Submitted:
12 August 2024
Posted:
14 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
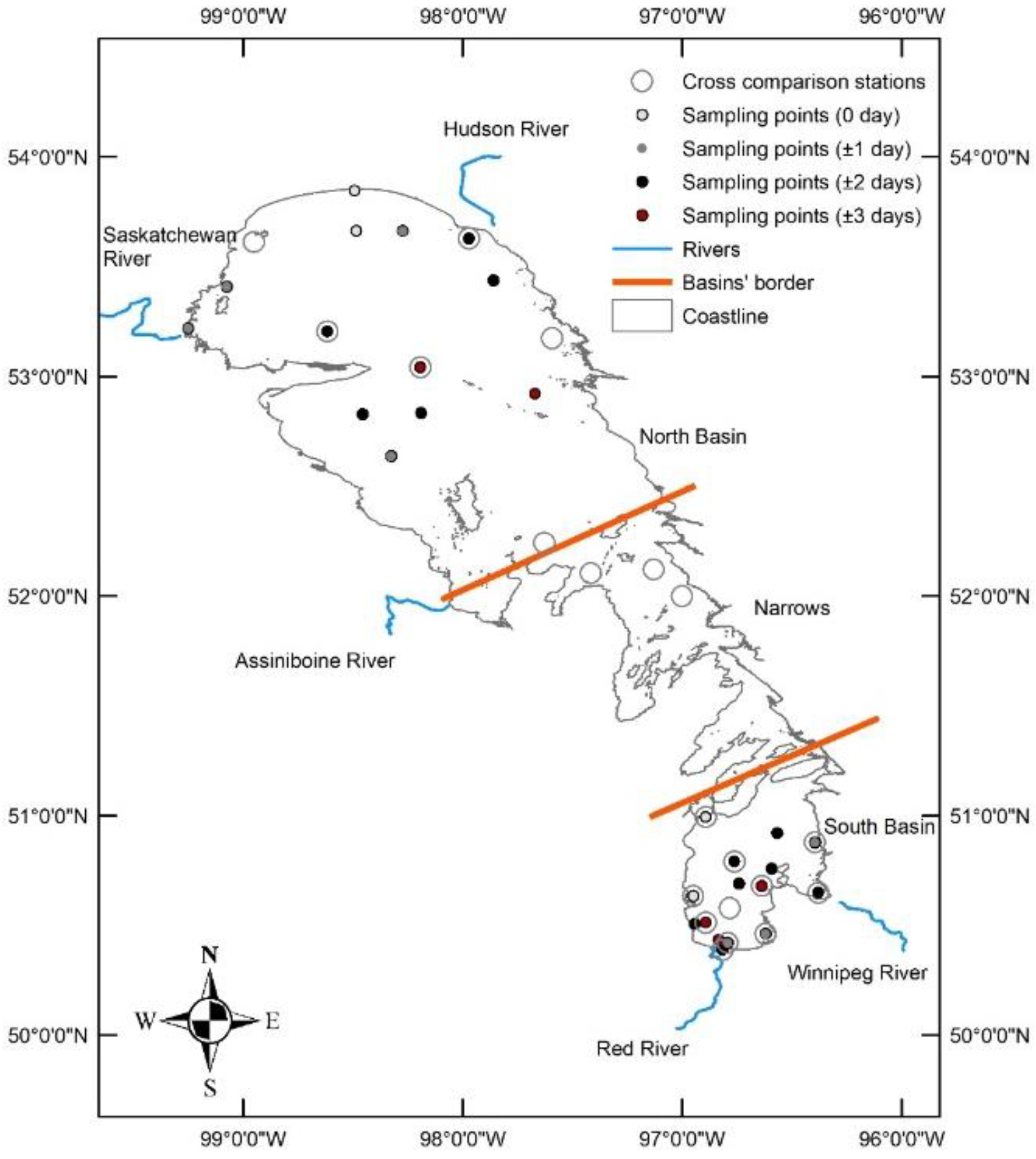
2.1. Case Study Area
2.2. LW Turbidity
2.3. Modeling Chl-a
2.3.1. In Situ Data Acquisition
2.3.2. Landsat Data
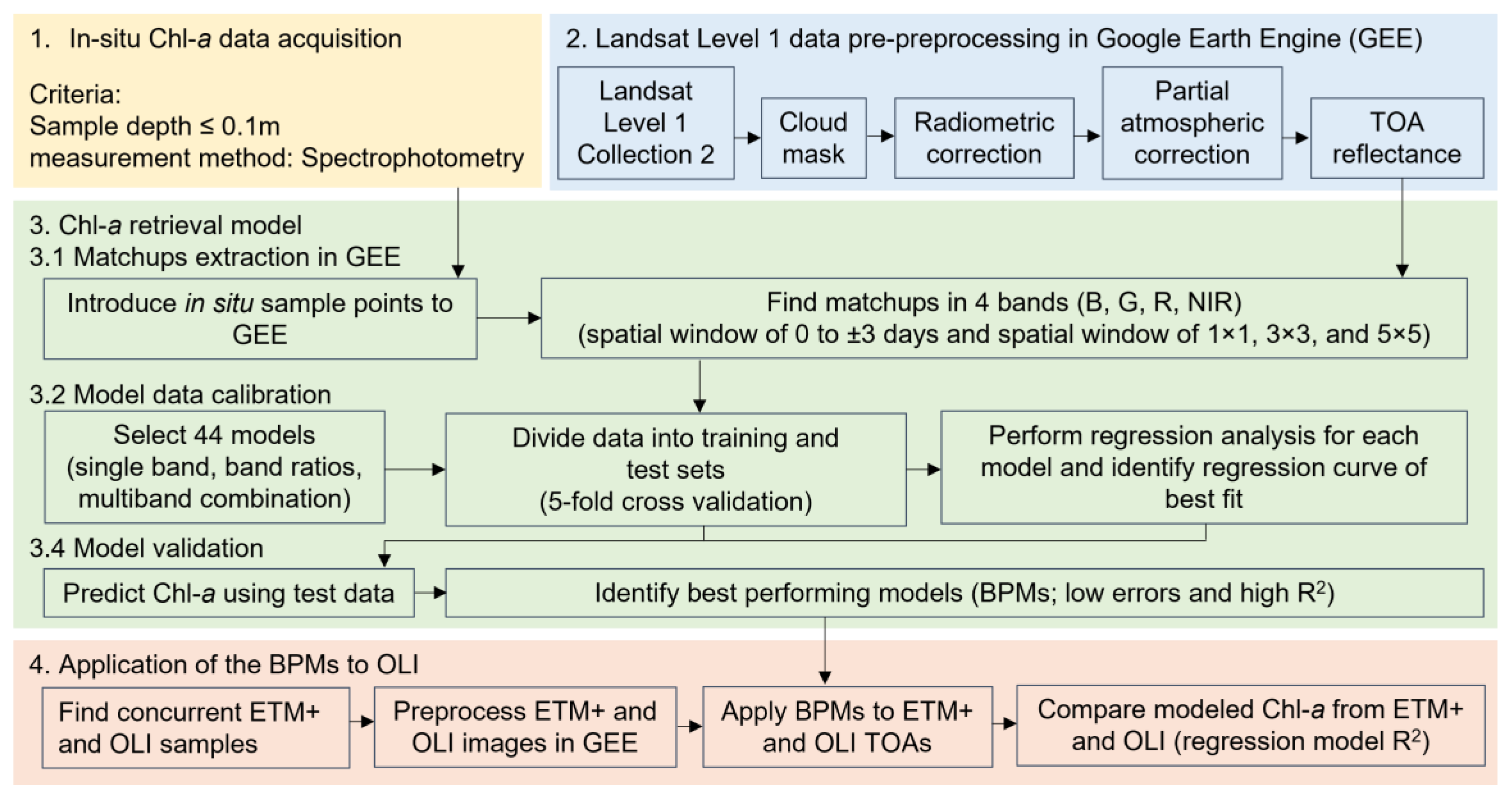
2.3.3. Pre-Processing Landsat Level 1 Data
2.3.4. Extracting Matchups
2.3.5. Model Calibration
2.3.6. Model Validation
2.4. Application of the BPMs to Landsat OLI
2.5. Comparing Chl-a Predictions Using Basin-Scale vs. Lake-Scale BPMs
3. Results
3.1. Spatial Heterogeneity in LW Turbidity
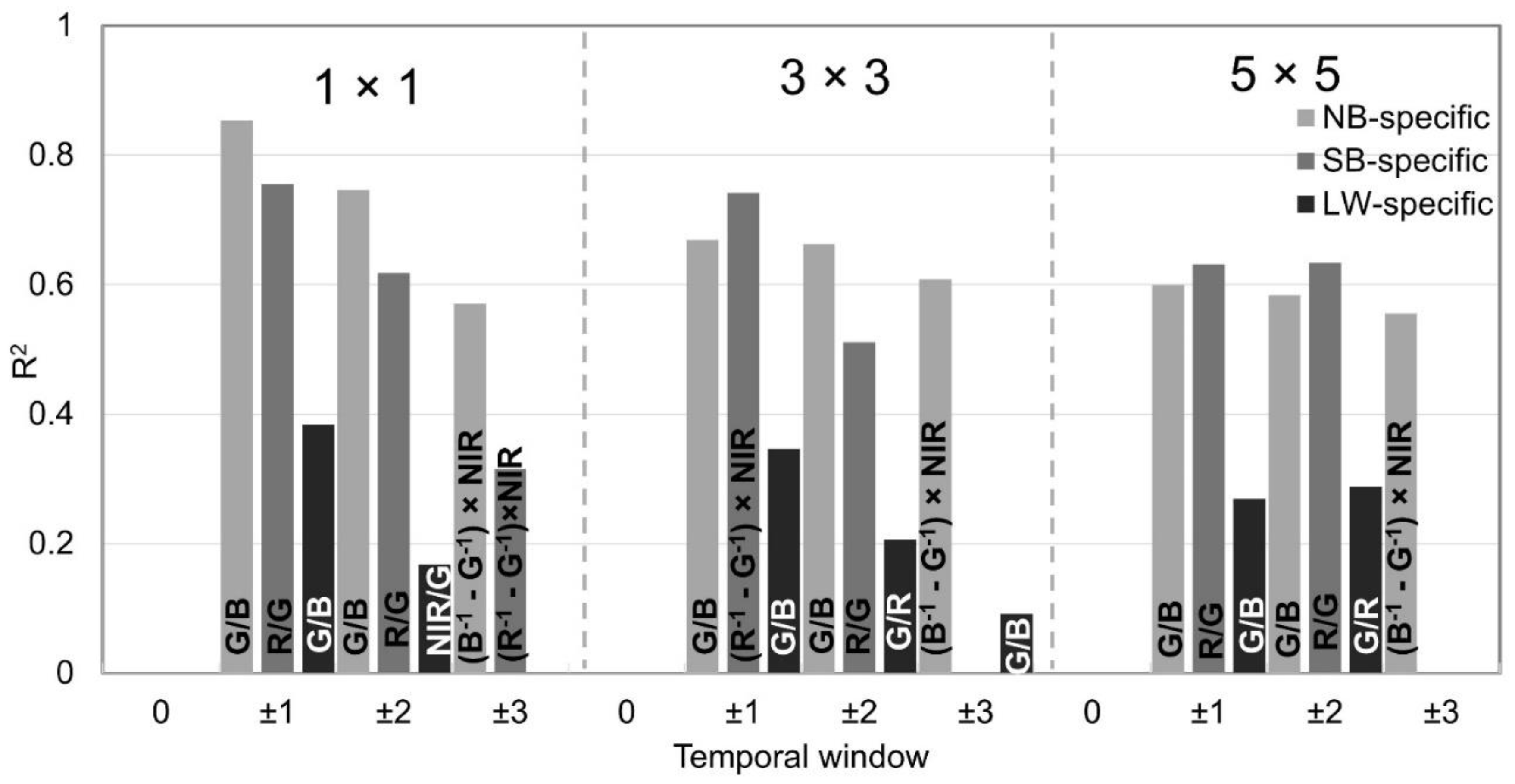
3.2. Best Performng Models
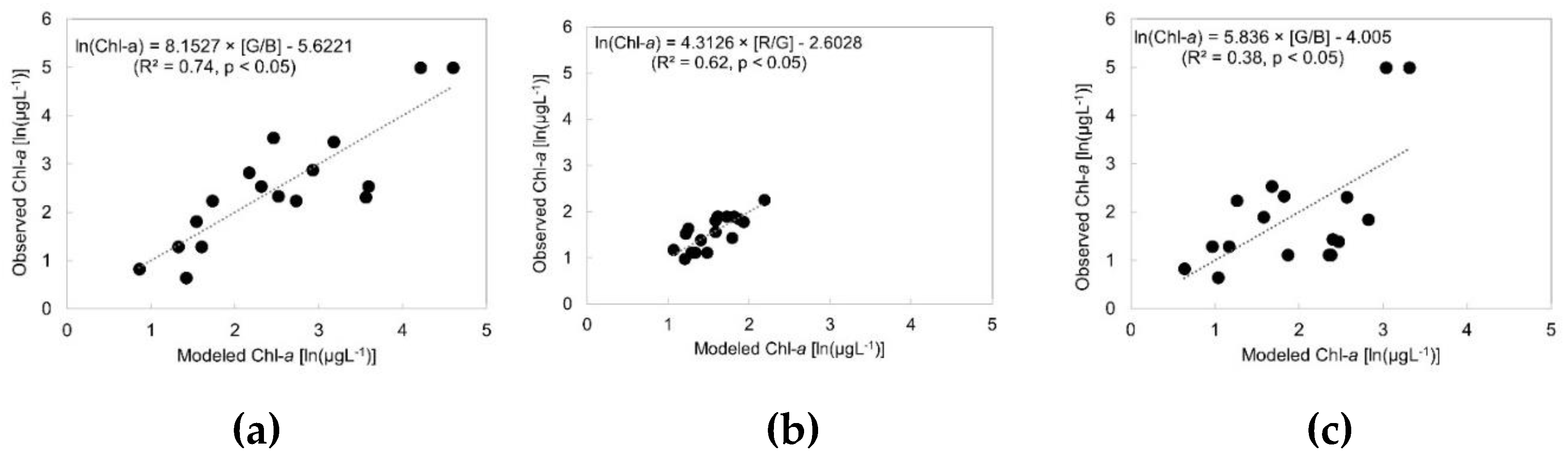
3.3. Best Chl-a Prediction Models
3.4. Application of the Models to Landsat OLI
3.5. Comparing Chl-a Predictions Using the Basin-Scale vs Lake-Scale BPMs
4. Discussion
4.1. Basin-Specific Chl-a Prediction Models
4.2. Application of the Models to Landsat OLI
4.3. Future Applications
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.; Visser, P.M. Cyanobacterial blooms. Nat. Rev. Microbiol. 2018, 16(8), 471–483. [Google Scholar] [CrossRef]
- Ho, J.C.; Michalak, A.M.; Pahlevan, N. Widespread global increase in intense lake phytoplankton blooms since the 1980s. Nature 2019, 574(7780), 667–670. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Feng, L.; Dai, Y.; Hu, C.; Gibson, L.; Tang, J.; Lee, Z.; Wang, Y.; Cai, X.; Liu, J.; Zheng, Y. Global mapping reveals increase in lacustrine algal blooms over the past decade. Nat. Geosci. 2022, 15(2), 130–134. [Google Scholar] [CrossRef]
- Randolph, K.; Wilson, J.; Tedesco, L.; Li, L.; Pascual, D.L.; Soyeux, E. Hyperspectral remote sensing of cyanobacteria in turbid productive water using optically active pigments, chlorophyll-a and phycocyanin. Remote. Sens. Environ. 2008, 112(11), 4009–4019. [Google Scholar] [CrossRef]
- Reid, J.L.; Bergman, J.N.; Kadykalo, A.N.; Taylor, J.J.; Twardek, W.; Rytwinski, T.; Chhor, A.D.; Frempong-Manso, A.; Martel, A.L.; Lapointe, N.W.R.; Bennett, J.R.; Nguyen, V.M.; Reid, A.J.; Marty, J.; Robinson, S.A.; Drake, A.R.; Winegardner, A.K.; Gregory-Eaves, I.S.; Taylor, M.K.; Smol, J.P.; Creed, I.F.; O’Connor, C.M.; Cooke, S.J. Developing a national level evidence-based toolbox for addressing freshwater biodiversity threats. Biol. Conserv 2022, 269, 109533. [Google Scholar] [CrossRef]
- An, K.G.; Lee, J.Y.; Kumar, H.K.; Lee, S.J.; Hwang, S.J.; Kim, B.H.; Park, Y.S.; Shin, K.H.; Park, S.; Um, H.Y. Control of algal scum using top-down biomanipulation approaches and ecosystem health assessments for efficient reservoir management. Water. Air. Soil. Pollut. 2010, 205(1-4), 3–24. [Google Scholar] [CrossRef]
- Weirich, C.A.; Miller, T.R. Freshwater harmful algal blooms: Toxins and children’s health. Curr. Probl. Pediatr. Adolesc. Health Care 2014, 44(1), 2–24. [Google Scholar] [CrossRef] [PubMed]
- Binding, C.E.; Pizzolato, L.; Zeng, C. EOLakeWatch; delivering a comprehensive suite of remote sensing algal bloom indices for enhanced monitoring of Canadian eutrophic lakes. Ecol. Indic. 2021, 121, 106999. [Google Scholar] [CrossRef]
- Guo, Q.; Wu, X.; Bing, Q.; Pan, Y.; Wang, Z.; Fu, Y.; Wang, D.; Liu, J. Study on retrieval of chlorophyll-a concentration based on Landsat OLI Imagery in the Haihe River, China. Sustainability 2016, 8(8), 1–15. [Google Scholar] [CrossRef]
- Kislik, C.; Dronova, I.; Grantham, T.E.; Kelly, M. Mapping algal bloom dynamics in small reservoirs using Sentinel-2 imagery in Google Earth Engine. Ecol. Indic. 2022, 140, 1–12. [Google Scholar] [CrossRef]
- Matthews, M.W. A current review of empirical procedures of remote sensing in inland and near-coastal transitional waters, Int. J. Remote Sens. 2011, 32(21), 6855–6899. [Google Scholar] [CrossRef]
- Sathyendranath, S.; Brewin, R.J.W.; Jackson, T.; Mélin, F.; Platt, T. Ocean-colour products for climate-change studies: What are their ideal characteristics? Remote Sens. Environ. 2017, 203, 125–138. [Google Scholar] [CrossRef]
- Kuhn, C.; de Matos Valerio, A.; Ward, N.; Loken, L.; Sawakuchi, H.O.; Kampel, M.; Richey, J.; Stadler, P.; Crawford, J.; Striegl, R.; Vermote, E.; Pahlevan, N.; Butman, D. Performance of Landsat-8 and Sentinel-2 surface reflectance products for river remote sensing retrievals of chlorophyll-a and turbidity, Remote Sens. Environ. 2019, 224, 104–118. [Google Scholar] [CrossRef]
- Watanabe, F.; Alcantara, E.; Rodrigues, T.; Rotta, L.; Bernardo, N.; IMAI, N. Remote sensing of the chlorophyll-a based on OLI/Landsat-8 and MSI/Sentinel-2A (Barra Bonita reservoir, Brazil), An. Acad. Bras. Cienc. 2017, 90(2 suppl 1), 1987–2000. [Google Scholar] [CrossRef]
- Tan, W.; Liu, P.; Liu, Y.; Yang, S.; Feng, S.; Tan, W.; Liu, P.; Liu, Y.; Yang, S.; Feng, S. A 30-year assessment of phytoplankton blooms in Erhai Lake using Landsat imagery: 1987 to 2016. Remote Sens. 2017, 9(12), 1265–1280. [Google Scholar] [CrossRef]
- Sass, G.Z.; Creed, I.F.; Bayley, S.E.; Devito, K.J. Understanding variation in trophic status of lakes on the Boreal Plain: A 20-year retrospective using Landsat TM imagery. Remote Sens. Environ. 2007, 109(2), 127–141. [Google Scholar] [CrossRef]
- Ho, J.C.; Stumpf, R.P.; Bridgeman, T.B.; Michalak, A.M. Using Landsat to extend the historical record of lacustrine phytoplankton blooms: a Lake Erie case study. Remote Sens. Environ. 2017, 191, 273–285. [Google Scholar] [CrossRef]
- Sayers, M.J.; Bosse, K.R.; Shuchman, R.A.; Ruberg, S.A.; Fahnenstiel, G.L.; Leshkevich, G.A.; Stuart, D.G.; Johengen, T.H.; Burtner, A.M.; Palladino, D. Spatial and temporal variability of inherent and apparent optical properties in western Lake Erie: Implications for water quality remote sensing. J. Great Lakes Res. 2019, 45(3), 490–507. [Google Scholar] [CrossRef]
- Environment Canada Manitoba Water Stewardship: State of Lake Winnipeg: 1999–2007, Manitoba Water Stewardship and Environment Canada, Winnipeg., 2011.
- Ulrich, A.E.; Malley, D.F.; Watts, P.D. Lake Winnipeg Basin: advocacy, challenges and progress for sustainable phosphorus and eutrophication control. Sci. Total Environ. 2016, 542, 1030–1039. [Google Scholar] [CrossRef] [PubMed]
- Zar, J.H. (Ed.) : Biostatistical Analysis, fourth ed. Prentice Hall, Upper Saddle River, NJ, 1999.
- Wheater, C.P.; Cook, P.A. (Eds.) : Using statistics to understand the environment, The Taylor & Francis e-Library, London and New York, 2005.
- APHA (American Public Health Association), AWWA (American Water Works Association), and WPCF (Water Pollution Control Federation), 1998. Standard Methods for the Examination of Water and Wastewater (20th Edition). APHA, New York, New York.
- Campbell, J.W. The lognormal distribution as a model for bio-optical variability in the sea. J. Geophys. Res. 1995, 100, 13237–13254. [Google Scholar] [CrossRef]
- Guanter, L.; Ruiz-Verdu, A.; Odermatt, D.; Giardino, C.; Simis, S.; Heege, T.; Domínguez- Gómez, J.A.; Moreno, J. Atmospheric correction of ENVISAT/MERIS data over inland waters: validation for European Lakes. Remote Sens. Environ. 2008, 114, 467–480. [Google Scholar] [CrossRef]
- Lobo, F.L.; Costa, M.P.F.; Novo, E.M.L.M. Time-series analysis of Landsat-MSS/TM/OLI images over Amazonian waters impacted by gold mining activities. Remote Sens. Environ. 2015, 157, 170–184. [Google Scholar] [CrossRef]
- Chander, G.; Markham, B.; Helder, D. Summary of current radiometric calibration coefficients for Landsat MSS, TM, ETM+, and EO-1 ALI sensors. Remote Sens. Environ. 2009, 113, 893–903. [Google Scholar] [CrossRef]
- Gilabert, M.A.; Conese, C.; Maselli, F. An atmospheric correction method for the automatic retrieval of surface reflectances from TM images. Int. J. Remote Sens. 1994, 15, 2065–2086. [Google Scholar] [CrossRef]
- Bucholtz, A. Rayleigh-scattering calculations for the terrestrial atmosphere. Appl. Opt. 1995, 34, 2765–2773. [Google Scholar] [CrossRef] [PubMed]
- Young, A.T. Revised depolarization corrections for atmospheric extinction. Appl. Opt. 1980, 19(20), 3427. [Google Scholar] [CrossRef] [PubMed]
- Vermote, E.; Tanré, D.; Deuzé, J.L.; Herman, M.; Morcrette, J.J.; Kotchenova, S.Y. 2006, Second simulation of a satellite signal in the solar spectrum - Vector (6SV). MODIS land surface reflectance science computing facility, user manual part two.
- Bodhaine, B.A.; Wood, N.B.; Dutton, E.G.; Slusser, J.R. On Rayleigh optical depth calculations, J. Atmos. Ocean. Tech. 1999, 16, 1854–1861. [Google Scholar] [CrossRef]
- Hansen, J.E.; Travis, L.D. Light scattering in planetary atmospheres. Space Sci. Rev. 1974, 16, 527–610. [Google Scholar] [CrossRef]
- Sturm, B. 1981. The atmospheric correction of remotely sensed data and the quantitative determination of suspended matter in marine water surface layers. In Remote Sensing in Meteorology, Oceanography and Hydrology, edited by A. P. Cracknell (Chichester: Ellis Horwood Limited), Chapter 11.
- Jorge, D.S.; Barbosa, C.C.; De Carvalho, L.A.; Affonso, A.G.; Lobo, F.D.L.; Novo, E.M.D.M. SNR (signal-to-noise ratio) impact on water constituent retrieval from simulated images of optically complex Amazon lakes. Remote Sens 2017, 9(7), 1–18. [Google Scholar] [CrossRef]
- Binding, C.E.; Greenberg, T.A.; McCullough, G.; Watson, S.B.; Page, E. An analysis of satellite-derived chlorophyll and algal bloom indices on Lake Winnipeg. J. Great Lakes Res. 2018, 44(3), 436–446. [Google Scholar] [CrossRef]
- Laiolo, L.; Matear, R.; Soja-Woźniak, M.; Suggett, D.J.; Hughes, D.J.; Baird, M.E.; Doblin, M.A. Modelling the impact of phytoplankton cell size and abundance on inherent optical properties (IOPs) and a remotely sensed chlorophyll-a product. J. Mar. Syst. 2021, 213, 1–12. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. John Lu, Z.Q., 2010. The elements of statistical learning: data mining, inference, and prediction. 2nd Edition, Springer. 2017. NY. 745p.
- Yates, L.A.; Aandahl, Z.; Richards, S.A.; Brook, B.W. Cross validation for model selection: a review with examples from ecology. Ecol. Monogr. 2023, 93(1), e1557. [Google Scholar] [CrossRef]
- Dallosch, M.A.; Creed, I.F. Optimization of Landsat chl-a retrieval algorithms in Freshwater Lakes through classification of optical water types. J. Remote Sens. 2021, 13(22), 4607. [Google Scholar]
- Ali, G.; English, C. Phytoplankton blooms in Lake Winnipeg linked to selective water-gatekeeper connectivity. Scientific reports 2019, 9(1), 8395. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, R.; Zhang, M.; Duan, H.; Loiselle, S.; Xu, J. Fourteen-year record (2000–2013) of the spatial and temporal dynamics of floating algae blooms in Lake Chaohu, observed from time series of MODIS images. Remote Sens. 2015, 7(8), 10523–10542. [Google Scholar] [CrossRef]
- Ha, N.T.T.; Koike, K.; Nhuan, M.T.; Canh, B.D.; Thao, N.T.P.; Parsons, M. Landsat 8/OLI two bands ratio algorithm for chlorophyll-a concentration mapping in hypertrophic waters: An application to West Lake in Hanoi (Vietnam). IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10(11), 4919–4929. [Google Scholar] [CrossRef]
- Keith, D.J.; Yoder, J.A.; Freeman, S.A. Spatial and Temporal Distribution of Coloured Dissolved Organic Matter (CDOM) in Narragansett Bay, Rhode Island: Implications for Phytoplankton on Coastal Waters. Estuar. Coast Shelf Sci. 2002, 55, 705–717. [Google Scholar] [CrossRef]
- Vincent, R.K.; Qin, X.; McKay, R.M.L.; Miner, J.; Czajkowski, K.; Savino, J.; Bridgeman, T. Phycocyanin detection from LANDSAT TM data for mapping cyanobacterial blooms in Lake Erie. Remote Sens. Environ. 2004, 89(3), 381–392. [Google Scholar] [CrossRef]
- Maeda, E.E.; Lisboa, F.; Kaikkonen, L.; Kallio, K.; Koponen, S.; Brotas, V.; Kuikka, S. Temporal patterns of phytoplankton phenology across high latitude lakes unveiled by long-term time series of satellite data, Remote Sens. Environ. 2018, 221, 609–620. [Google Scholar] [CrossRef]
- Nas, B.; Karabork, H.; Ekercin, S.; Berktay, A. Mapping chlorophyll-a through in situ measurements and Terra ASTER satellite data. Environ. Monit. Assess. 2009, 157, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Keith, D.; Rover, J.; Green, J.; Zalewsky, B.; Charpentier, M.; Thursby, G.; Bishop, J. Monitoring algal blooms in drinking water reservoirs using the Landsat-8 Operational Land Imager. Int. J. Remote Sens. 2018, 39(9), 2818–2846. [Google Scholar] [CrossRef]
- Paltsev, A.; Creed, I.F. Multi-decadal changes in phytoplankton biomass in northern temperate lakes as seen through the prism of landscape properties. Glob. Change Biol. 2022, 28, 2272–2085. [Google Scholar] [CrossRef]
- Paltsev, A.; Creed, I.F. Are northern lakes in relatively intact temperate forests showing signs of increasing phytoplankton biomass? Ecosyst 2022, 25, 727–755. [Google Scholar] [CrossRef]
- Palmer, S.C.; Kutser, T.; Hunter, P.D. Remote sensing of inland waters: Challenges, progress and future directions. Remote Sens. Environ. 2015, 157, 1–8. [Google Scholar] [CrossRef]
- Ruddick, K.G.; Gons, H.J.; Rijkeboer, M.; Tilstone, G. Optical remote sensing of chlorophyll-a in case 2 waters by use of an adaptive two-band algorithm with optimal error properties. Appl. Opt. 2001, 40(21), 3575–3585. [Google Scholar] [CrossRef]
- Dierssen, H.M. Perspectives on empirical Approaches for ocean color remote sensing of chlorophyll in a changing climate. Proc. Natl. Acad. Sci. USA 2010, 107(40), 17073–17078. [Google Scholar] [CrossRef]
- Tzortziou, M.; Subramaniam, A.; Herman, J.R.; Gallegos, C.L.; Neale, P.J.; Harding Jr, L.W. Remote sensing reflectance and inherent optical properties in the mid Chesapeake Bay. Estuar. Coast Shelf Sci. 2007, 72(1-2), 16–32. [Google Scholar] [CrossRef]
- Le, C.; Hu, C.; English, D.; Cannizzaro, J.; Kovach, C. Climate-driven chlorophyll-a changes in a turbid estuary: Observations from satellites and implications for management. Remote Sens. Environ. 2013, 130, 11–24. [Google Scholar] [CrossRef]
- Moradi, M.; Kabiri, K. Spatio-temporal variability of red-green chlorophyll-a index from MODIS data–Case study: Chabahar Bay, SE of Iran. Cont. Shelf Res. 2019, 184, 1–9. [Google Scholar] [CrossRef]
- Mishra, S.; Mishra, D.R. Normalized difference chlorophyll index: A novel model for remote estimation of chlorophyll-a concentration in turbid productive waters. Remote Sens. Environ. 2012, 117, 394–406. [Google Scholar] [CrossRef]
- Gilerson, A.A.; Gitelson, A.A.; Zhou, J.; Gurlin, D.; Moses, W.; Ioannou, I.; Ahmed, S.A. Algorithms for remote estimation of chlorophyll-a in coastal and inland waters using red and near infrared bands. Opt. Express 2010, 18(23), 24109–24125. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Dall'Olmo, G.; Moses, W.; Rundquist, D.C.; Barrow, T.; Fisher, T.R.; Gurlin, D.; Holz, J. A simple semi-analytical model for remote estimation of chlorophyll-a in turbid waters: Validation. Remote Sens. Environ. 2008, 112(9), 3582–3593. [Google Scholar] [CrossRef]
- Teillet, P.M.; Ren, X. Spectral band difference effects on vegetation indices derived from multiple satellite sensor data. Can. J. Remote Sens. 2008, 34, 159–173. [Google Scholar] [CrossRef]
- Boucher, J.; Weathers, K.C.; Norouzi, H.; Steele, B. Assessing the effectiveness of Landsat 8 chlorophyll a retrieval algorithm for regional freshwater monitoring. Ecol. Appl. 2018, 28(4), 1044–1054. [Google Scholar] [CrossRef]
- Thomalla, S.J.; Fauchereau, N.; Swart, S.; Monterio, P.M.S. Regional scale characteristics of the seasonal cycle of chlorophyll in the Southern Ocean. Biogeosciences 2011, 8, 2849–2866. [Google Scholar] [CrossRef]
- Neil, C.; Spyrakos, E.; Hunter, P.D.; Tyler, A.N. A global approach for chlorophyll-a retrieval across optically complex inland waters based on optical water types. Remote Sens. Environ. 2019, 229, 159–178. [Google Scholar] [CrossRef]
- Zhao, D.; Huang, J.; Li, Z.; Yu, G.; Shen, H. Dynamic monitoring and analysis of chlorophyll-a concentrations in global lakes using Sentinel-2 images in Google Earth Engine. Sci. Total Environ. 2024, 912, 169152. [Google Scholar] [CrossRef]
- Henson, S.A.; Sarmiento, J.L.; Dunne, J.P.; Bopp, L.; Lima, I.; Doney, S.C.; John, J.; Beaulieu, C. Detection of anthropogenic climate change in satellite records of ocean chlorophyll and productivity. Biogeosciences 2010, 7(2), 621–640. [Google Scholar] [CrossRef]
- Mirtl, M.T.; Borer, E.; Djukic, I.; Forsius, M.; Haubold, H.; Hugo, W.; Jourdan, J.; Lindenmayer, D.; McDowell, W.H.; Muraoka, H.; Orenstein, D.E.; Pauw, J.C.; Peterseil, J.; Shibata, H.; Wohner, C.; Yu, X.; Haase, P. Genesis, goals and achievements of long-term ecological research at the global scale: a critical review of ILTER and future directions, Sci. Total Environ. 2018, 626, 1439–1462. [Google Scholar] [CrossRef]
- Salmaso, N.; Anneville, O.; Straile, D.; Viaroli, P. European large perialpine lakes under anthropogenic pressures and climate change: present status, research gaps and future challenges. Hydrobiologia 2018, 824(1), 1–32. [Google Scholar] [CrossRef]
- Salgado-Hernanz, P.M.; Racault, M.-F.; Font-Muñoz, J.S.; Basterretxea, G. Trends in phytoplankton phenology in the Mediterranean Sea based on ocean-colour remote sensing. Remote Sens. Environ. 2019, 221, 50–64. [Google Scholar] [CrossRef]
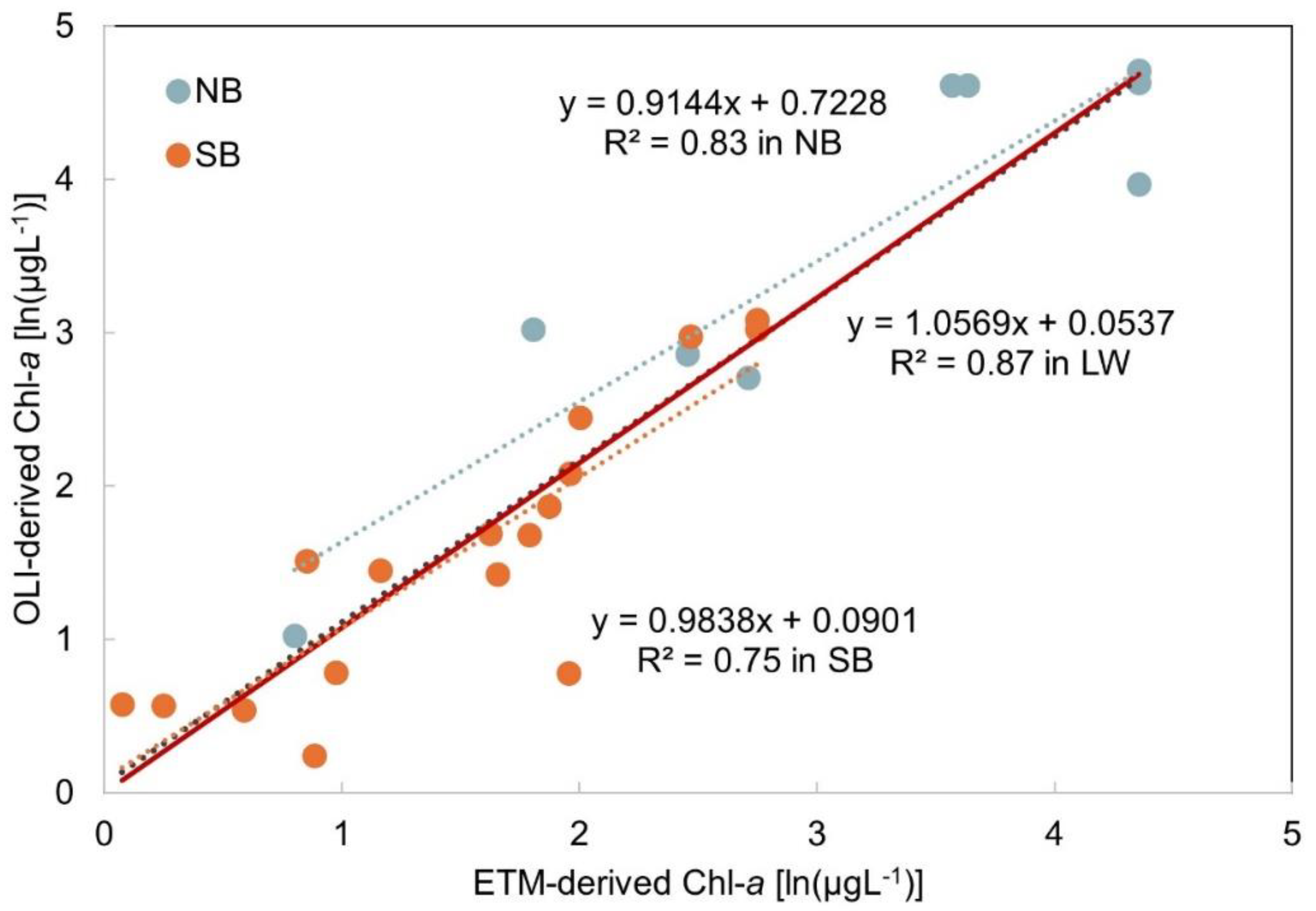
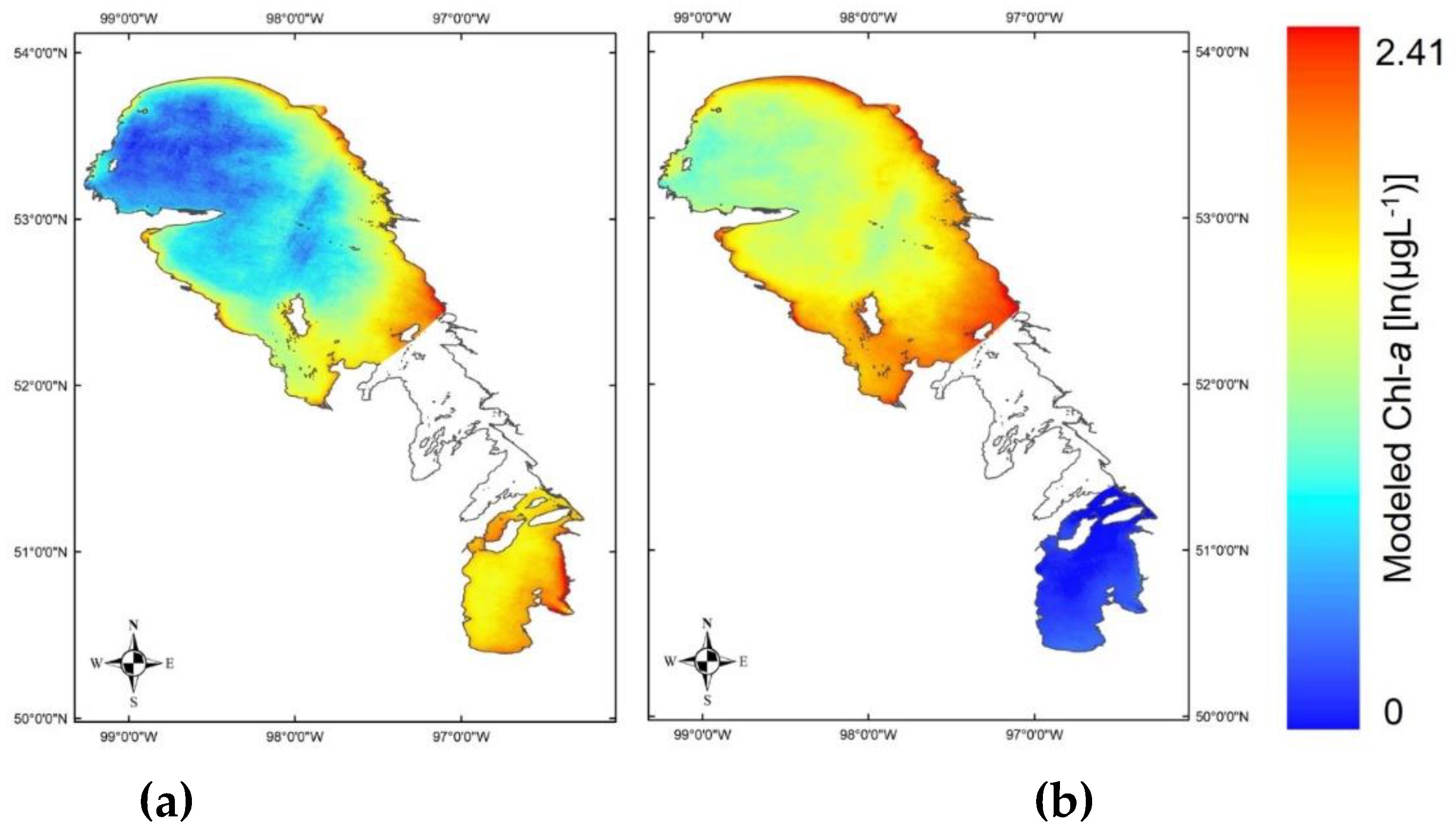
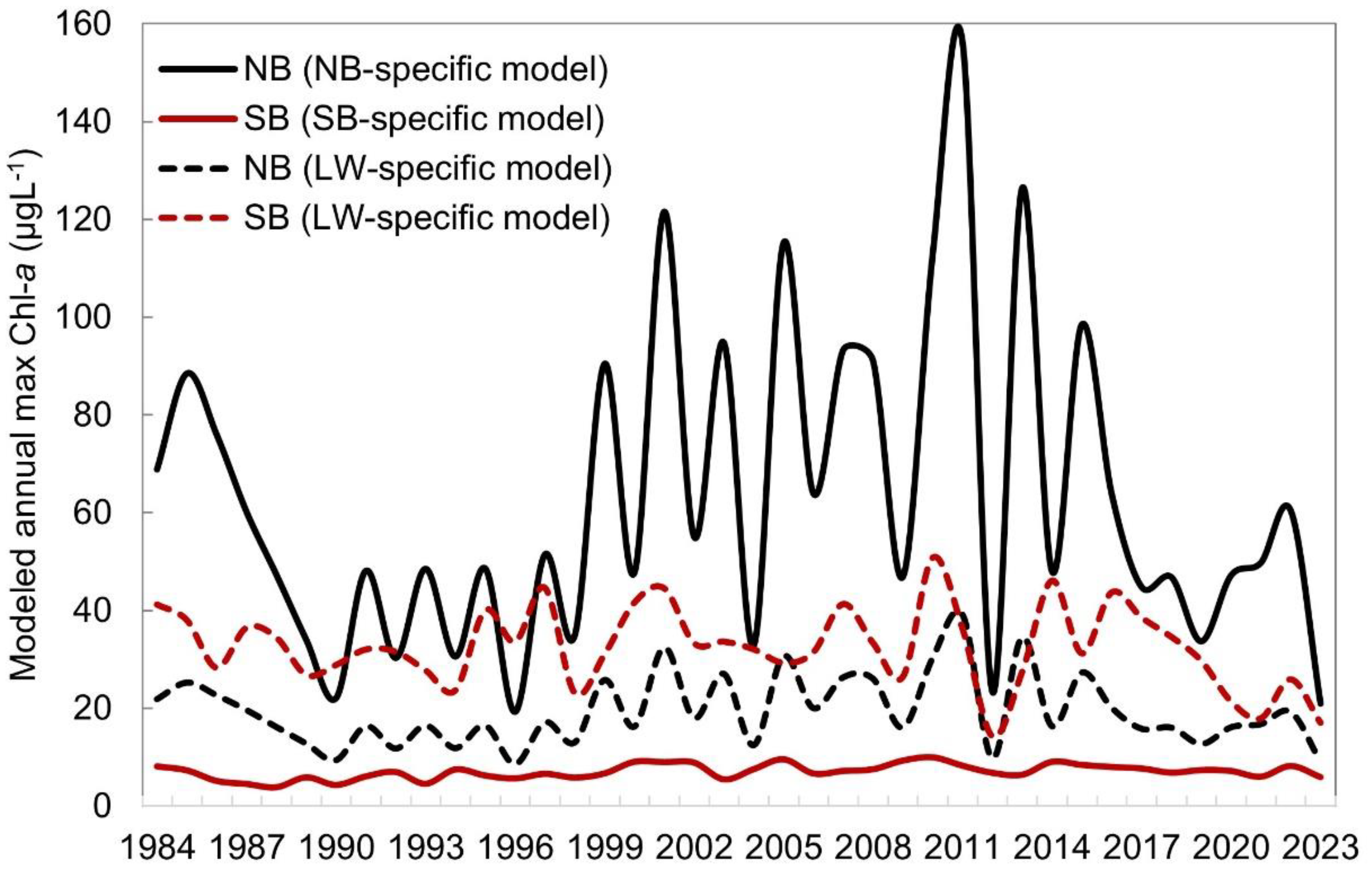
| Basin | Temporal window (days) | # of matchups | In situ Chl-a (µgL-1) | # of stations | # of samples | # of images | Landsat sensors | Landsat scenes (path/row) | Year | Month |
|---|---|---|---|---|---|---|---|---|---|---|
| NB | 0 | 2 | 10.1 - 147 | 2 | 2 | 1 | 5 | 3322 | 2011 | 9 |
| ±1 | 10 | 1.91 - 147 | 6 | 8 | 6 | 5, 7 | 3223, 3322/23 | 2010, 2011 | 7, 8, 9, 10 | |
| ±2 | 17 | 1.91 - 147 | 11 | 13 | 8 | 5, 7 | 3223, 3322/23 | 2010, 2011 | 7, 8, 9, 10 | |
| ±3 | 23 | 1.91 - 147 | 13 | 18 | 9 | 5, 7 | 3223, 3322/23 | 2010, 2011 | 7, 8, 9, 10 | |
| SB | 0 | 3 | 3.05 – 4.01 | 2 | 2 | 2 | 5, 7 | 3124/25 | 2010 | 7, 8 |
| ±1 | 7 | 3.05 – 6.68 | 5 | 5 | 5 | 5, 7 | 3025, 3124/25 | 2010, 2011 | 7, 8 | |
| ±2 | 20 | 2.67 – 9.55 | 12 | 15 | 8 | 5, 7 | 3025, 3124/25 | 2010, 2011 | 7, 8, 10 | |
| ±3 | 30 | 2.67 - 147 | 15 | 24 | 11 | 5, 7 | 3025, 3124/25 | 2010, 2011 | 7, 8, 9, 10 | |
| LW | 0 | 5 | 3.05 - 147 | 4 | 4 | 3 | 5, 7 | 3322, 3124/25 | 2010, 2011 | 7, 8, 9 |
| ±1 | 17 | 1.91 - 147 | 11 | 13 | 11 | 5, 7 | 3223, 3025, 3124/25, 3322/23 | 2010, 2011 | 7, 8, 9, 10 | |
| ±2 | 27 | 1.91 - 147 | 23 | 28 | 16 | 5, 7 | 3223, 3025, 3124/25, 3322/23 | 2010, 2011 | 7, 8, 9, 10 | |
| ±3 | 57 | 1.91 - 147 | 28 | 42 | 20 | 5, 7 | 3223, 3025, 3124/25, 3322/23 | 2010, 2011 | 7, 8, 9, 10 |
| All months | May | June | July | Aug | Sep | Oct | All months |
|---|---|---|---|---|---|---|---|
| NB | 5.5a (723) | - | 5.39a (205) | 5.53a (176) | 3.81a (95) | 5.55a (207) | 9.44a (41) |
| SB | 15.00b (396) | 10.60a (20) | 8.06b (107) | 19,00b (58) | 14.90b (73) | 19.75b (58) | 20.50a,b (80) |
| Narrows | 16.1b (243) | 5.50b (2) | 9.10b (78) | 21.80b (62) | 17.45b (70) | 17.90b (23) | 14.70b (8) |
| Basin | Model | Calibration R2 | RMSE (µgL-1) | RMSLE (µgL-1) | NRMSE | MAE (µgL-1) | MAPE (%) | |
|---|---|---|---|---|---|---|---|---|
| NB | G/B | 0.74 | 20.53 | 0.65 | 0.88 | 14.51 | 55.43 | |
| SB | R/G | 0.62 | 1.14 | 0.20 | 0.24 | 1 | 22.81 | |
| LW | G/B | 0.38 | 21.57 | 0.87 | 1.29 | 13.57 | 64.27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





