Submitted:
13 August 2024
Posted:
13 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
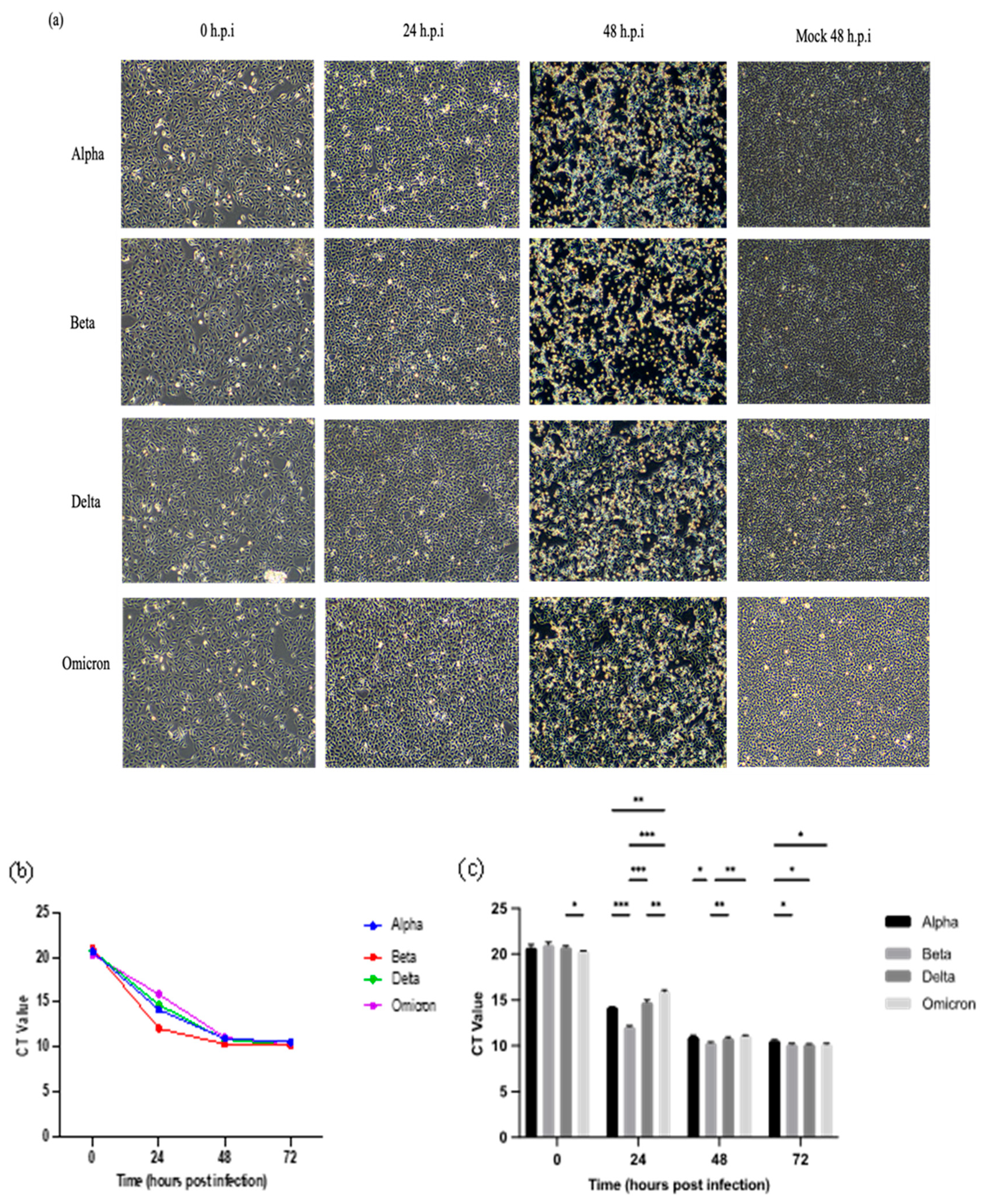
2.1. Morphology Changes of Vero E6 Cells Infected with Malaysian SARS-CoV-2 Variants
2.2. Viral RNA Multiplication of SARS-CoV-2 Variants in Vero E6 Cells
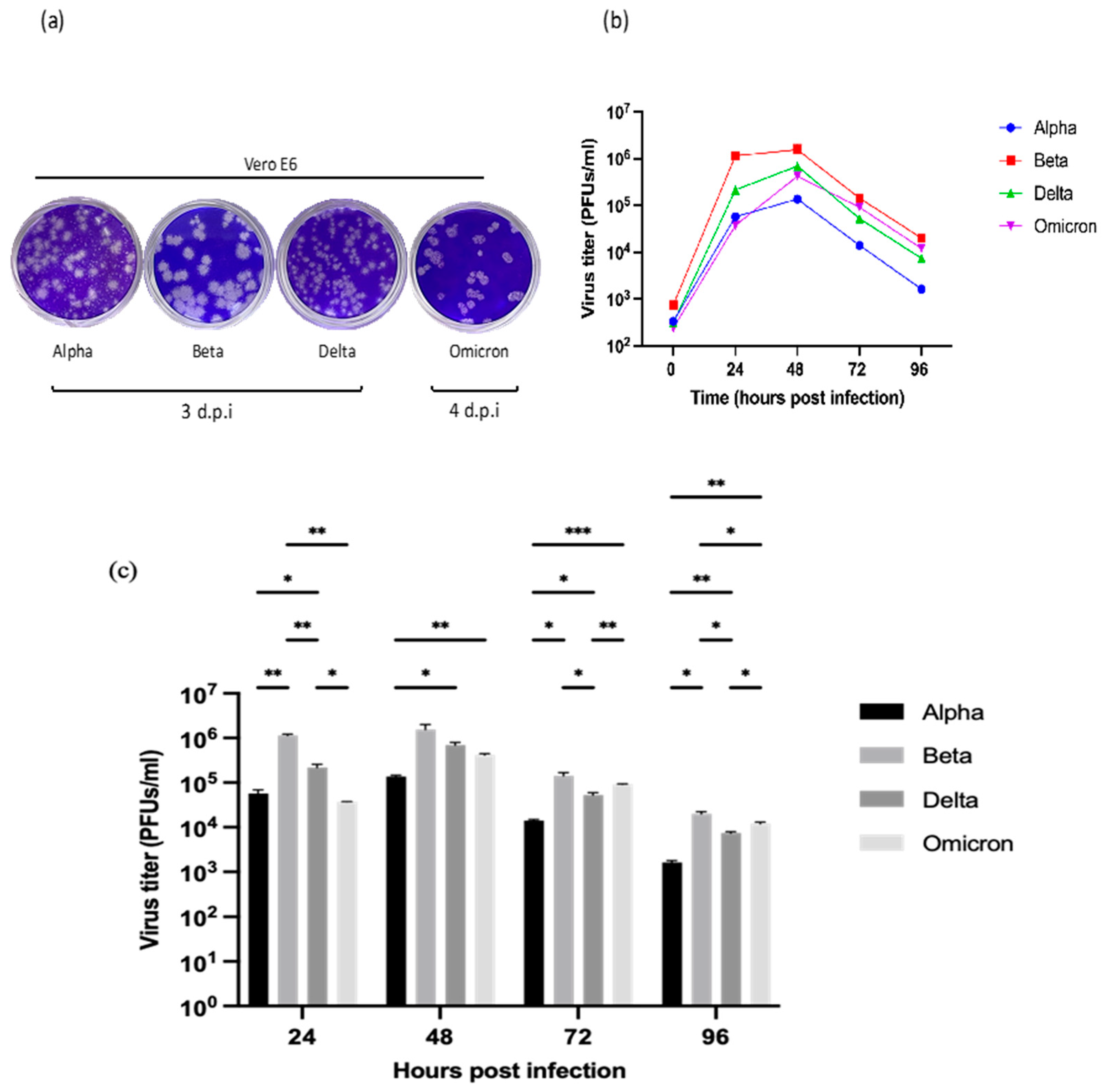
2.3. Plaque Formation and Phenotypes
2.4. Replication Kinetics of SARS-CoV-2 Variants in Vero E6 Cells
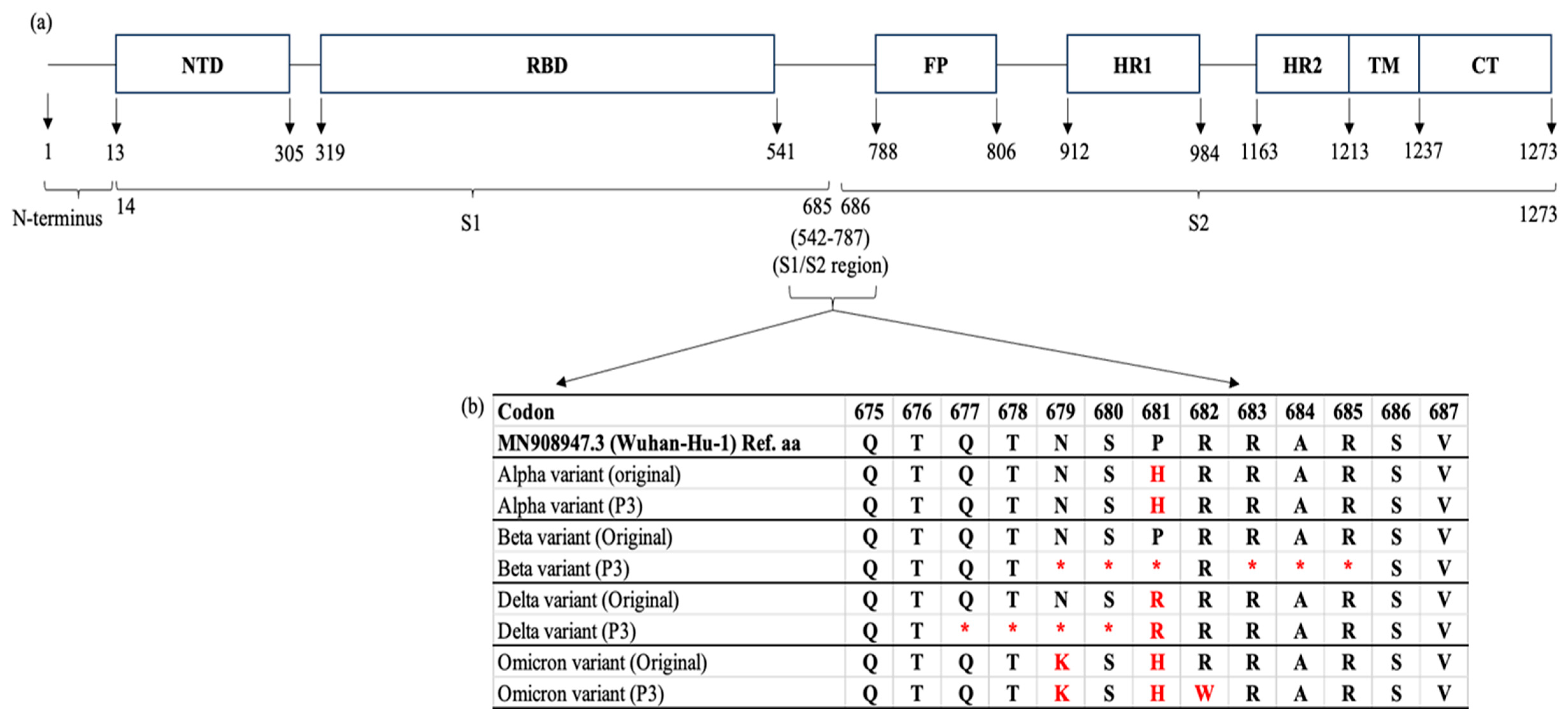
2.5. Whole-Genome Sequencing of SARS-CoV-2 Alpha, Beta, Delta, and Omicron Variants
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Virus Propagation, Isolation and Preparation of Virus Stocks
4.3. Plaque Assay
4.4. Infectivity Assay
4.5. RNA Extraction and Nucleic Acids Amplification
4.6. Whole Genome Sequencing
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rahimi, G.; Rahimi, B.; Panahi, M.; Abkhiz, S.; Saraygord-Afshari, N.; Milani, M.; Alizadeh, E. An overview of Betacoronaviruses-associated severe respiratory syndromes, focusing on sex-type-specific immune responses. International Immunopharmacology 2021, 92, 107365. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.Y.; Wong, S.Y.; Liew, N.W.C.; Joseph, N.; Zakaria, Z.; Nurulfiza, I.; Soe, H.J.; Kairon, R.; Amin-Nordin, S.; Chee, H.Y. Whole genome sequencing analysis of SARS-CoV-2 from Malaysia: From alpha to Omicron. Frontiers in Medicine 2022, 9, 1001022. [Google Scholar] [CrossRef] [PubMed]
- Azami, N.A.M.; Perera, D.; Thayan, R.; AbuBakar, S.; Sam, I.C.; Salleh, M.Z.; Isa, M.N.M.; Ab Mutalib, N.S.; Aik, W.K.; Suppiah, J.; et al. Malaysia COVID-19 Genomics Surveillance Consortium. SARS-CoV-2 genomic surveillance in Malaysia: displacement of B.1.617.2 with AY lineages as the dominant Delta variants and the introduction of Omicron during the fourth epidemic wave. International Journal of Infectious Diseases 2022, 125, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Amicone, M.; Borges, V.; Alves, M.J.; Isidro, J.; Zé-Zé, L.; Duarte, S.; ... & Gordo, I. Mutation rate of SARS-CoV-2 and emergence of mutators during experimental evolution. Evolution Medicine and Public Health 2022, 10, 142–155. [CrossRef]
- Peacock, T.P.; Penrice-Randal, R.; Hiscox, J.A.; Barclay, W.S. SARS-CoV-2 one year on: evidence for ongoing viral adaptation. Journal of General Virology 2021, 102. [Google Scholar] [CrossRef] [PubMed]
- Luan, B.; Wang, H.; Huynh, T. Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations. FEBS Letters 2021, 595, 1454–1461. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; ... & de Oliveira, T. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [CrossRef]
- Focosi, D.; Maggi, F. Neutralising antibody escape of SARS-CoV-2 spike protein: risk assessment for antibody-based Covid-19 therapeutics and vaccines. Reviews in Medical Virology 2021, 31, e2231. [Google Scholar] [CrossRef]
- Dhawan, M.; Sharma, A.; Priyanka Thakur, N.; Rajkhowa, T.K.; Choudhary, O.P. Delta variant (B.1.617.2) of SARS-CoV-2: Mutations, impact, challenges and possible solutions. Human Vaccines & Immunotherapeutics 2022, 18, 2068883. [Google Scholar] [CrossRef]
- Fan, Y.; Li, X.; Zhang, L.; Wan, S.; Zhang, L.; Zhou, F. SARS-CoV-2 Omicron variant: recent progress and future perspectives. Signal Transduction and Targeted Therapy 2022, 7, 141. [Google Scholar] [CrossRef]
- Hatta, Y.; Hershberger, K.; Shinya, K.; Proll, S.C.; Dubielzig, R.R.; Hatta, M.; ... & Kawaoka, Y. Viral replication rate regulates clinical outcome and CD8 T cell responses during highly pathogenic H5N1 influenza virus infection in mice. PLOS Pathogens 2010, 6, e1001139. [CrossRef]
- Lee, L.Y.; Rozmanowski, S.; Pang, M.; Charlett, A.; Anderson, C.; Hughes, G.J.; ... & Eyre, D.W. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectivity by viral load, S gene variants and demographic factors, and the utility of lateral flow devices to prevent transmission. Clinical Infectious Diseases 2022, 74, 407–415. [CrossRef]
- Banerjee, A.; Nasir, J.A.; Budylowski, P.; Yip, L.; Aftanas, P.; Christie, N.; ... & Mubareka, S. Isolation, sequence, infectivity, and replication kinetics of severe acute respiratory syndrome coronavirus 2. Emerging Infectious Diseases 2020, 26, 2054. [CrossRef]
- Chen, Y.; Liu, M.Q.; Luo, Y.; Jiang, R.D.; Si, H.R.; Zhu, Y.; ... & Yang, X.L. Genetic mutation of SARS-CoV-2 during consecutive passages in permissive cells. Virologica Sinica 2021, 36, 1073–1076. [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nature Reviews Molecular Cell Biology 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Emeny, J.M.; Morgan, M.J. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. Journal of General Virology 1979, 43, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Rosli, S.N.Z.; Dimeng, S.R.; Shamsuddin, F.; Mohd Ali, M.R.; Muhamad Hendri, N.A.; Suppiah, J.; Mohd Zain, R.; Thayan, R.; Ahmad, N. Vero CCL-81 and Calu-3 cell lines as alternative hosts for isolation and propagation of SARS-CoV-2 isolated in Malaysia. Biomedicines 2023, 11, 1658. [Google Scholar] [CrossRef] [PubMed]
- Mautner, L.; Hoyos, M.; Dangel, A.; Berger, C.; Ehrhardt, A.; Baiker, A. Replication kinetics and infectivity of SARS-CoV-2 variants of concern in common cell culture models. Virology Journal 2022, 19, 76. [Google Scholar] [CrossRef] [PubMed]
- Puhach, O.; Meyer, B.; Eckerle, I. SARS-CoV-2 viral load and shedding kinetics. Nature Reviews Microbiology 2023, 21, 147–161. [Google Scholar] [CrossRef]
- Goh, K.; Tang, C.; Norton, D.; Manivannan, L. Molecular determinants of plaque size as an indicator of dengue virus attenuation. Scientific Reports 2016, 6, 26100. [Google Scholar] [CrossRef]
- Pyke, A.T.; Nair, N.; van den Hurk, A.F.; Burtonclay, P.; Nguyen, S.; Barcelon, J.; ... & Prow, N.A. Replication kinetics of B.1.351 and B.1.1.7 SARS-CoV-2 variants of concern including assessment of a B.1.1.7 mutant carrying a defective ORF7a gene. Viruses 2021, 13, 1087. [CrossRef]
- Prince, T.; Dong, X.; Penrice-Randal, R.; Randle, N.; Hartley, C.; Goldswain, H.; ... & Hiscox, J.A. Analysis of SARS-CoV-2 in nasopharyngeal specimens from patients infected with variant B.1.1.7 compared with the wild-type strain in Liverpool, UK. Journal of Clinical Microbiology 2022, 60, e02592-21. [CrossRef]
- Jeong, G.U.; Yoon, G.Y.; Moon, H.W.; Lee, W.; Hwang, I.; Kim, H.; ... Kwon, Y.C. Comparison of plaque size, thermal stability, and replication rate among SARS-CoV-2 variants of concern. Viruses 2022, 14, 55. [CrossRef]
- Guruprasad, K. Mutations in human SARS-CoV-2 spike proteins, potential drug binding and epitope sites for COVID-19 therapeutics development. Current Research in Structural Biology 2022, 4, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Plante, K.S.; Plante, J.A.; Xie, X.; Zhang, X.; Ku, Z.; An, Z.; Scharton, D.; Schindewolf, C.; Widen, S.G.; Menachery, V.D.; Shi, P.-Y.; Weaver, S.C. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 2022, 602, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Lista, M.J.; Winstone, H.; Wilson, H.D.; Dyer, A.; Pickering, S.; Galao, R.P.; De Lorenzo, G.; Cowton, V.M.; Furnon, W.; Suarez, N.; Orton, R.; Palmarini, M.; Patel, A.H.; Snell, L.; Nebbia, G.; Swanson, C.; Neil, S.J.D. The P681H mutation in the spike glycoprotein of the Alpha variant of SARS-CoV-2 escapes IFITM restriction and is necessary for type I interferon resistance. Journal of Virology 2022, 96, e0125022. [Google Scholar] [CrossRef] [PubMed]
- Tanneti, N.S.; Patel, A.K.; Tan, L.H.; Marques, A.D.; Perera, R.A.P.M.; Sherrill-Mix, S.; Kelly, B.J.; Renner, D.M.; Collman, R.G.; Rodino, K.; Lee, C.; Bushman, F.D.; Cohen, N.A.; Weiss, S.R. Comparison of SARS-CoV-2 variants of concern in primary human nasal cultures demonstrates Delta as most cytopathic and Omicron as fastest replicating. mBio 2024, 15, e0312923. [Google Scholar] [CrossRef] [PubMed]
- Tandel, D.; Sah, V.; Singh, N.K.; Potharaju, P.S.; Gupta, D.; Shrivastava, S.; Sowpati, D.T.; Harshan, K.H. SARS-CoV-2 variant Delta potently suppresses innate immune response and evades interferon-activated antiviral responses in human colon epithelial cells. Microbiology Spectrum 2022, 10, e01604-22. [Google Scholar] [CrossRef] [PubMed]
- Shuai, H.; Chan, J.F.W.; Hu, B.; ... & Yuen, K.Y. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 2022, 603, 693–699. [CrossRef]
- Zhao, H.; Lu, L.; Peng, Z.; Chen, L.L.; Meng, X.; Zhang, C.; Ip, J.D.; Chan, W.M.; Chu, A.W.; Chan, K.H.; Jin, D.Y.; Chen, H.; Yuen, K.Y.; To, K.K. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerging Microbes & Infections 2022, 11, 277–283. [Google Scholar] [CrossRef]
- Bojkova, D.; Rothenburger, T.; Ciesek, S.; Wass, M.N.; Michaelis, M.; Cinatl, J. SARS-CoV-2 Omicron variant virus isolates are highly sensitive to interferon treatment. Cell Discovery 2022, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.T.M.; ... & Sato, K. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [CrossRef]
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Saito, A.; Nasser, H.; Tan, T.S.; Ngare, I.; Kimura, I.; Uriu, K.; Kosugi, Y.; Yue, Y.; Shimizu, R.; Ito, J.; Torii, S.; Yonekawa, A.; Shimono, N.; Nagasaki, Y.; Minami, R.; Toya, T.; Sekiya, N.; ... & Sato, K. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host & Microbe 2021, 29, 1124–1136. [CrossRef]
- Qu, P.; Evans, J.P.; Kurhade, C.; Zeng, C.; Zheng, Y.-M.; Xu, K.; Shi, P.-Y.; Xie, X.; Liu, S.-L. Determinants and mechanisms of the low fusogenicity and high dependence on endosomal entry of Omicron subvariants. mBio 2023, 14, e03176-22. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, H.; Yang, J.; ... & Luo, M. Mutation N856K in spike reduces fusogenicity and infectivity of Omicron BA.1. Signal Transduction and Targeted Therapy 2023, 8, 75. [CrossRef]
- Park, S.B.; Khan, M.; Chiliveri, S.C.; ... & Gu, H. SARS-CoV-2 Omicron variants harbor spike protein mutations responsible for their attenuated fusogenic phenotype. Communications Biology 2023, 6, 556. [CrossRef]
- Koch, J.; Uckeley, Z.M.; Doldan, P.; Stanifer, M.; Boulant, S.; Lozach, P.Y. TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. The EMBO Journal 2021, 40, e107821. [Google Scholar] [CrossRef] [PubMed]
- Aiewsakun, P.; Phumiphanjarphak, W.; Ludowyke, N.; Purwono, P.B.; Manopwisedjaroen, S.; Srisaowakarn, C.; Ekronarongchai, S.; Suksatu, A.; Yuvaniyama, J.; Thitithanyanont, A. Systematic exploration of SARS-CoV-2 adaptation to Vero E6, Vero E6/TMPRSS2, and Calu-3 cells. Genome Biology and Evolution 2023, 15, evad035. [Google Scholar] [CrossRef] [PubMed]
- Vu, M.N.; Lokugamage, K.G.; Plante, J.A.; Scharton, D.; Bailey, A.O.; Sotcheff, S.; Swetnam, D.M.; Johnson, B.A.; Schindewolf, C.; Alvarado, R.E.; Crocquet-Valdes, P.A.; Debbink, K.; Weaver, S.C.; Walker, D.H.; Russell, W.K.; Routh, A.L.; Plante, K.S.; Menachery, V.D. QTQTN motif upstream of the furin-cleavage site plays a key role in SARS-CoV-2 infection and pathogenesis. Proceedings of the National Academy of Sciences of the United States of America 2022, 119, e2205690119. [Google Scholar] [CrossRef]
- Romeu, A.R. Probable human origin of the SARS-CoV-2 polybasic furin cleavage motif. BMC Genomic Data 2023, 24, 71. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zheng, H.; Lin, H.; Li, M.; Yuan, R.; Peng, J.; Xiong, Q.; Sun, J.; Li, B.; Wu, J.; Yi, L.; Peng, X.; Zhang, H.; Zhang, W.; Hulswit, R.J.G.; Loman, N.; Rambaut, A.; Ke, C.; Bowden, T.A.; Pybus, O.G.; Lu, J. Identification of common deletions in the spike protein of severe acute respiratory syndrome coronavirus 2. Journal of Virology 2020, 94, e00790-20. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.D.; Williamson, M.K.; Lewis, S.; ... & Balloux, F. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Medicine 2020, 12, 68. [CrossRef]
 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





