Submitted:
14 August 2024
Posted:
15 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Method
2.2.1. The Process of Gene Cloning and Subsequent Bioinformatics Analysis
2.2.2. Expression Analysis of the EuSIP5 Gene in E. ulmoides
2.2.3. Vector Construction
2.2.4. Subcellular Localization
2.2.5. Genetic Transformation and Identifcation of Tobacco
2.2.6. Quantitative Fluorescence Real-Time PCR (qRT-PCR)
2.2.7. Drought Stress Treatment of Transgenic Tobacco
2.2.8. Expression Analysis of Genes Related to Drought Resistance in Transgenic Tobacco
2.2.9. Determination of Protective Enzyme Activity
2.2.10. Statistical Analysis
3. Results
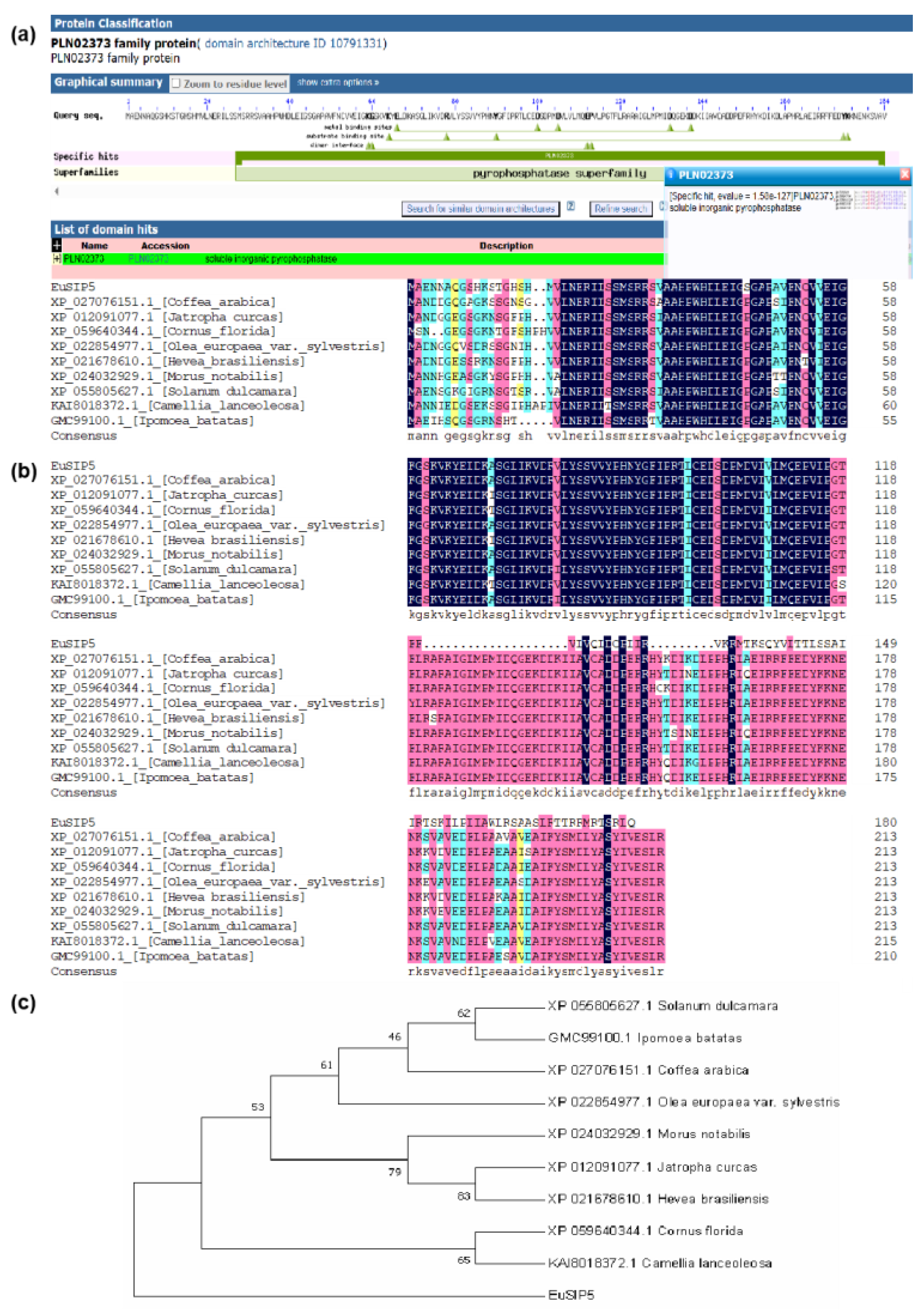
3.1. Cloning and Analysis of the EuSIP5 Gene
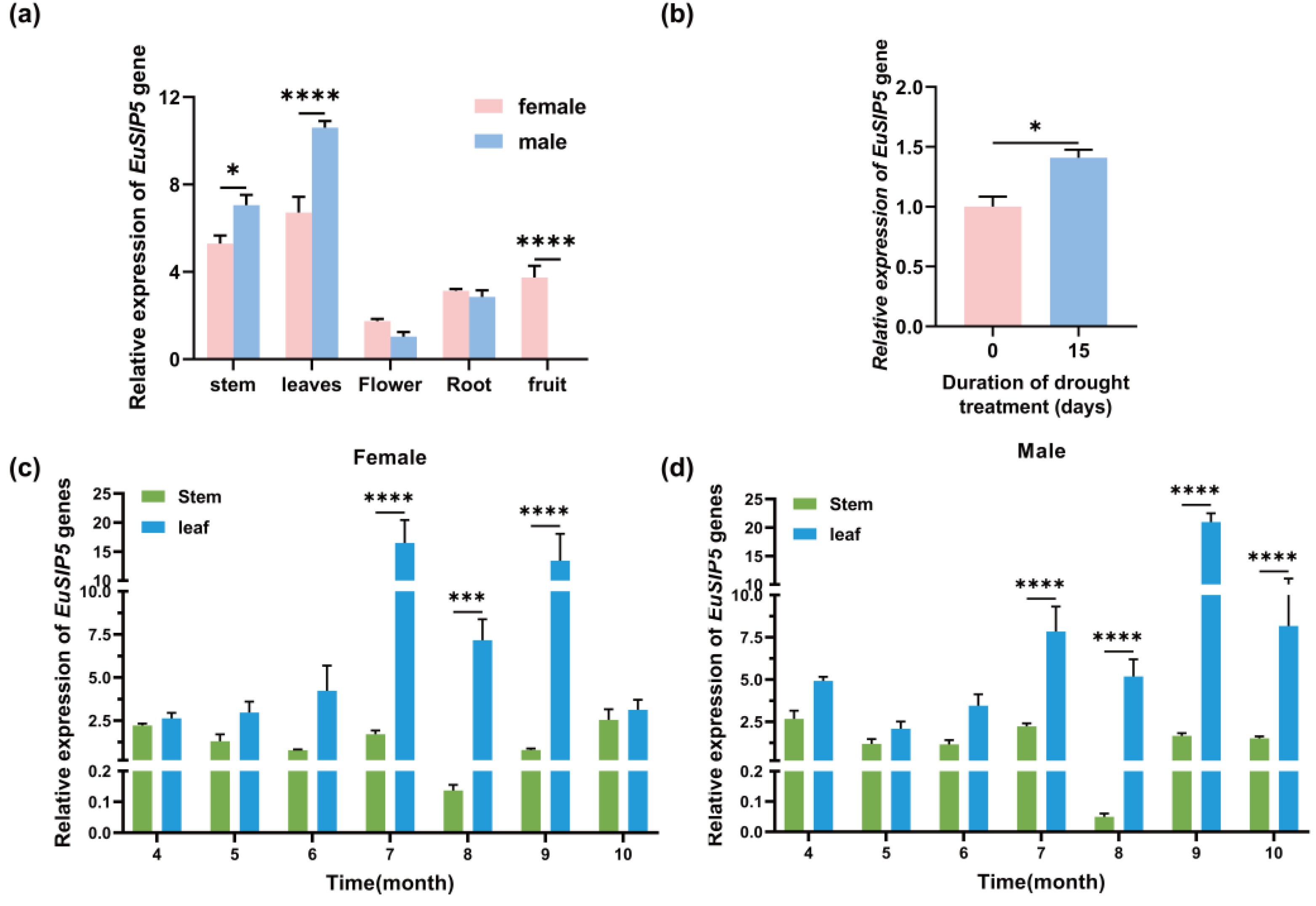
3.2. Spatio-Temporal Expression Characteristics of the EuSIP5 Gene
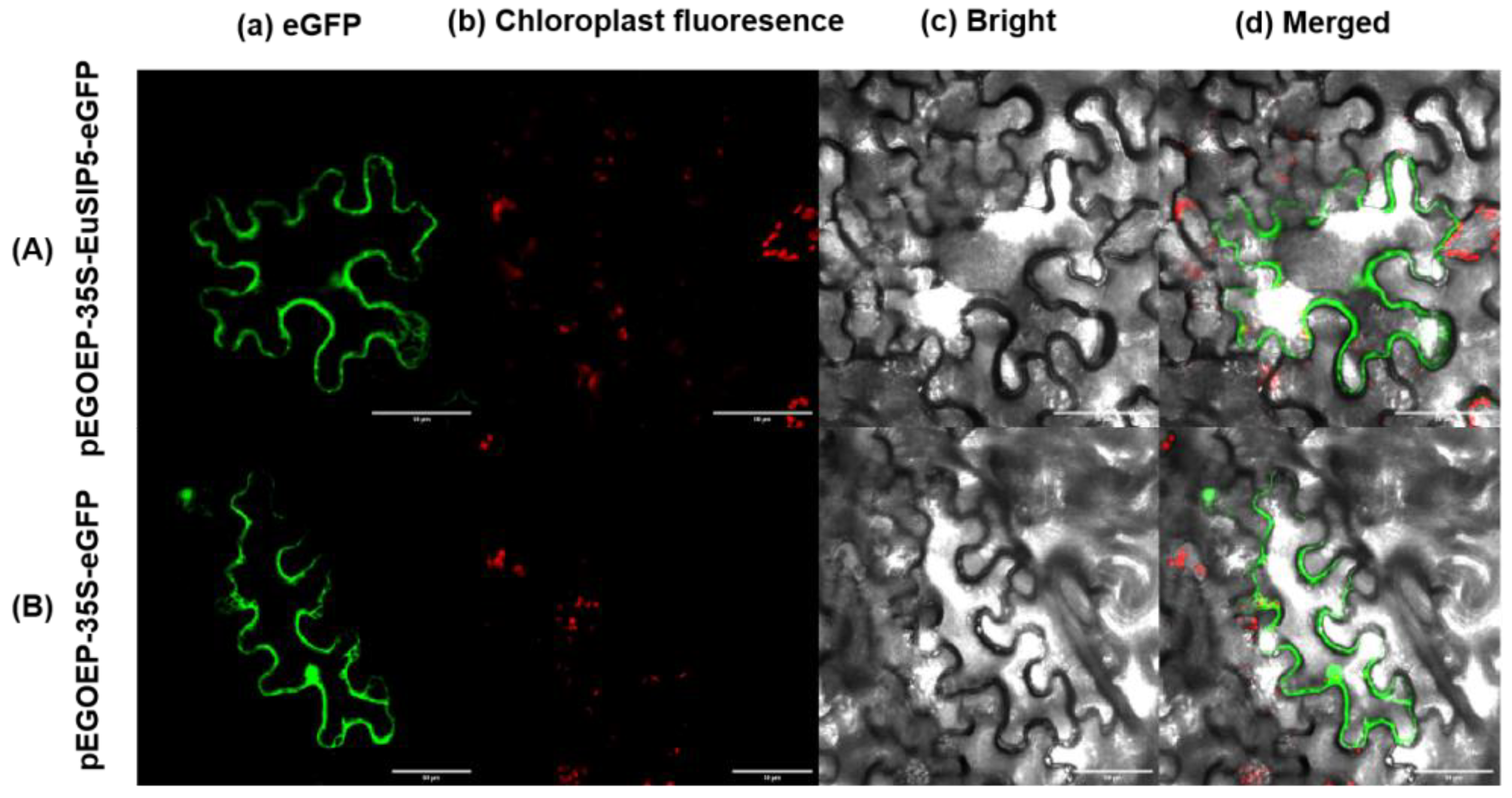
3.3. Subcellular Localization
3.4. Genetically Modified Tobacco and Identification of Transgenic Positive Plants
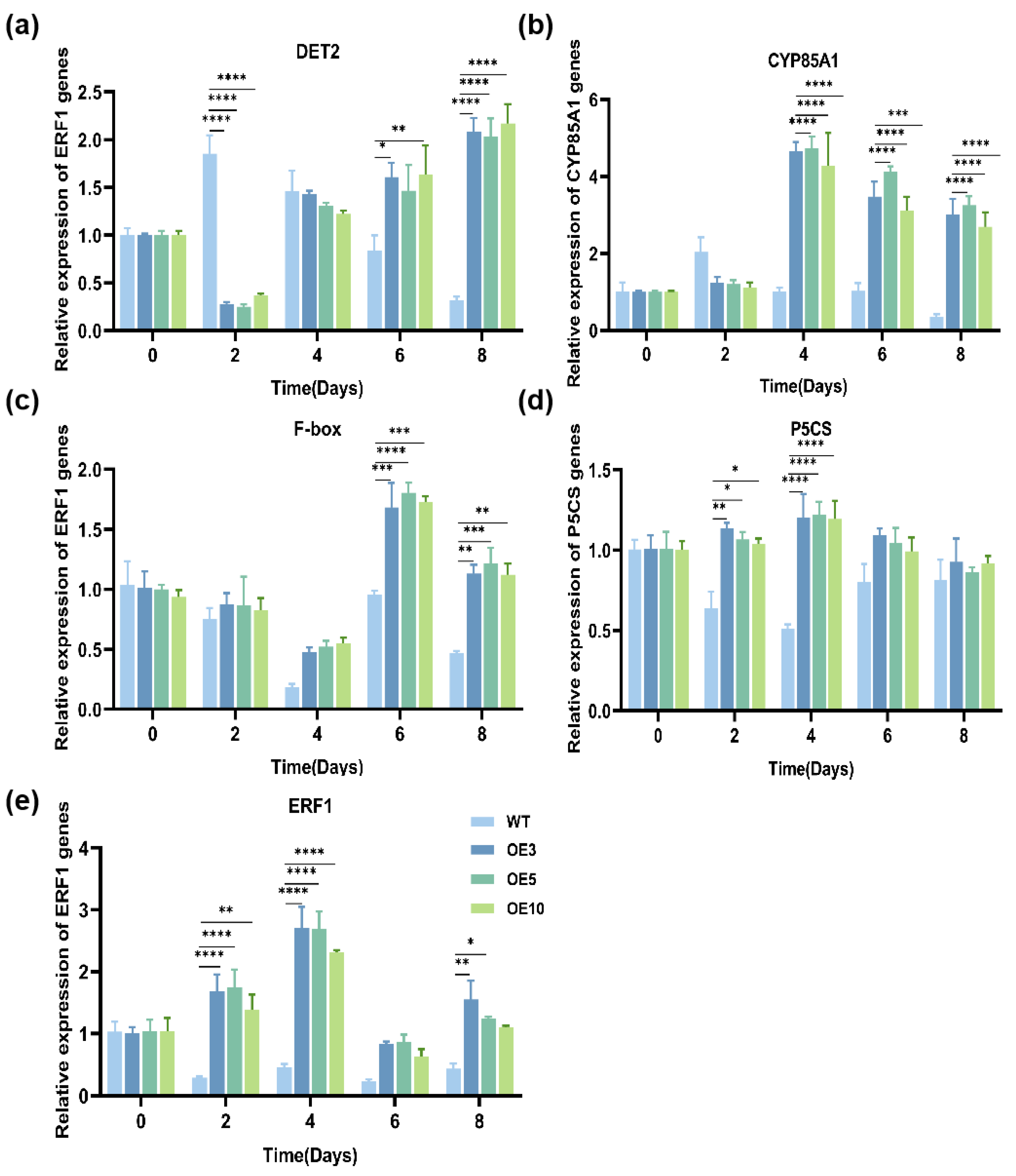
3.5. Effect of Trans-EuSIP5 Gene on Drought Tolerance in Tobacco
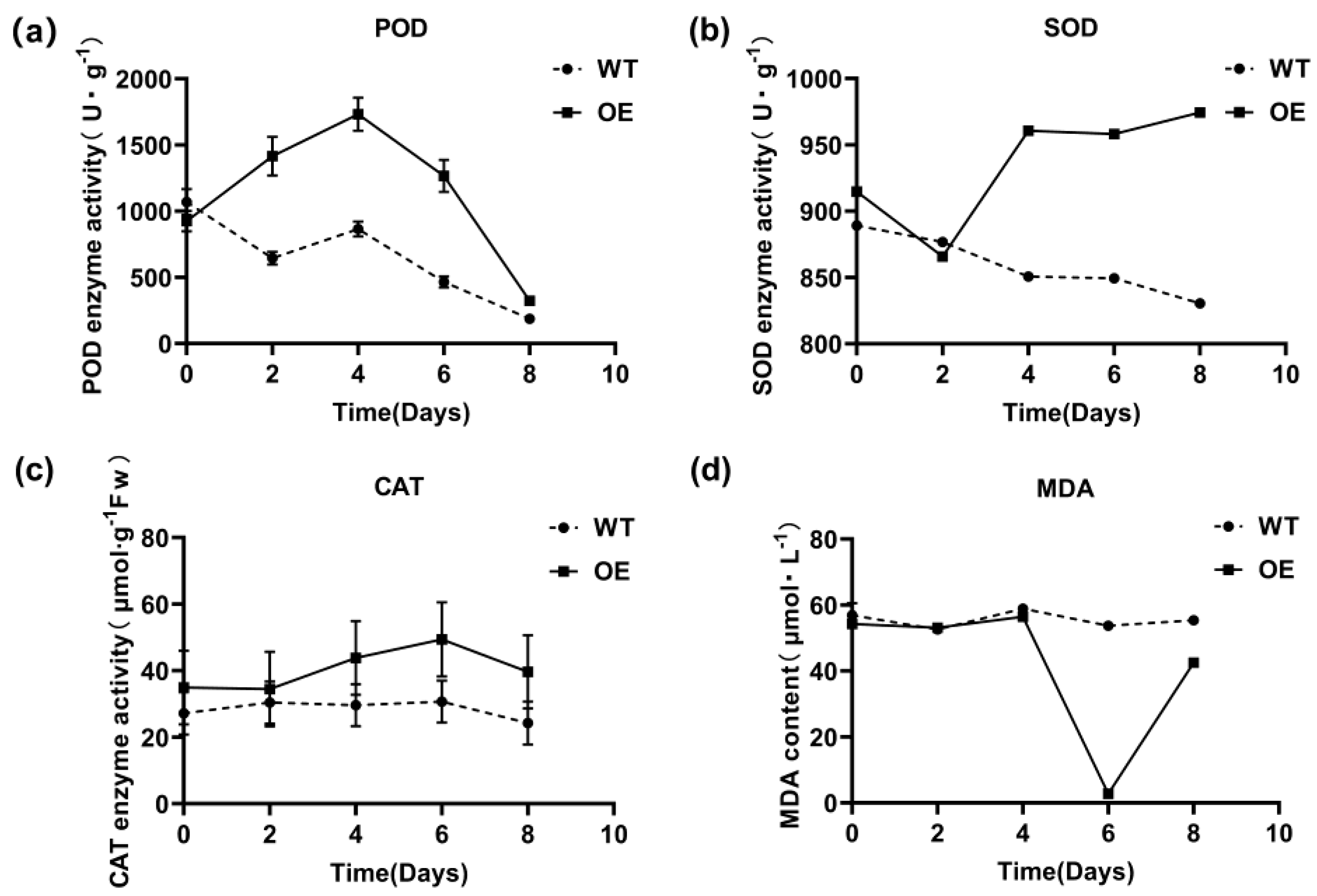
3.6. Effect of Trans-EuSIP5 on the Activity of Tobacco Protective Enzymes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science (New York, N.Y.) 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Razi, K.; Muneer, S. Drought stress-induced physiological mechanisms, signaling pathways and molecular response of chloroplasts in common vegetable crops. Critical Reviews in Biotechnology 2021, 41, 669–691. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.X.; Shi, J.W.; Li, R.; An, Y.C.; Yang, B.L. Effects of extreme drought on plant species in Karst area of Guizhou Province, Southwest China. Chinese Journal of Applied Ecology 2011, 22, 1127–1134. [Google Scholar] [CrossRef]
- Ren, N.; Gong, W.W.; Zhao, Y.C.; Zhao, D.G.; Xu, Y.W. Innovation in sweet rice wine with high antioxidant activity: Eucommia ulmoides leaf sweet rice wine. 2023, 9. [Google Scholar] [CrossRef]
- Wang, J.; Chen, X.; Huang, X.; Zhao, Y.; Zhao, D. Cloning and Function Identification of a Phytoene Desaturase Gene from Eucommia ulmoides. 2023, 92, 1377–1389. [Google Scholar] [CrossRef]
- Sasaki, Y.; Chiba, A.; Murakami, M.; Sekihashi, K.; Tanaka, M.; Takahoko, M.; Moribayashi, S.; Kudou, C.; Hara, Y.; Nakazawa, Y.; et al. Antimutagenicity of Tochu tea (an aqueous extract of Eucommia ulmoides leaves): 2. Suppressing effect of Tochu tea on the urine mutagenicity after ingestion of raw fish and cooked beef. Mutation Research/Genetic Toxicology 1996, 371, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Park, S.A.; Choi, M.S.; Kim, M.J.; Jung, U.J.; Kim, H.J.; Park, K.K.; Noh, H.J.; Park, H.M.; Park, Y.B.; Lee, J.S.; et al. Hypoglycemic and hypolipidemic action of Du-zhong (Eucommia ulmoides Oliver) leaves water extract in C57BL/KsJ-db/db mice. Journal of Ethnopharmacology 2006, 107, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Yen, G.C.; Hsieh, C.L. Reactive Oxygen Species Scavenging Activity of Du-zhong (Eucommia ulmoides Oliv.) and Its Active Compounds. Journal of Agricultural and Food Chemistry 2000, 48, 3431–3436. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Li, B.; Zhao, Y.C.; Zhao, D.G. Cloning and characterization of the DIR1 promoter from Eucommia ulmoides Oliv and its response to hormonal and abiotic stress. Plant Cell, Tissue and Organ Culture (PCTOC) 2021, 146, 313–322. [Google Scholar] [CrossRef]
- Zhao, Y.C.; Liu, Y.Q.; Dong, X.; Liu, J.J.; Zhao, D.G. Identification of a novel laccase gene EuLAC1 and its potential resistance against Botrytis cinerea. Transgenic Research 2022, 31, 215–225. [Google Scholar] [CrossRef]
- Dong, X.; Zhao, Y.C.; Ran, X.; Guo, L.X.; Zhao, D.G. Overexpression of a New Chitinase Gene EuCHIT2 Enhances Resistance to Erysiphe cichoracearum DC. in Tobacco Plants. International Journal of Molecular Sciences 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhao, D.G. Cloning, Characterization, and Functional Analysis of EuTIL1, a Gene-Encoding Temperature-Induced Lipocalin in Eucommia ulmoides Oliv. Horticulturae 2023, 9. [Google Scholar] [CrossRef]
- Lu, Y.X.; Dong, X.; Huang, X.Z.; Zhao, D.G.; Zhao, Y.C.; Peng, L. Combined analysis of the transcriptome and proteome of Eucommia ulmoides Oliv. (Duzhong) in response to Fusarium oxysporum. 2022, 10. [Google Scholar] [CrossRef]
- Wang, L.; Wang, R.; Li, Y.; Zhao, Y.; Zhao, D. Overexpression of β-1,4-Glucanase Gene EuEG1 Improves Micrografting of Eucommia ulmoides. 2023, 92, 3063–3075. [Google Scholar] [CrossRef]
- Gloria, S.B.; Agustín, H.; Guillermo, L.L.; José Román, P.C.; Plácido, N.; Aurelio, S. Inorganic Pyrophosphatase Defects Lead to Cell Cycle Arrest and Autophagic Cell Death through NAD+ Depletion in Fermenting Yeast*. Journal of Biological Chemistry 2013, 288, 13082–13092. [Google Scholar] [CrossRef]
- Serrano-Bueno, G.; Hernández, A.; López-Lluch, G.; Pérez-Castiñeira, J.R.; Navas, P.; Serrano, A. Inorganic pyrophosphatase defects lead to cell cycle arrest and autophagic cell death through NAD+ depletion in fermenting yeast. Journal of Biological Chemistry 2013, 288, 13082–13092. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Zeng, R.Z.; Xiao, X.Z. Advances on the Research of Pyrophosphatase in Plants. Life Science Research 2004, 83–87. [Google Scholar] [CrossRef]
- Xian, J.H.; Zhang, M.P.; Sun, C.Y.; Wang, Y.F.; Wang, K.Y.; Chen, J.; Zhao, M.Z.; Wang, Y. Research Progress of Soluble Pyrophosphatase. Jilin Agriculture University, 2019.
- Huang, H.; Patskovsky, Y.; Toro, R.; Farelli, J.D.; Pandya, C.; Almo, S.C.; Allen, K.N.; Dunaway-Mariano, D. Divergence of structure and function in the haloacid dehalogenase enzyme superfamily: Bacteroides thetaiotaomicron BT2127 is an inorganic pyrophosphatase. Biochemistry 2011, 50, 8937–8949. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, Y.R.; Wang, X.L.; Fang, T.; Zhen, S.H.; Lu, J.W.; Zhang, J.; Fu, J.J. Cloning and Expression Analysis of Soluble Inorganic Pyrophosphatase Family Genes in Maize (Zea mays L.). Journal of Plant Genetic Resources 2021, 22, 455–465. [Google Scholar] [CrossRef]
- Zhu, J.H.; Xu, J.; Yu, X.H.; Chang, W.J.; Zhang, Z.L. Prokaryotic Expression for Three Soluble Inorganic Pyrophosphatase Genes from Hevea brasiliensis. Chinese Journal of Tropical Crops 2013, 34, 41–45. [Google Scholar]
- George, G.M.; van der Merwe, M.J.; Nunes-Nesi, A.; Bauer, R.; Fernie, A.R.; Kossmann, J.; Lloyd, J.R. Virus-induced gene silencing of plastidial soluble inorganic pyrophosphatase impairs essential leaf anabolic pathways and reduces drought stress tolerance in Nicotiana benthamiana. Plant physiology 2010, 154, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Song, Q.; Wei, H.; Wang, Y.; Lin, M.; Sun, K.; Zhang, Y.; Yang, J.; Li, C.; Luo, K.J.N.P. The AP2/ERF transcription factor PtoERF15 confers drought tolerance via JA-mediated signaling in Populus. 2023, 240, 1848–1867. [Google Scholar] [CrossRef]
- Wang, H.; Ni, D.; Shen, J.; Deng, S.; Xuan, H.; Wang, C.; Xu, J.; Zhou, L.; Guo, N.; Zhao, J.J.F.i.P.S. Genome-wide identification of the AP2/ERF gene family and functional analysis of GmAP2/ERF144 for drought tolerance in soybean. 2022, 13, 848766. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Yu, M.; Zhang, S.; Song, T.; Zhang, M.; Zhou, H.; Wang, Y.; Xiang, J.; Zhang, X. Transcriptomic Identification of Wheat AP2/ERF Transcription Factors and Functional Characterization of TaERF-6-3A in Response to Drought and Salinity Stresses. International Journal of Molecular Sciences 2022, 23. [Google Scholar] [CrossRef]
- Ma, Z.; Hu, L.; Jiang, W. Understanding AP2/ERF Transcription Factor Responses and Tolerance to Various Abiotic Stresses in Plants: A Comprehensive Review. International Journal of Molecular Sciences 2024, 25. [Google Scholar] [CrossRef]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. The Plant Cell 1997, 9, 1859–1868. [Google Scholar]
- Van Moerkercke, A.; Duncan, O.; Zander, M.; Šimura, J.; Broda, M.; Vanden Bossche, R.; Lewsey, M.G.; Lama, S.; Singh, K.B.; Ljung, K.J.P.o.t.N.A.o.S. A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels. 2019, 116, 23345–23356. [Google Scholar] [CrossRef]
- Yang, D.; Ni, R.; Yang, S.; Pu, Y.; Qian, M.; Yang, Y.; Yang, Y. Functional Characterization of the Stipa purpurea P5CS Gene under Drought Stress Conditions. International Journal of Molecular Sciences 2021, 22. [Google Scholar] [CrossRef]
- Duan, F.M.; Ding, J.; Lee, D.S.; Lu, X.L.; Feng, Y.Q.; Song, W.W. Overexpression of SoCYP85A1, a spinach cytochrome p450 gene in transgenic tobacco enhances root development and drought stress tolerance. Frontiers in plant science 2017, 8, 1909. [Google Scholar] [CrossRef]
- Zheng, B.F.; Ma, Y.Z.; Lu, H.D.; An, W.K.; Zhang, F.C. Effects of Spraying Brassinolide on Expression of Steroid 5-Alpha Reductase Gene(Gh DET2) in Cotton under Drought Stress. Genomics and Applied Biology 2018, 37, 859–866. [Google Scholar] [CrossRef]
- An, J.; Li, Q.X.; Yang, J.J.; Zhang, G.Q.; Zhao, Z.X.; Wu, Y.Z.; Wang, Y.; Wang, W. Wheat F-box protein TaFBA1 positively regulates plant drought tolerance but negatively regulates stomatal closure. Frontiers in Plant Science 2019, 10, 1242. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.H.; Zhao, Y.; Zhao, Y.; Feng, C.; Zhang, Y.H.; Wang, F.W.; Li, X.W.; Gao, H.T.; Liu, W.C.; Jing, Y. Soybean F-box-like protein GmFBL144 interacts with small heat shock protein and negatively regulates plant drought stress tolerance. Frontiers in plant science 2022, 13, 823529. [Google Scholar] [CrossRef] [PubMed]
- Navarro-De la Sancha, E.; Coello-Coutiño, M.P.; Valencia-Turcotte, L.G.; Hernández-Domínguez, E.E.; Trejo-Yepes, G.; Rodríguez-Sotres, R. Characterization of two soluble inorganic pyrophosphatases from Arabidopsis thaliana. Plant Science 2007, 172, 796–807. [Google Scholar] [CrossRef]
- Schulze, S.; Mant, A.; Kossmann, J.; Lloyd, J.R. Identification of an Arabidopsis inorganic pyrophosphatase capable of being imported into chloroplasts. FEBS letters 2004, 565, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Luna, F.M.; Navarro de la Sancha, E.; Valencia-Turcotte, L.G.; Vázquez-Santana, S.; Rodríguez-Sotres, R. Evidence for a non-overlapping subcellular localization of the family I isoforms of soluble inorganic pyrophosphatase in Arabidopsis thaliana. Plant Science 2016, 253, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Qiao, X.; Zhu, X.X.; Khan, W.; Wu, J.; Zhang, S.L. Expression and evolutionary analysis of soluble inorganic pyrophosphatase gene family in pear and four other Rosaceae species. Plant Systematics and Evolution 2020, 306, 46. [Google Scholar] [CrossRef]
- Wang, J.; Kang, L.Y.; Liu, Z.B.; Lv, J.H.; Liu, Y.H.; Zou, X.X. Research Progress on the Impact of Drought on Plant. Hunan Agricultural Sciences, 2017; 123-126+130. [Google Scholar] [CrossRef]
- Li, B.; Zeng, Q.; Zhao, D.; Zhao, D.G. Cloning and Function Analysis of EuERD16 Gene in Eucommia ulmoides. Genomics and Applied Biology 2023, 42, 373–383. [Google Scholar] [CrossRef]
- Chen, S.Y. Membrane lipid peroxidation and plant adversity stresses. Bulletin of Botany 1989, 212–215. [Google Scholar]
- Li, M.Q. Physiological and Molecular Regulatory Mechanisms of Soybean in Response to Drought. Shenyang Agricultural University, 2023.
- Wang, Q.M. Effects of Drought Stress on Protective Enzymes Activities and Membrane Lipid Peroxidation in Leaves of Soybean Seedlings. Journal of Agro-Environment Science 2006, 918–921. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Zheng, A.Z.; Liu, C.P.; Shen, Z.G. Effect of Cadmium on MDA Content, POD and SOD Activities of Brassica Pekinensis and Brassica Chinensis. Hubei Agricultural Sciences 2005, 67–69. [Google Scholar]
- Song, X.G.; She, X.P. The Generation and the Role of Hydrogen Peroxide in Plant. Journal of Lianyungang Teachers College 2010, 27, 99–103. [Google Scholar] [CrossRef]
- Türkan, İ.; Bor, M.; Özdemir, F.; Koca, H. Differential responses of lipid peroxidation and antioxidants in the leaves of drought-tolerant P. acutifolius Gray and drought-sensitive P. vulgaris L. subjected to polyethylene glycol mediated water stress. Plant Science 2005, 168, 223–231. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




