Submitted:
13 August 2024
Posted:
14 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Ethics Approval
2.3. Sample Processing
2.4. RNA Isolation from Endometrial Samples
2.6. Statistical Analysis
3. Results
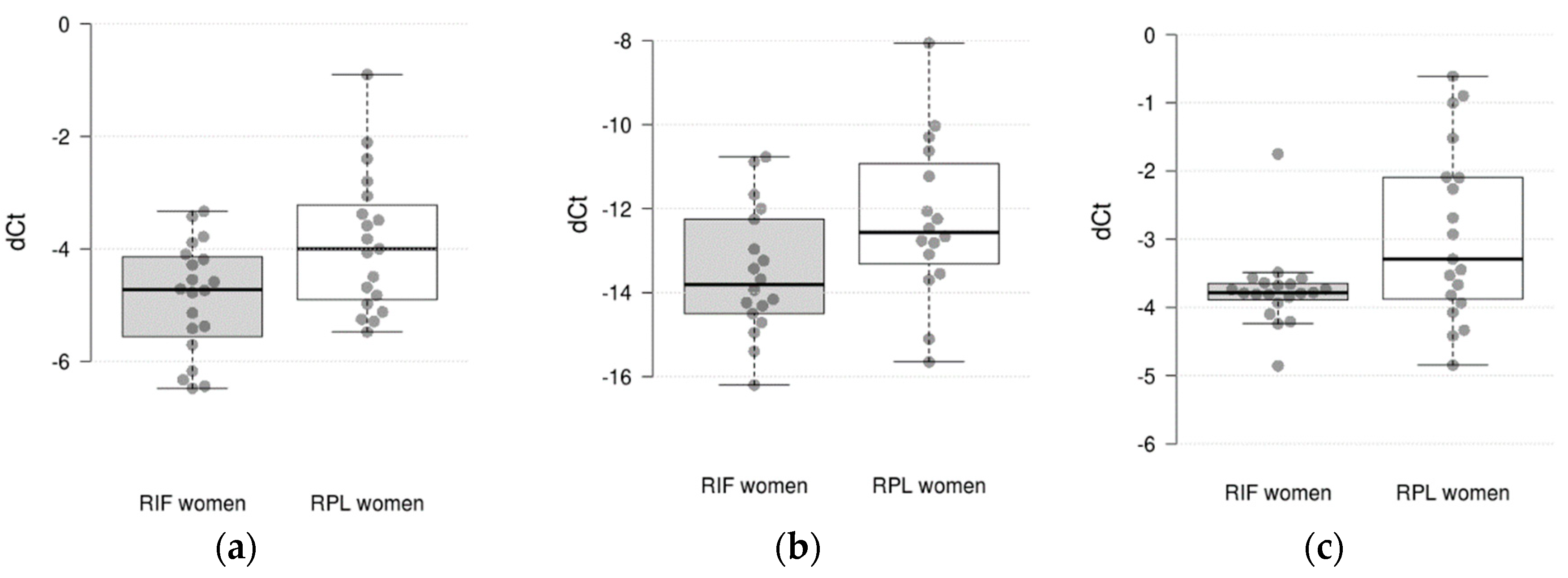
3.1. The Level of mRNA in Endometrial Samples of RIF Patients in Comparison with RPL Patients
3.2. Associations with Clinical and Laboratory Characteristics
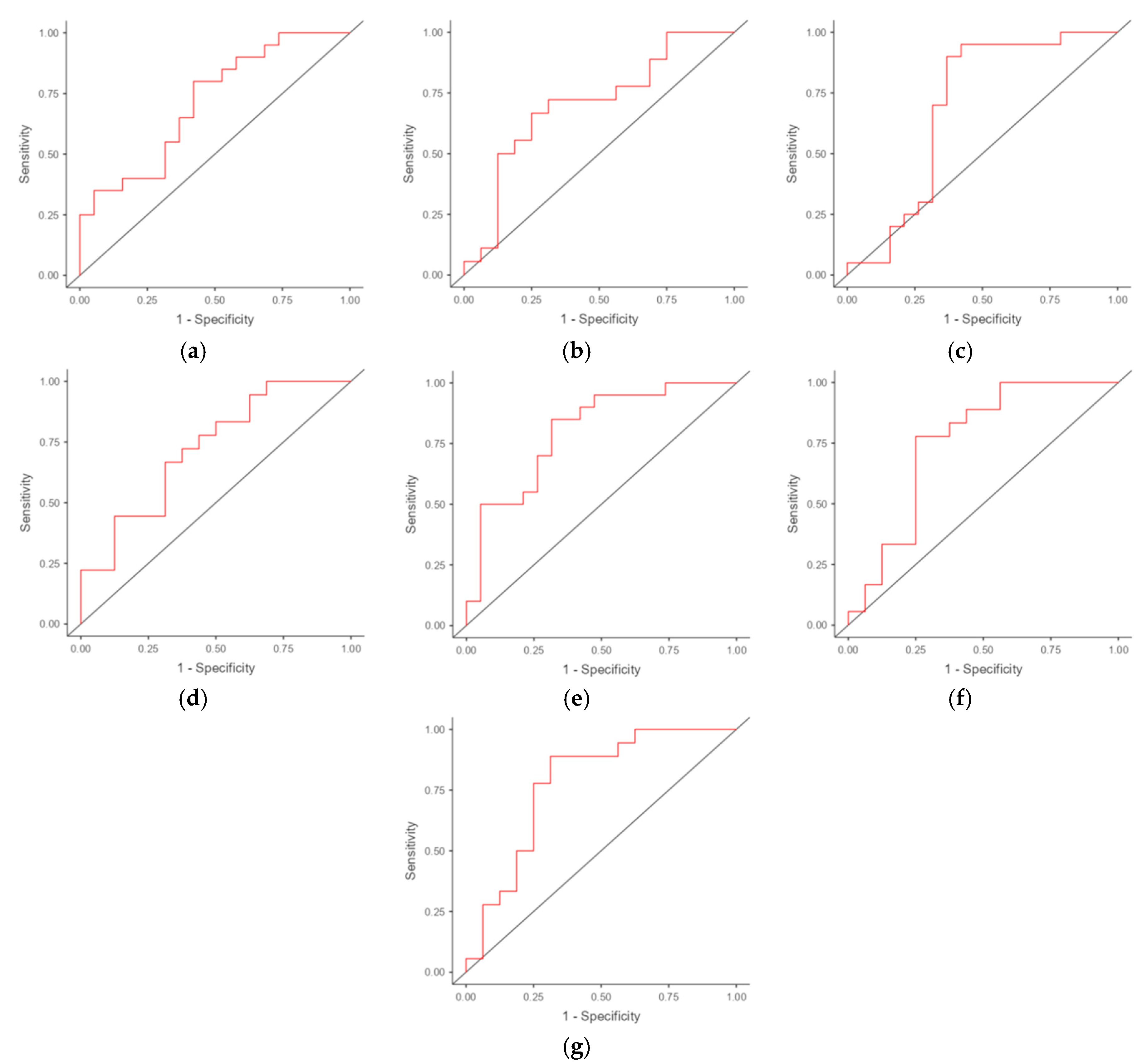
3.3. ROC Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, X.; Yu, Q. An update on the progress of transcriptomic profiles of human endometrial receptivity. Biol Reprod. 2018, 98, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Harrity, C.; Shkrobot, L.; Walsh, D.; Marron, K. ART implantation failure and miscarriage in patients with elevated intracellular cytokine ratios: response to immune support therapy. Fertil Res Pract. 2018, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Lessey, B.A.; Young, S.L. What exactly is endometrial receptivity? Fertil Steril. 2019, 111, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhou, W.; Sacco, M.; Downing, P.; Dimitriadis, E.; Zhao, F. Using organoids to investigate human endometrial receptivity. Front. Endocrinol. 2023, 14, 1158515. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, R.; Taketani, T.; Mihara, Y.; Sato, S.; Okada, M.; Tamura, I.; Jozaki, K.; Kajimura, T.; Asada, H.; Tamura, H.; Takasaki, A.; Sugino, N. Thin endometrium transcriptome analysis reveals a potential mechanism of implantation failure. Reprod Med Biol. 2017, 16, 206–227. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Lu, H.; Yu, X.; Dong, L.; Mi, L.; Wang, J.; Zheng, X.; Feng, K. The effect of icariin for infertile women with thin endometrium: A protocol for systematic review. Medicine (Baltimore). 2020, 99, e19111. [Google Scholar] [CrossRef] [PubMed]
- Azizi, R.; Aghebati-Maleki, L.; Nouri, M.; Marofi, F.; Negargar, S.; Yousefi, M. Stem cell therapy in Asherman syndrome and thin endometrium: stem cell-based therapy. Biomed. Pharmacother. 2018, 102, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Yao, G.; Wang, Y.; Xu, H.; Ji, X.; He, Y.; Zhu, Q.; Chen, Z.; Sun, Y. Transcriptomic changes during the pre-receptive to receptive transition in human endometrium detected by RNA-seq. J Clin Endocrinol Metab. 2014, 99, E2744–E2753. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Matthews, D. A.; Bessant, C. High Throughput Discovery of Protein Variants Using Proteomics Informed by Transcriptomics. Nucleic Acids Res. 2018, 46, 4893–4902. [Google Scholar] [CrossRef] [PubMed]
- Zong, L.; Zheng, S.; Meng, Y.; Tang, W.; Li, D.; Wang, Z.; Tong, X.; Xu, B. Integrated Transcriptomic Analysis of the miRNA-mRNA Interaction Network in Thin Endometrium. Front Genet. 2021, 16, 12–589408. [Google Scholar] [CrossRef] [PubMed]
- Enciso, M.; Carrascosa, J.P.; Sarasa, J.; Martínez-Ortiz, P.A.; Munné, S.; Horcajadas, JA.; Aizpurua, J. Development of a new comprehensive and reliable endometrial receptivity map (ER Map/ER Grade) based on RT-qPCR gene expression analysis. Hum Reprod. 2018, 33, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Messaoudi, S.; El Kasmi, I.; Bourdiec, A.; Crespo, K.; Bissonnette, L.; Le Saint, C.; Bissonnette, F.; Kadoch, I.-J. 15 years of transcriptomic analysis on endometrial receptivity: What have we learnt? Fertil. Res. Pract. 2019, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Altmäe, S.; Koel, M.; Võsa, U.; Adler, P.; Suhorutšenko, M.; Laisk-Podar, T.; Kukushkina, V.; Saare, M.; Velthut-Meikas, A.; Krjutškov, K.; Aghajanova, L.; Lalitkumar, P.G.; Gemzell-Danielsson, K.; Giudice, L.; Simón, C.; Salumets, A. Meta-signature of human endometrial receptivity: a meta-analysis and validation study of transcriptomic biomarkers. Sci Rep. 2017, 7, 10077. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Fan, Y.; Wang, J.; Shi, R. Dysfunctional intercellular communication and metabolic signaling pathways in thin endometrium. Front. Physiol. 2022, 13, 1050690. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kumar, A.; Zhang, F.; Lee, C.; Tang, Z. Complicated life, complicated VEGF-B. Trends Mol Med. 2012, 18, 119–27. [Google Scholar] [CrossRef] [PubMed]
- Albayrak, İ.G.; Azhari, F.; Çolak, E.N.; Balcı, B.K.; Ülgen, E.; Sezerman, U.; Baştu, E.; Günel, T. Endometrial gene expression profiling of recurrent implantation failure after in vitro fertilization. Mol Biol Rep. 2021, 48, 5075–5082. [Google Scholar] [CrossRef] [PubMed]
- Basatvat, S.; Russell, J.M.; Saare, M.; Thurston, L.M.; Salumets, A.; Fazeli, A. Potential innate immunity-related markers of endometrial receptivity and recurrent implantation failure (RIF). Reprod Biol. 2021, 21, 100569. [Google Scholar] [CrossRef] [PubMed]
- Lédée, N.; Petitbarat, M.; Rahmati, M.; Dubanchet, S.; Chaouat, G.; Sandra, O.; Perrier-d'Hauterive, S.; Munaut, C.; Foidart, J.M. New pre-conception immune biomarkers for clinical practice: interleukin-18, interleukin-15 and TWEAK on the endometrial side, G-CSF on the follicular side. J Reprod Immunol. 2011, 88, 118–23. [Google Scholar] [CrossRef] [PubMed]
- Murata, H.; Tanaka, S.; Tsuzuki-Nakao, T.; Kido, T.; Kakita-Kobayashi, M.; Kida, N.; Hisamatsu, Y.; Tsubokura, H.; Hashimoto, Y.; Kitada, M.; Okada, H. The transcription factor HAND2 up-regulates transcription of the IL15 gene in human endometrial stromal cells. J Biol Chem. 2020, 295, 9596–9605. [Google Scholar] [CrossRef] [PubMed]
- Bender Atik, R.; Christiansen, O.B.; Elson, J.; Kolte, A.M.; Lewis, S.; Middeldorp, S.; Nelen, W.; Peramo, B.; Quenby, S.; Vermeulen, N.; Goddijn, M.; ESHRE Guideline Group on RPL. ESHRE guideline: Recurrent pregnancy loss. Hum. Reprod. Open. 2018, 6, hoy004. [Google Scholar] [CrossRef]
- ESHRE Working Group on Recurrent Implantation Failure; Cimadomo D, de Los Santos MJ, Griesinger G, Lainas G, Le Clef N, McLernon DJ, Montjean D, Toth B, Vermeulen N, Macklon N. ESHRE good practice recommendations on recurrent implantation failure. Hum Reprod Open. 2023, 15, hoad023. [Google Scholar] [CrossRef]
- Königshoff, M.; Kramer, M.; Balsara, N.; Wilhelm, J.; Amarie, O.V.; Jahn, A.; Rose, F.; Fink, L.; Seeger, W.; Schaefer, L.; Günther, A.; Eickelberg, O. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. Journal of Clinical Investigation. 2009, 119, 772–787. [Google Scholar] [CrossRef] [PubMed]
- Goksuluk, D.; Korkmaz, S.; Zararsiz, G.; Karaagaoglu, A.E. Easy ROC: an interactive web-tool for ROC curve analysis using R language environment. The R Journal. 2016, 8, 213–230. [Google Scholar] [CrossRef]
- Muller, M.P.; Tomlinson, G.; Marrie, T.J.; Tang, P.; McGeer, A.; Low, D.E.; Detsky, A.S.; Gold, W.L. Can routine laboratory tests discriminate between severe acute respiratory syndrome and other causes of community-acquired pneumonia? Clin Infect Dis. 2005, 40, 1079–86. [Google Scholar] [CrossRef] [PubMed]
- Devesa-Peiro, A.; Sebastian-Leon, P.; Garcia-Garcia, F.; Arnau, V.; Aleman, A.; Pellicer, A.; Diaz-Gimeno, P. Uterine disorders affecting female fertility: what are the molecular functions altered in endometrium? Fertil Steril. 2020, 113, 1261–1274. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Yao, S.; Dong, Y.; Liu, D.; Wang, H.; Jiang, P.; Dai, C.; Lv, H.; Cao, C.; Zhou, Z.; Wang, L.; Gou, W.; Zhang, X.; Zhao, G.; Hu, Y. Down-regulation of PBK inhibits proliferation of human endometrial stromal cells in thin endometrium. Reprod Biol Endocrinol. 2022, 20, 25. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.A.; Moldenhauer, L.M.; Green, E.S.; Care, A.S.; Hull, M.L. Immune determinants of endometrial receptivity: a biological perspective. Fertil Steril. 2022, 117, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Gao, W.; Li, Y.; Wang, Y.; Liu, L.; Zhang, X. Potential Biomarkers and Endometrial Immune Microenvironment in Recurrent Implantation Failure. Biomolecules. 2023, 13, 406. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Eguren, A.; Bueno-Fernandez, C.; Gómez-Álvarez, M.; Francés-Herrero, E.; Pellicer, A.; Bellver, J.; Seli, E.; Cervelló, I. Evolution of biotechnological advances and regenerative therapies for endometrial disorders: a systematic review. Hum Reprod Update. 2024, dmae013. [Google Scholar] [CrossRef] [PubMed]
- Maziotis, E.; Kalampokas, T.; Giannelou, P.; Grigoriadis, S.; Rapani, A.; Anifantakis, M.; Kotsifaki, A.; Pantou, A.; Triantafyllidou, O.; Tzanakaki, D.; Neofytou, S.; Vogiatzi, P.; Bakas, P.; Simopoulou, M.; Vlahos, N. Commercially Available Molecular Approaches to Evaluate Endometrial Receptivity: A Systematic Review and Critical Analysis of the Literature. Diagnostics 2022, 12, 2611. [Google Scholar] [CrossRef]
- Bastu, E.; Demiral, I.; Gunel, T.; Ulgen, E.; Gumusoglu, E.; Hosseini, M.K.; Sezerman, U.; Buyru, F.; Yeh, J. Potential Marker Pathways in the Endometrium That May Cause Recurrent Implantation Failure. Reprod Sci. 2019, 26, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Macklon, N.S.; Brosens, J.J. The human endometrium as a sensor of embryo quality. Biol Reprod. 2014, 91, 98. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Wu, X.H. Decreased CD56+CD16-CD94+uNK cells in the mid-luteal phase in women with recurrent implantation failure are associated with IL-15 deficiency. Am J Reprod Immunol. 2023, 90, e13794. [Google Scholar] [CrossRef] [PubMed]
- Bashiri, A.; Halper, K.I.; Orvieto, R. Recurrent Implantation Failure-update overview on etiology, diagnosis, treatment and future directions. Reprod. Biol. Endocrinol. 2018, 16, 21. [Google Scholar] [CrossRef] [PubMed]
- Zargar, M.; Ghafourian, M.; Behrahi, F.; Nikbakht, R.; Salehi, A.M. Association of recurrent implantation failure and recurrent pregnancy loss with peripheral blood natural killer cells and interferon-gamma level. Obstet Gynecol Sci. 2024, 67, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, P.A.S.; Assone, T.; Prates, G.; Tedeschi, M.R.M.; Fonseca, L.A.M.; Casseb, J. The role of IFN-γ production during retroviral infections: an important cytokine involved in chronic inflammation and pathogenesis. Rev Inst Med Trop Sao Paulo. 2022, 30, 64–e64. [Google Scholar] [CrossRef] [PubMed]
- Meek, S.; Sutherland, L.; Wei, J.; Sturmey, R.; Binas, B.; Clinton, M.; Burdon, T. Hypoxanthine phosphoribosyltransferase (HPRT)-deficiency is associated with impaired fertility in the female rat. Mol Reprod Dev. 2020, 87, 930–933. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | RIF patients (N = 20) |
RPL patients (N = 19) |
|---|---|---|
| Age (years): Mean±SD | 34.65 ± 5.29 | 33.37 ± 5.22 |
| BMI (kg/m2): Mean±SD | 23.36 ± 4.24 | 25.69 ± 4.79 |
| Endometrium thickness (mm): Mean±SD | 6.51 ± 1.27 | 6.13 ± 1.86 |
| Fibrinogen, g/L: Mean±SD | 3.05 ± 0.51 | 3.10 ± 0.23 |
| Protrombin index, %: Mean±SD | 99.16 ± 8.22 | 97.57 ± 6.49 |
| Protrombin time, sec: Mean±SD | 13.04 ± 2.35* | 14.69 ± 1.78 |
| Activated Partial Thromboplastin Time, sec: Mean±SD | 34.04 ± 3.61 | 32.10 ± 2.20 |
| International Normalized Ratio: Mean±SD | 1.04 ± 0.12 | 1.21 ± 0.14 |
| Anti-Mullerian Hormone, ng/mL: Mean±SD | 2.60 ± 1.03 | 2.64 ± 0.32 |
| Luteinizing hormone, mIU/mL: Mean±SD | 8.34 ± 2.13 | 11.24 ± 3.26 |
| Follicle stimulating hormone, mIU/mL: Mean±SD | 6.19 ± 1.43 | 6.00 ± 1.07 |
| Prolactin, mIU/L: Mean±SD | 325.0 ± 152.4 | 333.6 ± 61.6 |
| Thyroid stimulating hormone, mIU/mL: Mean±SD | 2.84 ± 0.74 | nd |
| Chronic endometritis, yes/no | 20/0 | 16/3 |
| Chronic salpingoophoritis, yes/no | 10/10 | 10/9 |
| Pelvic organs surgeries, yes/no | 10/10 | 6/13 |
| Endometriosis, yes/no | 5/15 | 4/15 |
| Uterine fibroids, yes/no | 1/19 | 3/16 |
| Polyps, yes/no | 1/19 | 4/15 |
| Gene | Ct mean ± SD | ΔCt mean ± SE | ΔΔCt (95% CI) log2 fold change |
P value | ||
|---|---|---|---|---|---|---|
| RIF | RLP | RIF | RLP | |||
| C4BPA | 30.51 ± 3.75 | 29.69 ± 3.29 | -8.20 ± 0.61 | -9.65 ± 0.38 | 0.99 (-0.06; 2.13) | 0.059 |
| CXCL1 | 28.19 ± 4.85 | 27.84 ± 4.27 | -5.53 ± 0.65 | -6.82 ± 0.29 | 0.83 (-0.33; 1.97) | 0.175 |
| HAND2 | 24.36 ± 4.29 | 22.90 ± 3.52 | -1.69 ± 0.44 | -1.88 ± 0.17 | 0.35 (-0.24; 1.00) | 0.235 |
| HPRT1 | 25.59 ± 3.81 | 24.77 ± 4.23 | -2.92 ± 0.29 | -3.75 ± 0.13 | 0.57 (0.01; 1.54) | 0.041 |
| IFNG | 33.68 ± 2.49 | 33.58 ± 2.85 | -12.27 ± 0.48 | -13.52 ± 0.36 | 1.24 (0.01; 2.43) | 0.046 |
| IL15 | 26.54 ± 3.63 | 25.89 ± 3.89 | -3.88 ± 0.28 | -4.87 ± 0.22 | 0.92 (0.10; 1.65) | 0.023 |
| IL8 | 28.93 ± 4.19 | 28.16 ± 4.00 | -6.26 ± 0.67 | -7.55 ± 0.54 | 1.09 (-0.04; 2.55) | 0.070 |
| MMP10 | 32.07 ± 3.89 | 29.76 ± 5.55 | -9.41 ± 0.86 | -8.74 ± 0.66 | -0.04 (-2.70; 1.81) | 0.945 |
| TNC | 27.24 ± 4.15 | 25.07 ± 4.49 | -4.57 ± 0.45 | -4.04 ± 0.40 | -0.58 (-1.45; 0.78) | 0.396 |
| VEGFB | 25.64 ± 3.46 | 24.20 ± 3.41 | -2.97 ± 0.19 | -3.18 ± 0.21 | 0.34 (-0.23; 0.77) | 0.214 |
| GAPDH | 22.90 ± 4.09 | 21.24 ± 4.45 | - | - | - | - |
| YWHAZ | 22.43 ± 3.47 | 20.81 ± 3.89 | - | - | - | - |
| Group | Parameter 1 | Parameter 2 | Spearman rho | P value |
|---|---|---|---|---|
| RPL group | CXCL1 | Protrombine time | -0.456 | 0.04959 |
| CXCL1 | Activated Partial Thromboplastin Time | 0.587 | 0.0083 | |
| MMP10 | Activated Partial Thromboplastin Time | -0.494 | 0.0317 | |
| RIF group | VEGFB | Prolactin | -0.446 | 0.049 |
| VEGFB | Thyroid stimulating hormone | -0.526 | 0.017 | |
| HAND2 | Thyroid stimulating hormone | -0.671 | 0.001 | |
| HAND2 | Fibrinogen | -0.465 | 0.038 |
| Potential markers and combinations | AUC (95% CI) | Optimal cut-off point | Sensitivity (95% CI) | Specificity(95% CI) |
|---|---|---|---|---|
| IL15 | 0.713 (0.550-0.876) | -4.091 | 0.800 (0.563-0.943) | 0.579 (0.335-0.797) |
| IFNG | 0.701 (0.516-0.886) | -13.238 | 0.667 (0.410-0.867) | 0.750 (0.476-0.927) |
| HPRT1 | 0.692 (0.505-0.879) | -3.568 | 0.900 (0.683-0.988) | 0.632 (0.384-0.837) |
| IL15 + IFNG | 0.722 | - | 0.667 | 0.688 |
| IL15 + HPRT1 | 0.800 | - | 0.850 | 0.684 |
| IFNG + HPRT1 | 0.753 | - | 0.778 | 0.750 |
| IL15 + IFNG + HPRT1 | 0.778 | - | 0.889 | 0.688 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





