed under atmospheric pressure conditions after 1, 3, and 5 days of cultivation. As detailed in
Table 3, methane production markedly increased with increasing concentrations of H2O2 or CdCl2. For instance, exposure to 0.75 mM CdCl
2 resulted in methane production 1.45 times higher than the control after five days, which further increased to 2.44 times higher with 3 mM CdCl
2. Similarly, exposure to 0.75 mM H₂O₂ led to a 1.74-fold enhancement in methane production relative to the control condition, while increasing the H₂O₂ concentration to 3 mM further augmented methane production by a factor of 3.06. It is noteworthy that supplementation of the mPD culture medium with BHT to scavenge intracellular ROS under different oxidative stress conditions led to a corresponding decrease in methane production by the strain. For example, addition of antioxidant BHT (5 mM) to the fungal mycelium exposed to 3 mM H
2O
2 for five days resulted in a significant reduction in methane production from 2.63 mmol/g to 2.23 mmol/g. Concurrently, increasing concentrations of H
2O
2 or CdCl
2 correlated with elevated levels of intracellular ROS (
Table S4), accompanied by reduced fungal cell viability and biomass (Figures S2, S3). For instance, exposure to 3 mM H
2O
2 led to a decrease in biomass by 45.27 mg relative to the control, accompanied by significant trypan blue staining of the cells. These findings collectively underscore that oxidative stress induced by H
2O
2 or CdCl
2 promotes methane production by
S. commune 20R-7-F01. Thus, oxidative damage induced by HHP likely contributes to the increased methane production by this fungus. Similar observations by Gu et al. and Samma et al. in alfalfa roots indicate that elevated ROS levels from metal stressors enhance methane emission [
17,
18].
4. Conclusions
In conclusion, our study demonstrates that Schizophyllum commune 20R-7-F01, isolated from the subseafloor sediment approximately 2 km below the seabed, exhibited enhanced methane (CH4) production capabilities under in situ temperature, high hydrostatic pressure and anaerobic conditions. Through comprehensive analyses encompassing biological activity assays, biomass quantification, transcriptomics, and metabolomics, we found that S. commune not only survived but significantly increased CH4 production under HHP conditions. Pathways related to carbohydrate metabolism, methylation, hydrolase activity, and the metabolism of cysteine and methionine, as well as activities of redox enzymes, were notably activated under HHP. Specifically, critical genes involved in fungal anaerobic CH4 synthesis, such as methyltransferase mct1 and dehalogenase dh3, were markedly upregulated. The observed enhancement in CH4 production under HHP was primarily attributed to pressure-induced oxidative stress, supported by comparative analyses of intracellular ROS levels and treatments involving cadmium chloride and hydrogen peroxide. These findings underscore the potential significant role of deep subseafloor sediment fungi in global methane generation, which has not been unaccounted for in previous estimations. Further elucidation of the mechanisms governing methane production by sediment fungi in the deep biosphere promises to advance our understanding of Earth’s additional sources of methane that contribute to global climate change.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Mengshi Zhao: designed the project, conducted the research work, analyzed and interpreted the data, drafted the manuscript. Dongxu Li and Jie Liu: participated in data analysis. Jiasong Fang: edited the manuscript. Changhong Liu: project administration, supervision, conceptualization, funding acquisition, manuscript riting-review & editing. The manuscript was reviewed and approved by all authors.
Funding
This work was supported by the National Natural Science Foundation of China (no. 41973073, 41773083, and 92251303), and the Science and Technology Innovation Program of Jiangsu Province (no.BK20220036).
Institutional Review Board Statement
The study protocol was approved by the Institutional Review Board at the University of Minnesota, and all participants provided written informed consent.
Data Availability Statement
No data was used for the research described in the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Crow, D.J., et al., Assessing the impact of future greenhouse gas emissions from natural gas production. 2019. 668: p. 1242-1258. [CrossRef]
- Liu, J., et al., A novel pathway of direct methane production and emission by eukaryotes including plants, animals and fungi: an overview. 2015. 115: p. 26-35. [CrossRef]
- Sakata, S., et al. Methane production from coal by a single methanogen. in AGU Fall Meeting Abstracts. 2017.
- Akob, D., et al., Enhanced microbial coalbed methane generation: A review of research, commercial activity, and remaining challenges. 2015. [CrossRef]
- Wang, Y., Y. Bao, and Y.J.E.R. Hu, Recent progress in improving the yield of microbially enhanced coalbed methane production. 2023. 9: p. 2810-2819. [CrossRef]
- Liu, Y. and W.B.J.A.o.t.n.Y.A.o.S. Whitman, Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. 2008. 1125(1): p. 171-189.
- Keppler, F., et al., Methane emissions from terrestrial plants under aerobic conditions. 2006. 439(7073): p. 187-191. [CrossRef]
- Tuboly, E., et al., Methane biogenesis during sodium azide-induced chemical hypoxia in rats. 2013. 304(2): p. C207-C214. [CrossRef]
- Lenhart, K., et al., Evidence for methane production by saprotrophic fungi. 2012. 3(1): p. 1046. [CrossRef]
- Ernst, L., et al., Methane formation driven by reactive oxygen species across all living organisms. 2022. 603(7901): p. 482-487. [CrossRef]
- Huang, X., et al., Methane production by facultative anaerobic wood-rot fungi via a new halomethane-dependent pathway. 2022. 10(5): p. e01700-22. [CrossRef]
- Doddapaneni, H., et al., A comparative genomic analysis of the oxidative enzymes potentially involved in lignin degradation by Agaricus bisporus. 2013. 55: p. 22-31. [CrossRef]
- Terashita, T., et al., Changes in carbohydrase activities during vegetative growth and development of fruit-bodies of Hypsizygus marmoreus grown in sawdust-based culture. 1998. 44: p. 234-236. [CrossRef]
- Valášková, V. and P.J.R.i.m. Baldrian, Estimation of bound and free fractions of lignocellulose-degrading enzymes of wood-rotting fungi Pleurotus ostreatus, Trametes versicolor and Piptoporus betulinus. 2006. 157(2): p. 119-124.
- Liu, C.H., et al., Exploration of cultivable fungal communities in deep coal-bearing sediments from∼ 1.3 to 2.5 km below the ocean floor. 2017. 19(2): p. 803-818.
- Zain Ul Arifeen, M., et al., The anaerobic survival mechanism of Schizophyllum commune 20R-7-F01, isolated from deep sediment 2 km below the seafloor. 2021. 23(2): p. 1174-1185.
- Samma, M.K., et al., Methane alleviates copper-induced seed germination inhibition and oxidative stress in Medicago sativa. 2017. 30: p. 97-111. [CrossRef]
- Gu, Q., et al., Methane alleviates alfalfa cadmium toxicity via decreasing cadmium accumulation and reestablishing glutathione homeostasis. 2018. 147: p. 861-871. [CrossRef]
- Basu, A., et al., Evaluating the antimicrobial, apoptotic, and cancer cell gene delivery properties of protein-capped gold nanoparticles synthesized from the edible mycorrhizal fungus Tricholoma crassum. 2018. 13: p. 1-16.
- Botella, C., et al., Dry weight model, capacitance and metabolic data as indicators of fungal biomass growth in solid state fermentation. 2019. 114: p. 144-153. [CrossRef]
- Chen, M.-F., et al., Losartan inhibits monocytic adhesion induced by ADMA via downregulation of chemokine receptors in monocytes. 2009. 65: p. 457-464. [CrossRef]
- Qian, H., et al., The effect of exogenous nitric oxide on alleviating herbicide damage in Chlorella vulgaris. 2009. 92(4): p. 250-257. [CrossRef]
- Cai, B., et al., Decreasing fructose-1, 6-bisphosphate aldolase activity reduces plant growth and tolerance to chilling stress in tomato seedlings. 2018. 163(2): p. 247-258.
- Liu, Z., et al., Comprehensive analysis of BpHSP genes and their expression under heat stresses in Betula platyphylla. 2018. 152: p. 167-176.
- Hammel, K.E., et al., Reactive oxygen species as agents of wood decay by fungi. 2002. 30(4): p. 445-453. [CrossRef]
- Yin, Y.-J., et al., The fight against Panax notoginseng root-rot disease using Zingiberaceae essential oils as potential weapons. 2018. 9: p. 1346. [CrossRef]
- Yang, Y., et al., Amelioration of nonalcoholic fatty liver disease by swertiamarin in fructose-fed mice. 2019. 59: p. 152782. [CrossRef]
- Kim, D., et al., Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions., 2013, 14, R36. [CrossRef]
- Trapnell, C., et al., Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. 2012. 7(3): p. 562-578. [CrossRef]
- Love, M.I., W. Huber, and S.J.G.b. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. 2014. 15: p. 1-21.
- Anders, S. and W.J.N.P. Huber, Differential expression analysis for sequence count data. 2010: p. 1-1. [CrossRef]
- Livak, K.J. and T.D.J.m. Schmittgen, Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. 2001. 25(4): p. 402-408.
- Mauerhofer, L.-M., et al., Hyperthermophilic methanogenic archaea act as high-pressure CH4 cell factories. 2021. 4(1): p. 289. [CrossRef]
- Wang, H., et al., Transcriptomic analysis reveals common adaptation mechanisms under different stresses for moderately piezophilic bacteria. 2021. 81: p. 617-629. [CrossRef]
- Li, X.-G., et al., Pressure-regulated gene expression and enzymatic activity of the two periplasmic nitrate reductases in the deep-sea bacterium Shewanella piezotolerans WP3. 2018. 9: p. 3173. [CrossRef]
- Ghafoor, K., et al., Effects of high hydrostatic pressure on structure and colour of red ginseng (Panax ginseng). 2012. 92(15): p. 2975-2982. [CrossRef]
- Scoma, A., et al., The polyextremophilic bacterium Clostridium paradoxum attains piezophilic traits by modulating its energy metabolism and cell membrane composition. 2019. 85(15): p. e00802-19. [CrossRef]
- Xie, Z., et al., Enhancing the adaptability of the deep-sea bacterium Shewanella piezotolerans WP3 to high pressure and low temperature by experimental evolution under H2O2 stress. 2018. 84(5): p. e02342-17. [CrossRef]
- Jian, H., et al., A transcriptome resource for the deep-sea bacterium Shewanella piezotolerans WP3 under cold and high hydrostatic pressure shock stress. 2016. 30: p. 87-91. [CrossRef]
- Bravim, F., et al., High hydrostatic pressure leads to free radicals accumulation in yeast cells triggering oxidative stress. 2016. 16(5): p. fow052. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).